UvSorA and UvSorB Involved in Sorbicillinoid Biosynthesis Contribute to Fungal Development, Stress Response and Phytotoxicity in Ustilaginoidea virens
Abstract
1. Introduction
2. Results
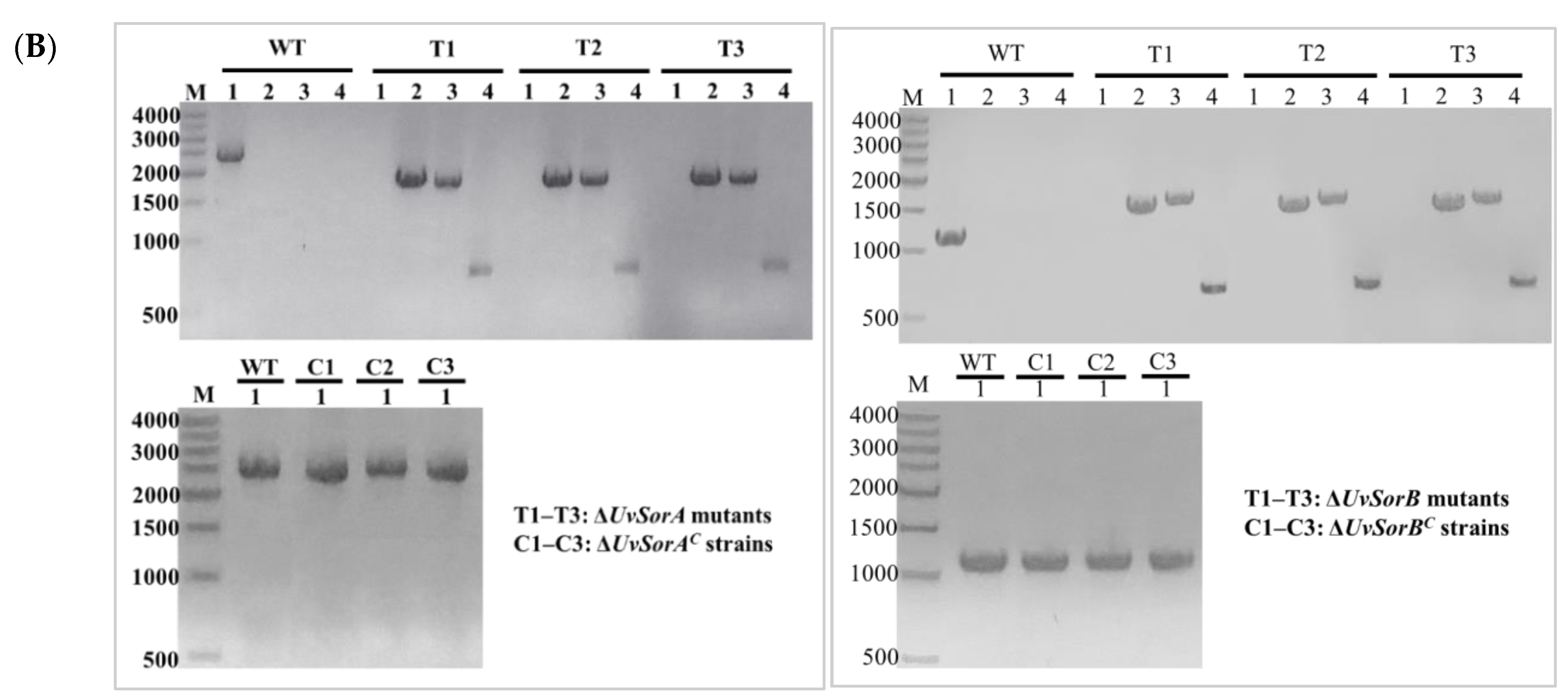
2.1. Identification and Characteriation of UvSorA and UvSorB in U. virens
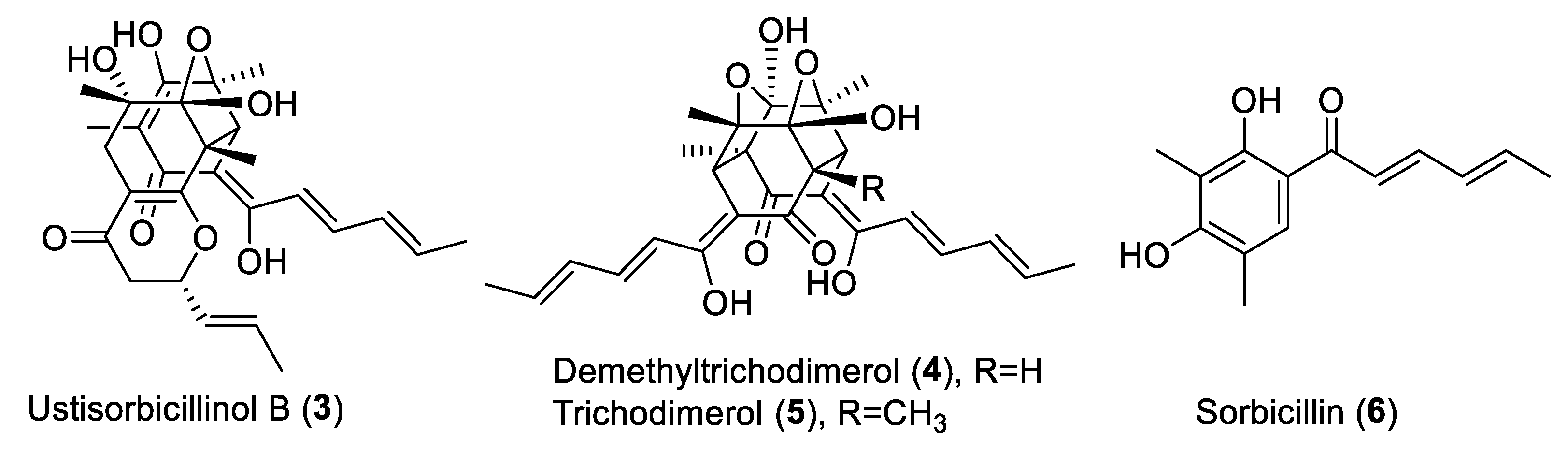
2.1.1. Identification of the Sorbicillinoid Biosynthesis Gene Cluster in U. virens
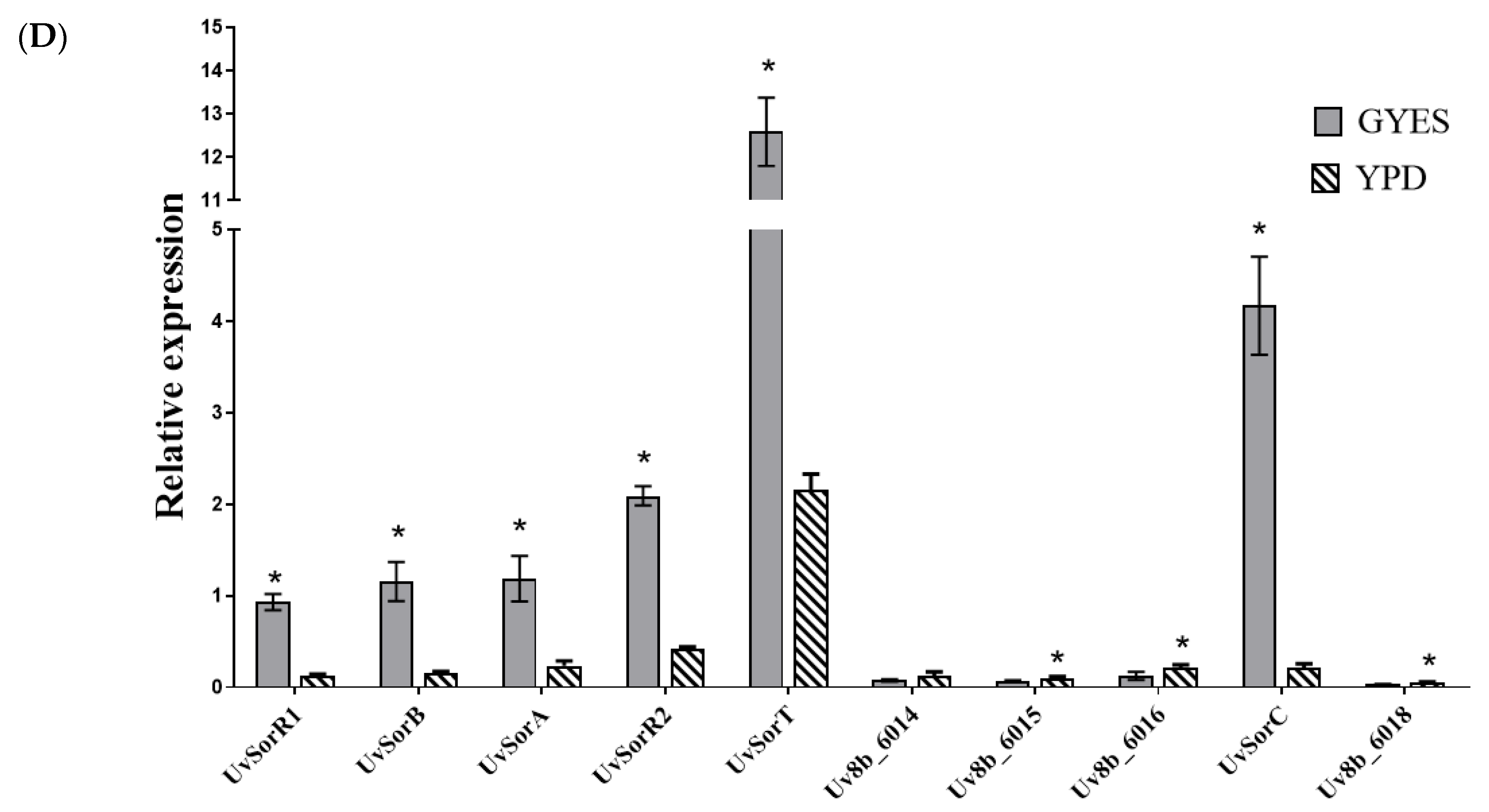
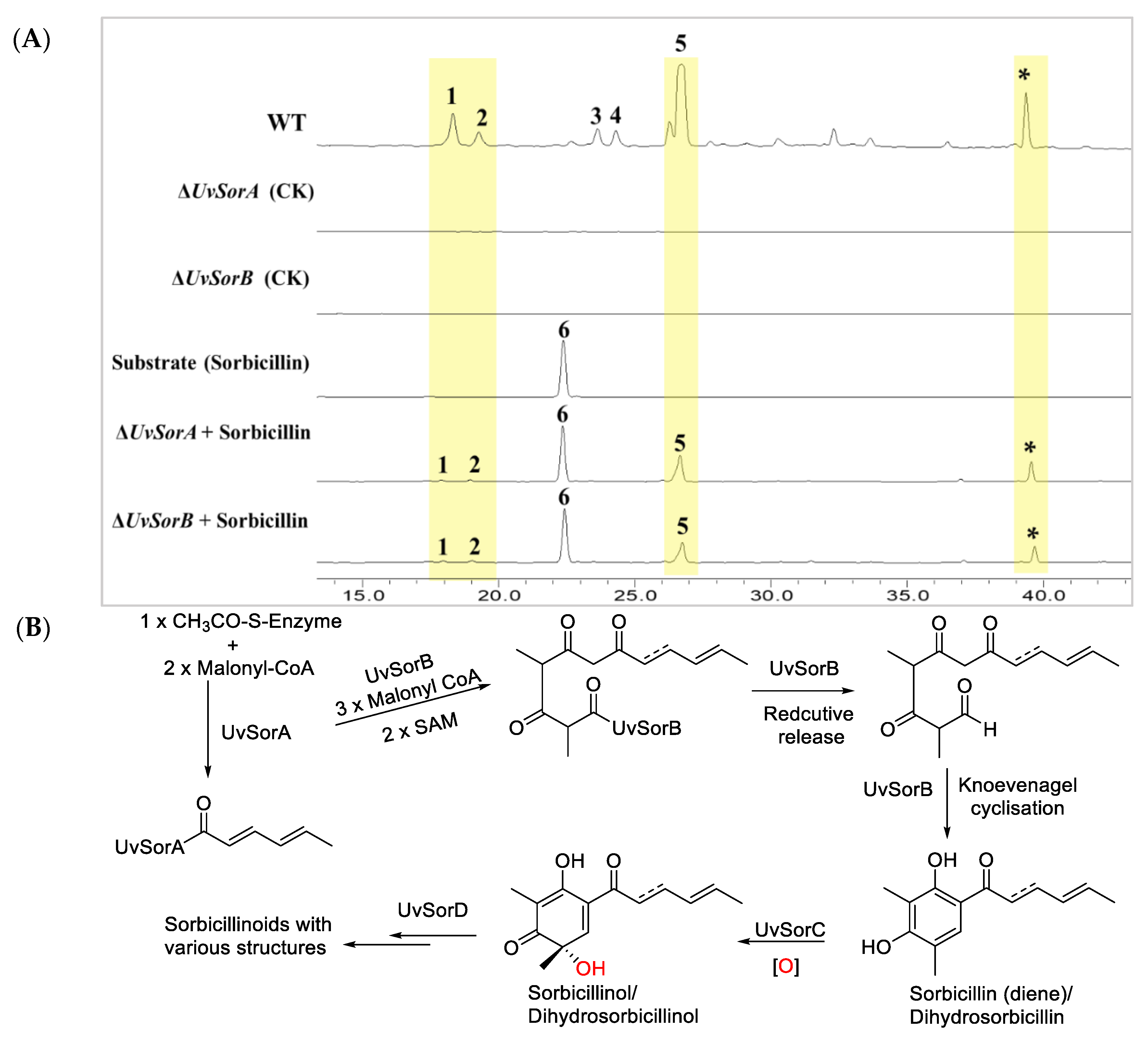
2.1.2. UvSorA and UvSorB as Polyketide Synthase Genes of Sorbicillinoid Biosynthesis
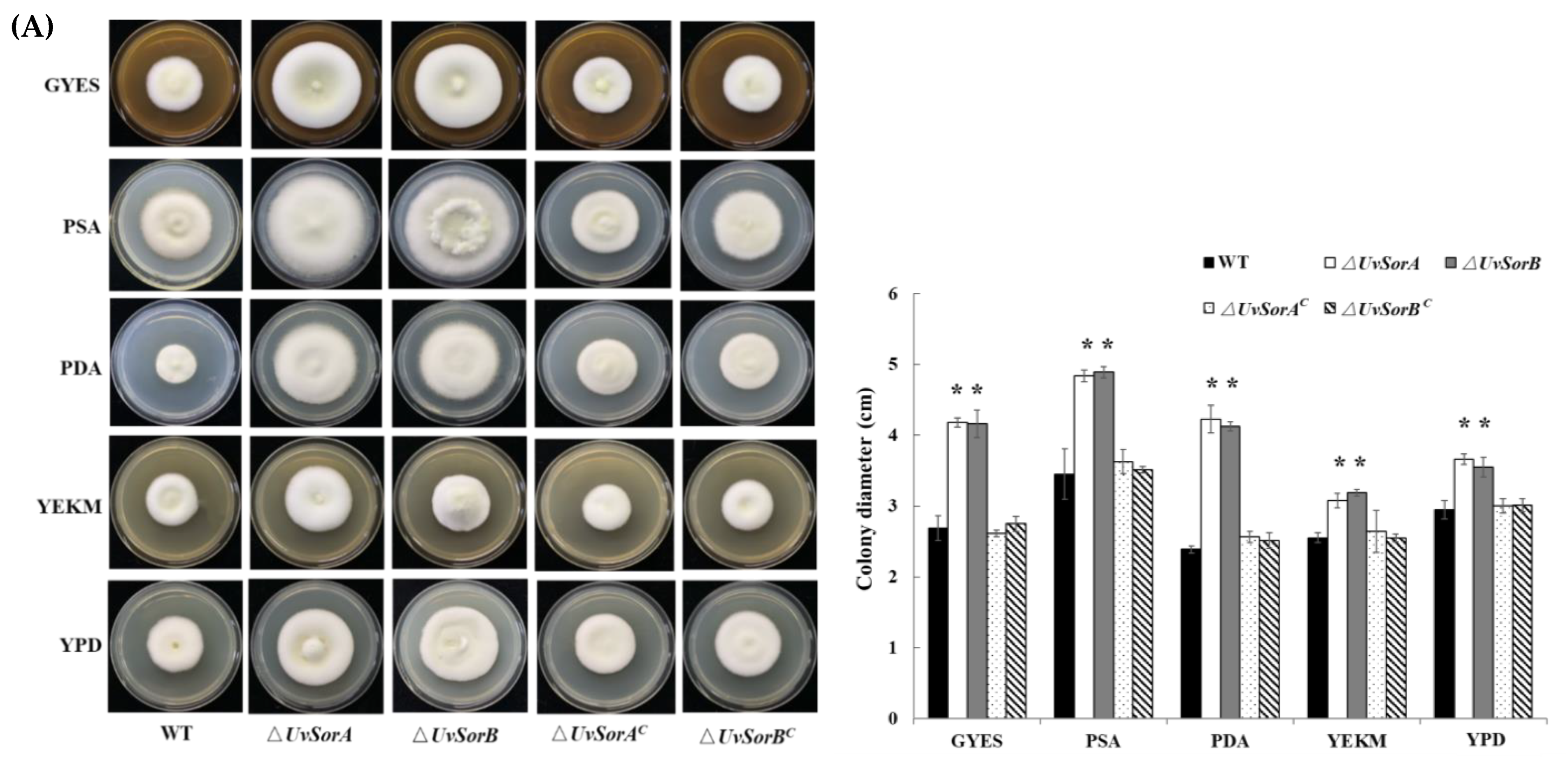
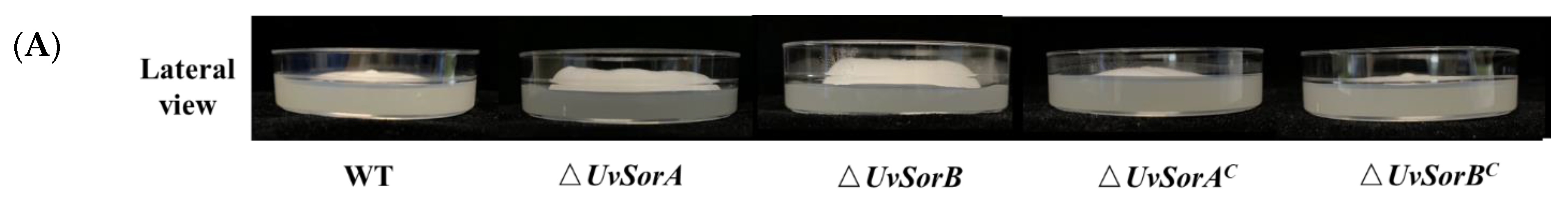
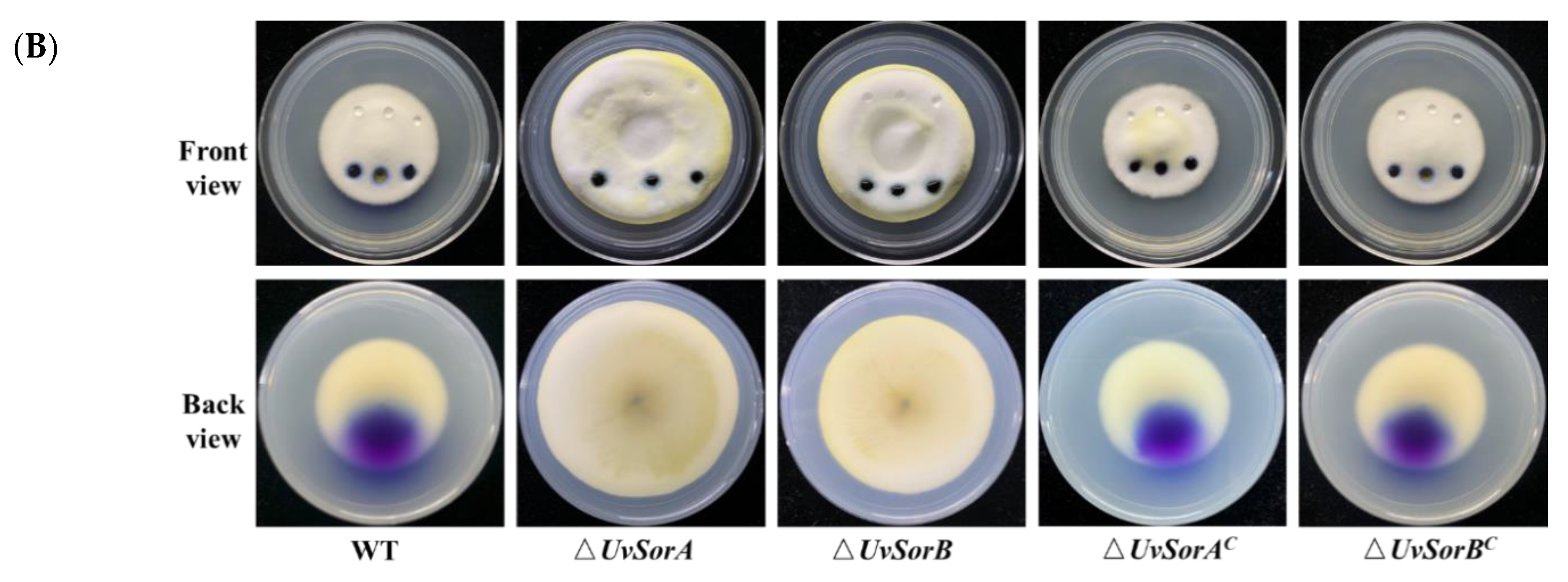
2.2. Deletion of UvSorA and UvSorB Increased Mycelial Growth, Sporulation and Hyphal Hydrophobicity
2.2.1. Deletion of UvSorA and UvSorB Increased Mycelial Growth and Sporulation
2.2.2. Deletion of UvSorA and UvSorB Increased Hyphal Hydrophobicity
2.3. Deletion of UvSorA and UvSorB Decreased Resistances to Osmotic, Metal Cation and Fungicide Stresses
2.3.1. Deletion of UvSorA and UvSorB Resulted in Defects in Response to Hyperosmotic and Metal Cation Stresses
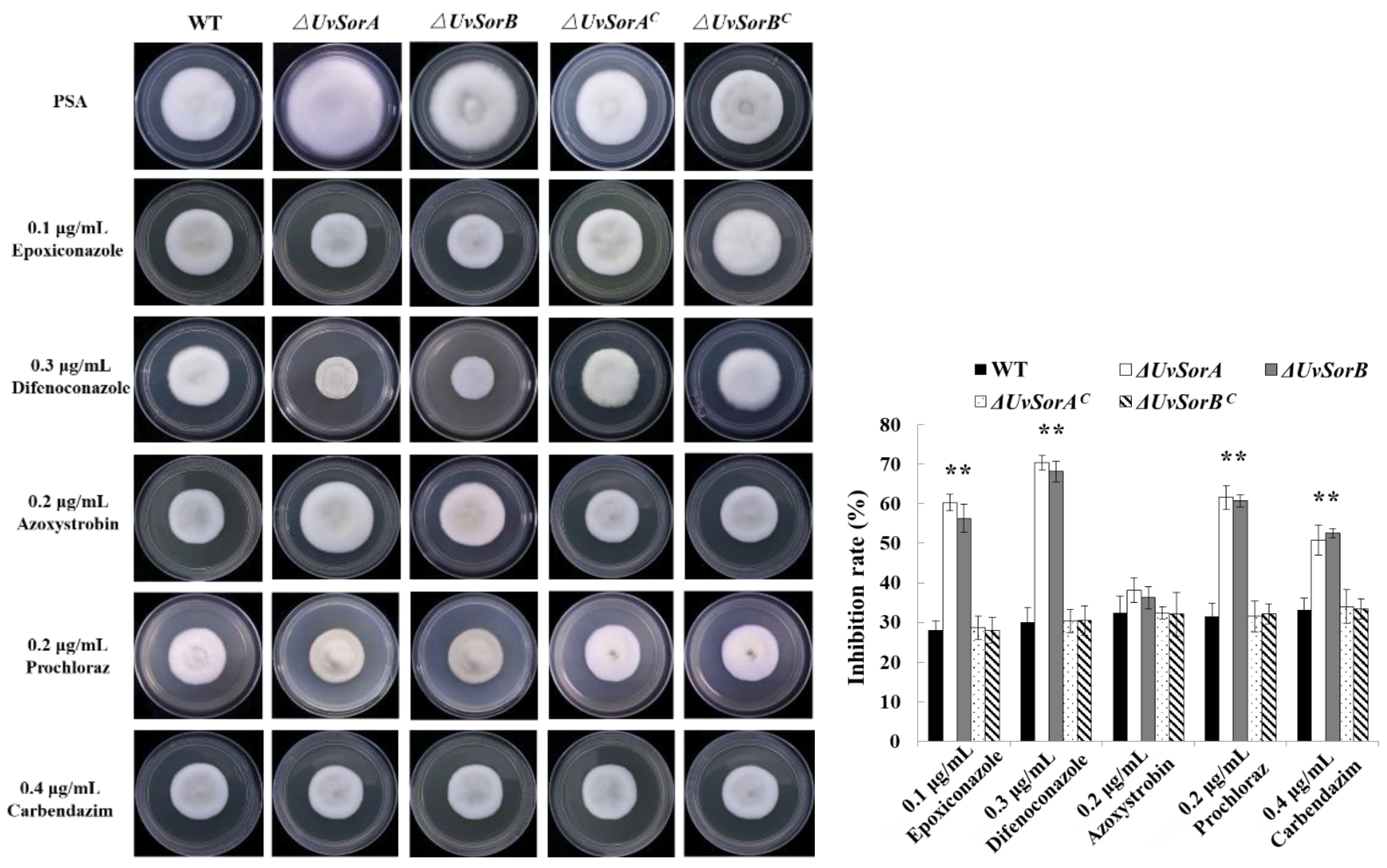
2.3.2. Deletion of UvSorA and UvSorB Increased Sensitivity to Fungicides
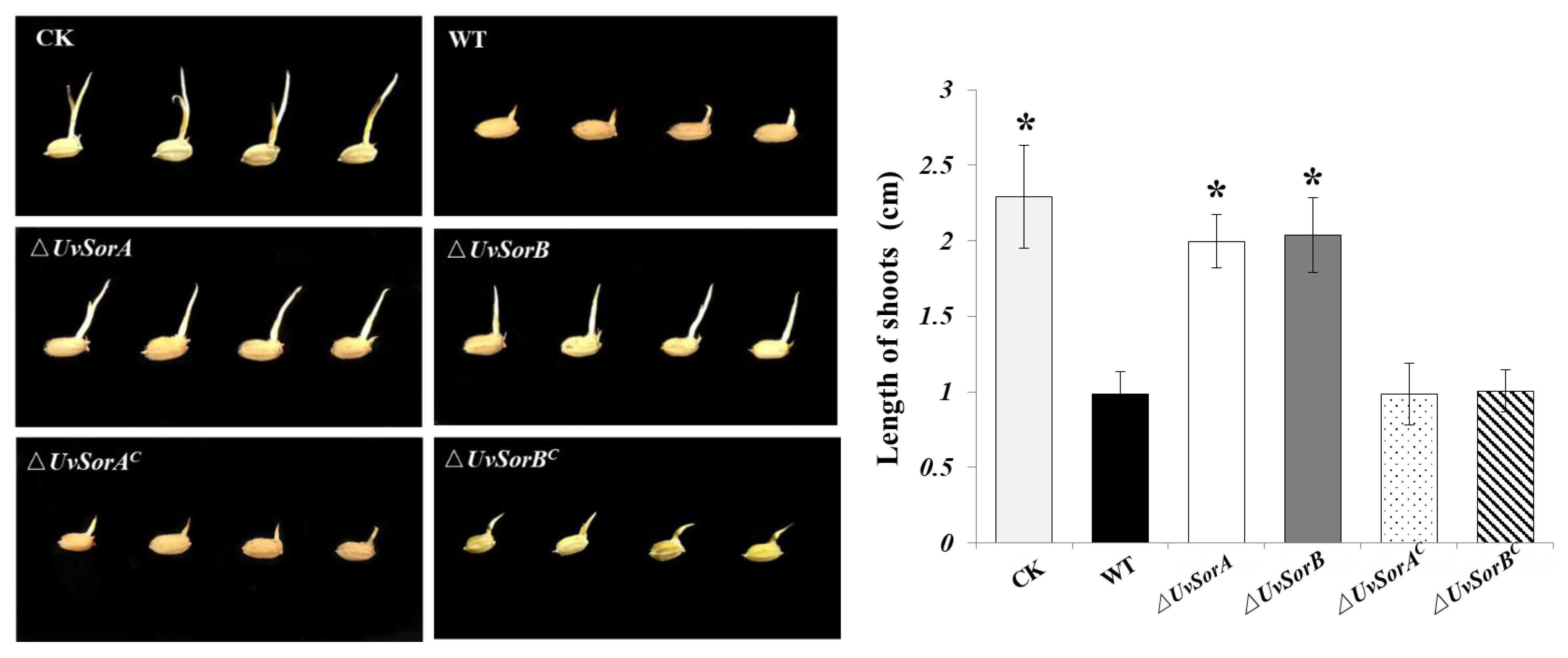
2.4. Culture Filtrates of ∆UvSorA and ∆UvSorB Showed a Decreased Inhibition on Germ Elongation of Rice Germinated Seeds
2.5. Deletion of UvSorA and UvSorB Resulted in Deficiency of Cell Wall Integrity
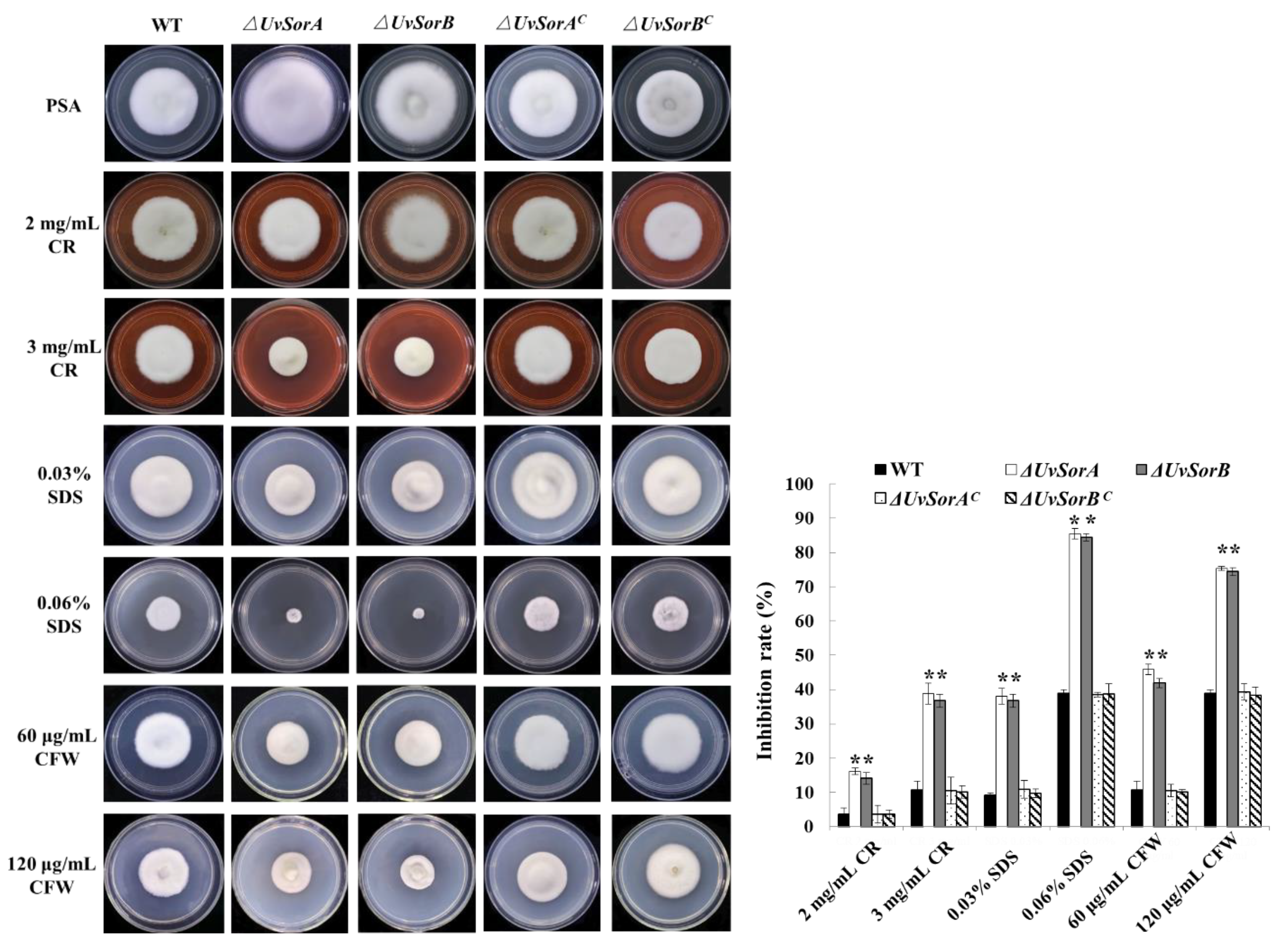
2.5.1. Deletion of UvSorA and UvSorB Resulted in Decrease of Tolerance to Cell Wall Damaging Agents
2.5.2. Deletion of UvSorA and UvSorB Resulted in Alterations of Microscopic and Submicroscopic Structures of Mycelia and Cell Walls
3. Discussion
3.1. Sorbicillinoid Biosythetic Gene Cluster
3.2. Biological Functions of Sorbicillinoids in Fungi
4. Materials and Methods
4.1. Fungal Strains, Plasmids and Culture Conditions
4.2. Bioinformatic Analysis
4.3. Targeted Gene Deletion and Complementation
4.4. RNA Preparation and qRT−PCR
