Fetal Programming of the Endocrine Pancreas: Impact of a Maternal Low-Protein Diet on Gene Expression in the Perinatal Rat Pancreas
Abstract
1. Introduction
2. Results
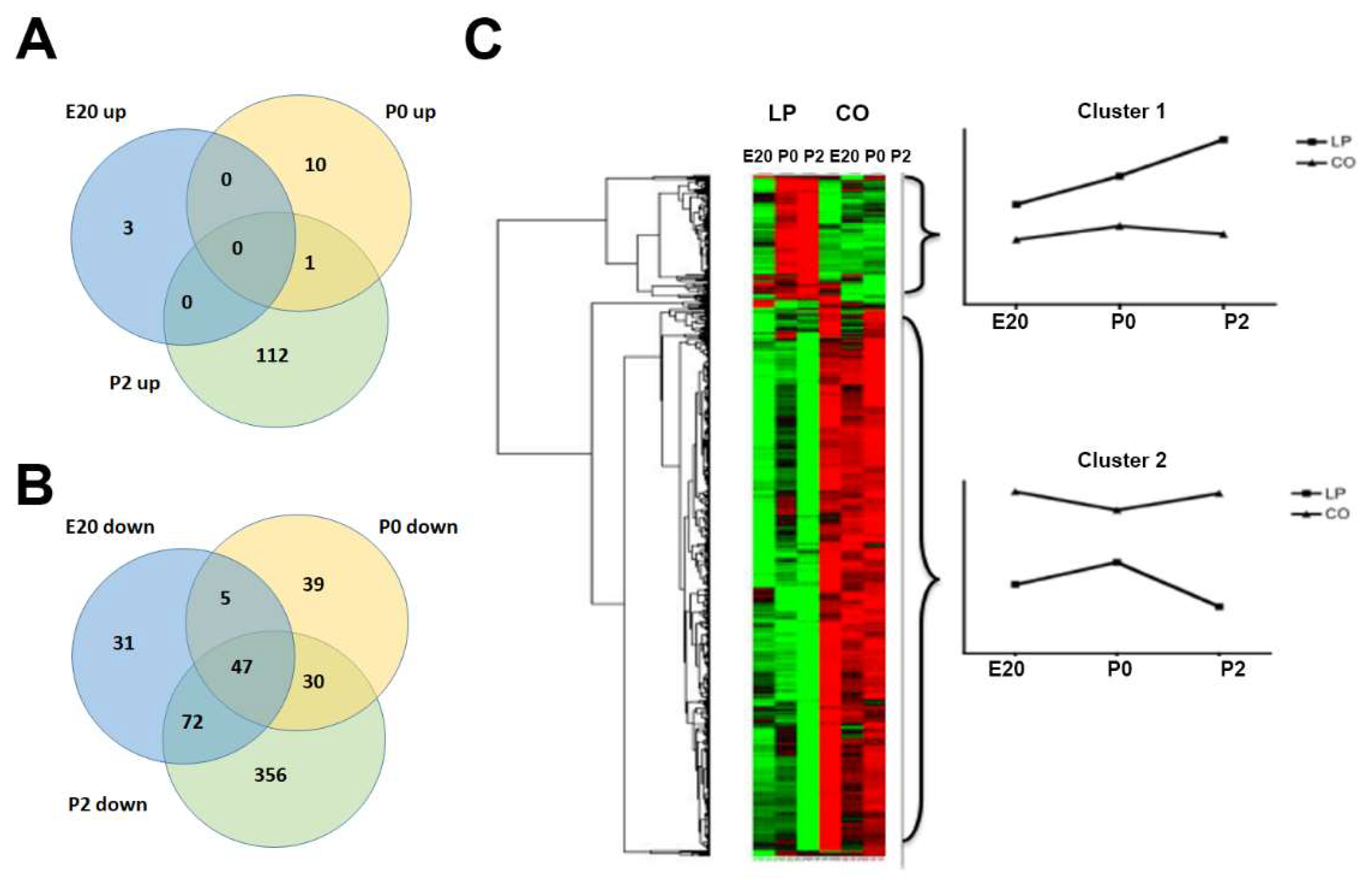