4.5. Chemical Analysis of Sorbicillinoids in WT Strain, Gene Deletion Mutants, and Complemented Strains of U. virens
4.6. Physiology Experiments
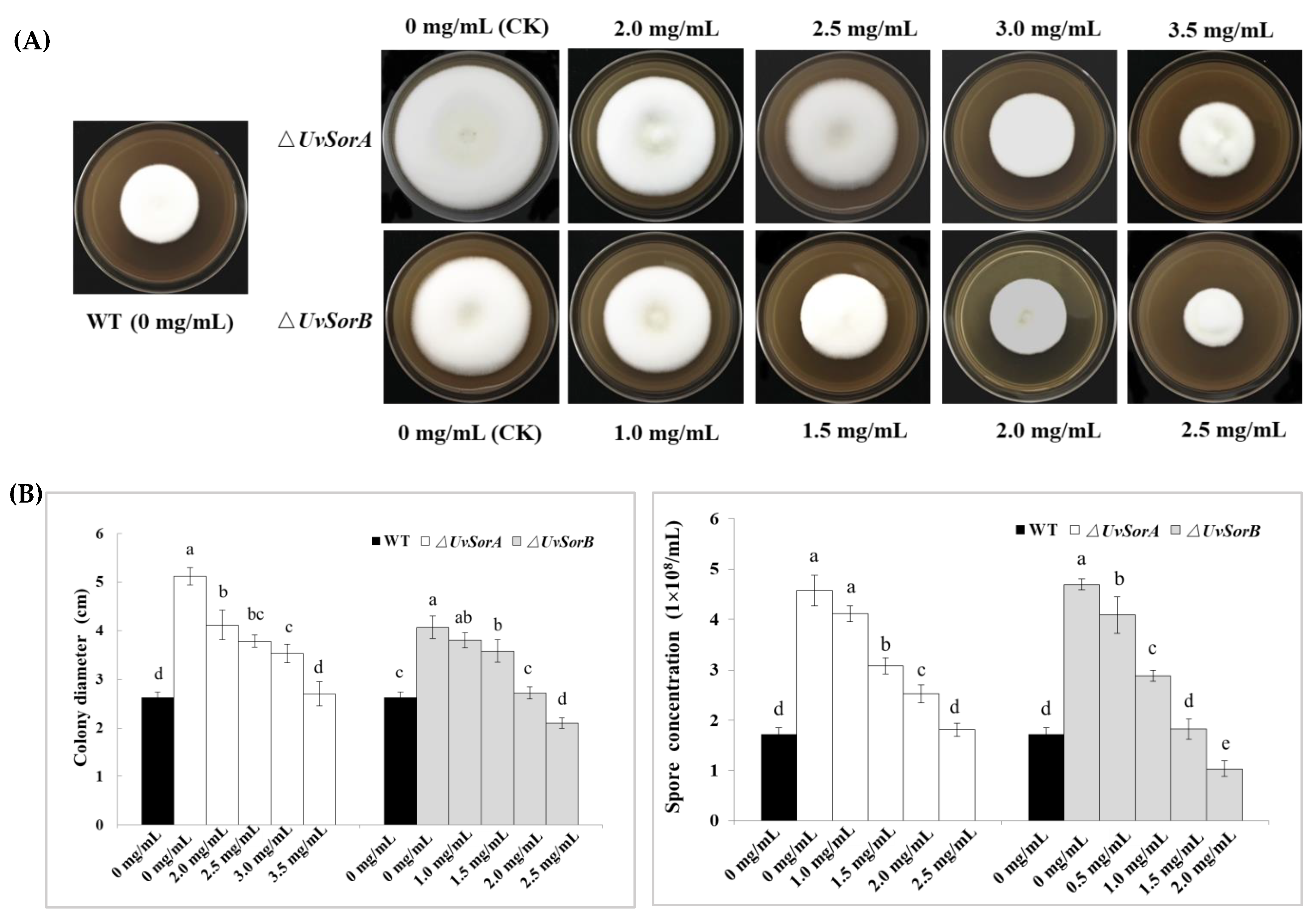
4.7. Feeding Sorbicillinoids in ΔUvSorA and ΔUvSorB Mutants
4.8. Microscopic Observation of Hyphal Morphology
4.9. Phytotoxic Assays with Culture Filtrates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.Y.; Keller, N.P. Spatial and temporal control of fungal natural product synthesis. Nat. Prod. Rep. 2014, 31, 1277–1286. [Google Scholar] [CrossRef]
- Keller, N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brankhage, A.A. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Mei, Y.-Z.; Zhu, Y.-L.; Huang, P.-W.; Yang, Q.; Dai, C.-C. Strategies for gene disruption and expression in filamentous fungi. Appl. Microbiol. Biotechnol. 2019, 103, 6041–6059. [Google Scholar] [CrossRef]
- Fan, J.; Yang, J.; Wang, Y.-Q.; Li, G.-B.; Li, Y.; Huang, F.; Wang, W.-M. Current understanding on Villosiclava virens, a unique flower-infecting fungus causing rice false smut disease. Mol. Plant Pathol. 2016, 17, 1321–1330. [Google Scholar] [CrossRef]
- Sun, W.; Fan, J.; Fang, A.; Li, Y.; Tariqjaveed, M.; Li, D.; Hu, D.; Wang, W.-M. Ustilaginoidea virens: Insights into an emerging rice pathogen. Annu. Rev. Phytopathol. 2020, 58, 363–385. [Google Scholar] [CrossRef]
- Hu, M.; Luo, L.; Wang, S.; Liu, Y.; Li, J. Infection processes of Ustilaginoidea virens during artificial inoculation of rice panicles. Eur. J. Plant Pathol. 2014, 139, 67–77. [Google Scholar] [CrossRef]
- Nakamura, K.; Izumiyama, N.; Ohtsubo, K.; Koiso, Y.; Iwasaki, S.; Sonoda, R.; Fujita, Y.; Yaegashi, H.; Sato, Z. “Lupinosis”-like lesions in mice caused by ustiloxin, produced by Ustilaginoieda virens: A morphological study. Nat. Toxins 1994, 2, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Lu, S.; Shan, T.; Wang, P.; Sun, W.; Chen, Z.; Wang, S. Chemistry and biology of mycotoxins from rice false smut pathogen. In Mycotoxins: Properties, Applications and Hazards; Melborn, B.J., Greene, J.C., Eds.; Nova Science Publishers: New York, NY, USA, 2012; pp. 109–130. [Google Scholar]
- Wang, B.; Liu, L.; Li, Y.; Zou, J.; Li, D.; Zhao, D.; Li, W.; Sun, W. Ustilaginoidin D induces hepatotoxicity and behaviour aberrations in zebrafish larvae. Toxicology 2021, 456, 152786. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Sun, W.; Meng, J.; Wang, A.; Wang, X.; Tian, J.; Fu, X.; Dai, J.; Liu, Y.; Lai, D.; et al. Bioactive bis-naphtho-γ-pyrones from rice false smut pathogen Ustilaginoidea virens. J. Agric. Food Chem. 2015, 63, 3501–3508. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J.; Lai, D.; Wang, W.; Dai, J.; Zhou, L.; Liu, Y. Ustiloxin G, a new cyclopeptide mycotoxin from rice false smut balls. Toxins 2017, 9, 54. [Google Scholar] [CrossRef]
- Meng, J.; Gu, G.; Dang, P.; Zhang, X.; Wang, W.; Dai, J.; Liu, Y.; Lai, D.; Zhou, L. Sorbicillinoids from the fungus Ustilaginoidea virens and their phytotoxic, cytotoxic, and antimicrobial activities. Front. Chem. 2019, 7, 435. [Google Scholar] [CrossRef]
- Koiso, Y.; Natori, M.; Iwasaki, S.; Sato, S.; Sonoda, R.; Fujita, Y.; Yaegashi, H.; Sato, Z. Ustiloxin: A phytotoxin and a mycotoxin from false smut balls on rice panicles. Tetrahedron Lett. 1992, 33, 4157–4160. [Google Scholar] [CrossRef]
- Koiso, Y.; Li, Y.; Iwasaki, S.; Hanaoka, K.; Kobayashi, T.; Sonoda, R.; Fujita, Y.; Yaegashi, H.; Sato, Z. Ustiloxins, antimitotic cyclic peptides from false smut balls on rice panicles caused by Ustilaginoidea virens. J. Antibiot. 1994, 47, 765–773. [Google Scholar] [CrossRef]
- Meng, J.; Sun, W.; Mao, Z.; Xu, D.; Wang, X.; Lu, S.; Lai, D.; Liu, Y.; Zhou, L.; Zhang, G. Main ustilaginoidins and their distribution in rice false smut balls. Toxins 2015, 7, 4023–4034. [Google Scholar] [CrossRef]
- Sun, W.; Wang, A.; Xu, D.; Wang, W.; Meng, J.; Dai, J.; Liu, Y.; Lai, D.; Zhou, L. New ustilaginoidins from rice false smut balls caused by Villosiclava virens and their phytotoxic and cytotoxic activities. J. Agric. Food Chem. 2017, 65, 5151–5160. [Google Scholar] [CrossRef]
- Meng, J.; Zhao, S.; Dang, P.; Zhou, Z.; Lai, D.; Zhou, L. Ustilaginoidin M1, a new bis-naphtho-γ-pyrone from the fungus Villosiclava virens. Nat. Prod. Res. 2021, 35, 1555–1560. [Google Scholar] [CrossRef]
- Xu, D.; Yin, R.; Zhou, Z.; Gu, G.; Zhao, S.; Xu, J.-R.; Liu, J.; Peng, Y.-L.; Lai, D.; Zhou, L. Elucidation of ustilaginoidin biosynthesis reveals a previously unrecognised class of ene-reductases. Chem. Sci. 2021, 12, 14883–114892. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Meng, J.; Zhang, X.; Xu, D.; Dai, J.; Zhou, L. Ustilobisorbicillinol A, a cytotoxic sorbyl-containing aromatic polyketide from Ustilagninoidea virens. Org. Lett. 2019, 21, 1311–1314. [Google Scholar] [CrossRef] [PubMed]
- Harned, A.M.; Volp, K.A. The sorbicillinoid family of natural products: Isolation, biosynthesis, and synthetic studies. Nat. Prod. Rep. 2011, 28, 1790–1810. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Wang, X.; Xu, D.; Fu, X.; Zhang, X.; Lai, D.; Zhou, L.; Zhang, G. Sorbicillinoids from fungi and their bioactivities. Molecules 2016, 21, 715. [Google Scholar] [CrossRef]
- Hou, X.; Zhang, X.; Xue, M.; Zhao, Z.; Zhang, H.; Xu, D.; Lai, D.; Zhou, L. Recent advances in sorbicillinoids from fungi and their bioactivities (covering 2016–2021). J. Fungi 2022, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Takagi, M.; Shin-ya, K. JBIR-124: A novel antioxidative agent from a marine sponge-derived fungus Penicillium citrinum SpI080624G1f01. J. Antibiot. 2012, 65, 45–47. [Google Scholar] [CrossRef]
- Du, L.; Zhu, T.; Li, L.; Cai, S.; Zhao, B.; Gu, Q. Cytotoxic Sorbicillinoids and bisorbicillinoids from a marine-derived fungus Trichoderma sp. Chem. Pharm. Bull. 2009, 57, 220–223. [Google Scholar] [CrossRef]
- Kahlert, L.; Bassiony, E.F.; Cox, R.J.; Skellam, E.J. Diels–Alder reactions during the biosynthesis of sorbicillinoids. Angew. Chem. Int. Ed. 2020, 59, 5816–5822. [Google Scholar] [CrossRef]
- Chen, G.; Chu, J. Characterization of two polyketide synthases involved in sorbicillinoid biosynthesis by Acremonium chrysogenum using the CRISPR/Cas9 System. Appl. Biochem. Biotechnol. 2019, 188, 1134–1144. [Google Scholar] [CrossRef]
- Kahlert, L.; Cox, R.J.; Skellam, E. The same but different: Multiple functions of the fungal flavin dependent monooxygenase SorD from Penicillium chrysogenum. Chem. Commun. 2020, 56, 10934–10937. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Muller, R.; Wohlleben, W.; et al. antiSMASH3.0 – a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, K.; Fang, A.; Han, Y.; Yang, J.; Xue, M.; Bao, J.; Hu, D.; Zhou, B.; Sun, X.; et al. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat. Commun. 2014, 5, 3849. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Rather, M.A.; Dar, M.S.; Aga, M.A.; Ahmad, N.; Manzoor, A.; Qayum, A.; Shah, A.; Mushtaq, S.; Ahmad, Z.; et al. Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorg. Med. Chem. Lett. 2017, 27, 2579–2582. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Wang, Y.; Han, Y.; Xu, J.-R.; Wang, C. HvHOG1 is important for hyphal growth and stress responses in the rice false smut fungus Ustilaginoidea virens. Sci. Rep. 2016, 6, 24824. [Google Scholar] [CrossRef]
- Liang, Y.; Han, Y.; Wang, C.; Jiang, C.; Xu, J.-R. Targeted deletion of the USTA andUvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system. Front. Plant Sci. 2018, 9, 699. [Google Scholar] [CrossRef]
- Wosten, H.; Richter, M.; Willey, J.M. Structural proteins involved in emergence of microbial aerial hyphae. Fungal genetic and biology 1999, 27, 153–160. [Google Scholar] [CrossRef]
- Craney, A.; Ahmed, S.; Nodwell, J. Towards a new science of secondary metabolism. J. Antibiot. 2013, 66, 387–400. [Google Scholar] [CrossRef]
- Schneider, P.; Misiek, M.; Hoffmeister, D. In vivo and in vitro production options for fungal secondary metabolites. Mol. Pharmaceut. 2008, 5, 234–242. [Google Scholar] [CrossRef]
- Mou, Y.; Luo, H.; Mao, Z.; Shan, T.; Sun, W.; Zhou, K.; Zhou, L. Enhancement of palmarumycins C12 and C13 production in liquid culture of endophytic fungus Berkleasmium sp. Dzf12 after treatments with metal ions. Int. J. Mol. Sci. 2013, 14, 979–998. [Google Scholar] [CrossRef]
- Luo, H.; Xu, D.; Xie, R.; Zhang, X.; Wang, J.; Dong, X.; Lai, D.; Zhou, L.; Liu, Y. Enhancement of botrallin and TMC-264 production in liquid culture of endophytic fungus Hyalodendriella sp. Ponipodef12 after treatments with metal ions. Electron. J. Biotechnol. 2016, 24, 12–20. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Yao, J.; Li, Y.-F.; Yang, X.; Wang, W.-X.; Zhang, A.-F.; Gao, T.-C. Frequency distribution of sensitivity of Ustilaginoidea virens to four EBI fungicides, prochloraz, difenoconazole, propiconazole and tebuconazole, and their efficacy in controlling rice false smut in Anhui Province of China. Phytoparasitica 2013, 41, 277–284. [Google Scholar] [CrossRef]
- Muniraju, K.M.; Pramesh, D.; Mallesh, S.B.; Mallikarjun, K.; Guruprasad, G.S. Novel fungicides for the management of false smut disease of rice caused by Ustilaginoidea virens. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2664–2669. [Google Scholar] [CrossRef]
- Singh, S.; Singh, N.; Kumar, V.; Kumar, V.; Datta, S.; Wani, A.B.; Singh, D.; Singh, K.; Singh, J. Toxicity, monitoring and biodegradation of the fungicide carbendazim. Environ. Chem. Lett. 2016, 14, 317–329. [Google Scholar] [CrossRef]
- Zhao, H.; Tao, X.; Song, W.; Xu, H.; Li, M.; Cai, Y.; Wang, J.; Duan, Y.; Zhou, M. Mechanism of Fusarium graminearum resistance to ergosterol biosynthesis inhibitors: G443S substitution of the drug target FgCYP51A. J. Agric. Food Chem. 2022, 70, 1788–1798. [Google Scholar] [CrossRef] [PubMed]
- Affourtit, C.; Heaney, S.P.; Moore, A.L. Mitochondrial electron transfer in the wheat pathogenic fungus Septoria tritici: On the role of alternative respiratory enzymes in fungicide resistance. Biochim. Biophys. Acta 2000, 1459, 291–298. [Google Scholar] [CrossRef]
- Bohnert, H.U.; Fudal, I.; Dioh, W.; Tharreau, D.; Notteghem, J.-L.; Lebrun, M.-H. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 2004, 16, 2499–2513. [Google Scholar] [CrossRef]
- Dagenais, T.R.T.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 2009, 22, 447–465. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M.; Manners, J.M. On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 2012, 13, 399–413. [Google Scholar] [CrossRef]
- Yago, J.I.; Lin, C.-H.; Chung, K.-R. The SLT2 mitogen-activated protein kinase-mediated signaling pathway governs conidiation, morphogenesis, fungal virulence and production of toxin and melanin in the tangerine pathotype of Alternaria alternata. Mol. Plant Pathol. 2011, 12, 653–665. [Google Scholar] [CrossRef]
- Niehaus, E.-M.; von Bargen, K.W.; Espino, J.J.; Pfannmuller, A.; Humpf, H.-U. Tudzynski, B. Characterization of the fusaric acid gene cluster in Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2014, 98, 1749–1762. [Google Scholar] [CrossRef]
- Ding, Z.; Li, M.; Sun, F.; Xi, P.; Sun, L.; Zhang, L.; Jiang, Z. Mitogen-activated protein kinases are associated with the regulation of physiological traits and virulence in Fusarium oxysporum f. sp. cubense. PLoS ONE 2015, 10, e0122634. [Google Scholar] [CrossRef] [PubMed]
- Guzman-Chavez, F.; Salo, O.; Nygard, Y.; Lankhorst, P.P.; Bovenberg, R.A.L.; Driessen, A.J.M. Mechanism and regulation of sorbicillin biosynthesis by Penicillium chrysogenum. Microb. Biotechnol. 2017, 10, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Derntl, C.; Guzman-Chavez, F.; Mello-de-Sousa, T.M.; Busse, H.J.; Driessen, A.J.M.; Mach, R.L.; Mach-Aigner, A.R. In vivo study of the sorbicillinoid gene cluster in Trichoderma reesei. Front. Microbiol. 2017, 8, 2037. [Google Scholar] [CrossRef] [PubMed]
- Druzhinina, I.S.; Kubicek, E.M.; Kubicek, C.P. Several steps of lateral gene transfer followed by events of ‘birth-and-death’ evolution shaped a fungal sorbicillinoid biosynthetic gene cluster. BMC Evol. Biol. 2016, 16, 269. [Google Scholar] [CrossRef]
- Dattenbock, C.; Tisch, D.; Schuster, A.; Monroy, A.A.; Hinterdobler, W.; Schmoll, M. Gene regulation associated with sexual development and female fertility in different isolates of Trichoderma reesei. Fungal Biol. Biotechnol. 2018, 5, 9. [Google Scholar] [CrossRef]
- Hinterdobler, W.; Schuster, A.; Tisch, D.; Ozkan, E.; Bazafkan, H.; Schinnerl, J.; Brecker, L.; Bohmdorfer, S.; Schmoll, M. The role of PKAc1 in gene regulation and trichodimerol production in Trichoderma reesei. Fungal Biol. Biotechnol. 2019, 6, 12. [Google Scholar] [CrossRef]
- Xu, D.; Xue, M.; Shen, Z.; Jia, X.; Hou, X.; Lai, D.; Zhou, L. Phytotoxic secondary metabolites from fungi. Toxins 2021, 13, 261. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gilson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2003, 2, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef]
- Yu, J.-H.; Hamari, Z.; Han, K.-H.; Seo, J.-A.; Reyes-Dominguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, G.; Xu, J.-R. Efficient approaches for generating GFP fusion and epitope-tagging constructs in filamentous fungi. Methods Mol. Biol. 2011, 722, 199–212. [Google Scholar] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Xu, D.; Hou, X.; Wei, P.; Fu, J.; Zhao, Z.; Jing, M.; Lai, D.; Yin, W.; Zhou, L. UvSorA and UvSorB Involved in Sorbicillinoid Biosynthesis Contribute to Fungal Development, Stress Response and Phytotoxicity in Ustilaginoidea virens. Int. J. Mol. Sci. 2022, 23, 11056. https://doi.org/10.3390/ijms231911056
Zhang X, Xu D, Hou X, Wei P, Fu J, Zhao Z, Jing M, Lai D, Yin W, Zhou L. UvSorA and UvSorB Involved in Sorbicillinoid Biosynthesis Contribute to Fungal Development, Stress Response and Phytotoxicity in Ustilaginoidea virens. International Journal of Molecular Sciences. 2022; 23(19):11056. https://doi.org/10.3390/ijms231911056
Chicago/Turabian StyleZhang, Xuping, Dan Xu, Xuwen Hou, Penglin Wei, Jiajin Fu, Zhitong Zhao, Mingpeng Jing, Daowan Lai, Wenbing Yin, and Ligang Zhou. 2022. "UvSorA and UvSorB Involved in Sorbicillinoid Biosynthesis Contribute to Fungal Development, Stress Response and Phytotoxicity in Ustilaginoidea virens" International Journal of Molecular Sciences 23, no. 19: 11056. https://doi.org/10.3390/ijms231911056
APA StyleZhang, X., Xu, D., Hou, X., Wei, P., Fu, J., Zhao, Z., Jing, M., Lai, D., Yin, W., & Zhou, L. (2022). UvSorA and UvSorB Involved in Sorbicillinoid Biosynthesis Contribute to Fungal Development, Stress Response and Phytotoxicity in Ustilaginoidea virens. International Journal of Molecular Sciences, 23(19), 11056. https://doi.org/10.3390/ijms231911056