2.1. Gene Regulation in Normal Perinatal Rat Pancreas from E20 to P2
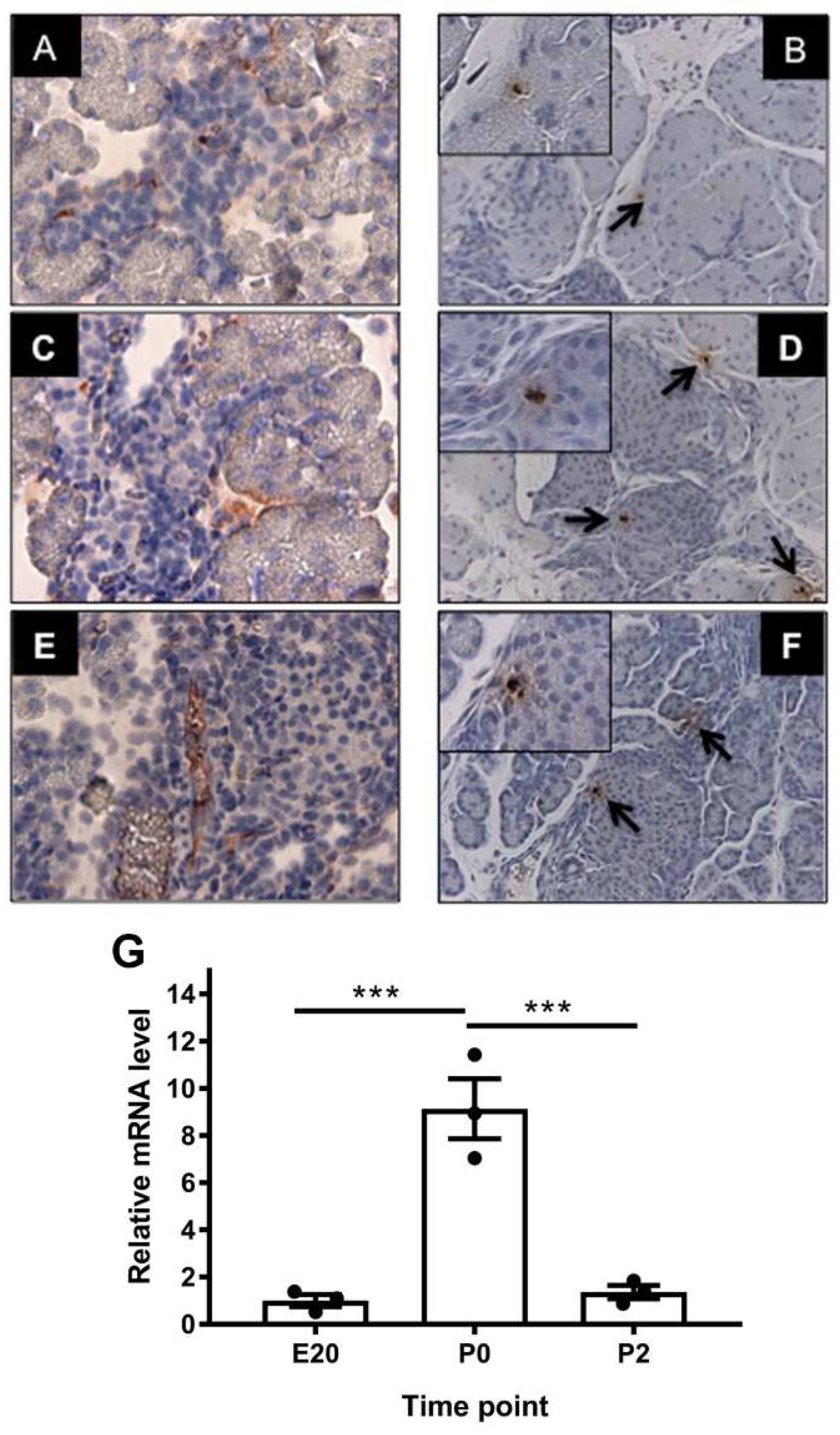
2.2. Localization of Alpha-Feto Protein (Afp) in Perinatal Rat Pancreas
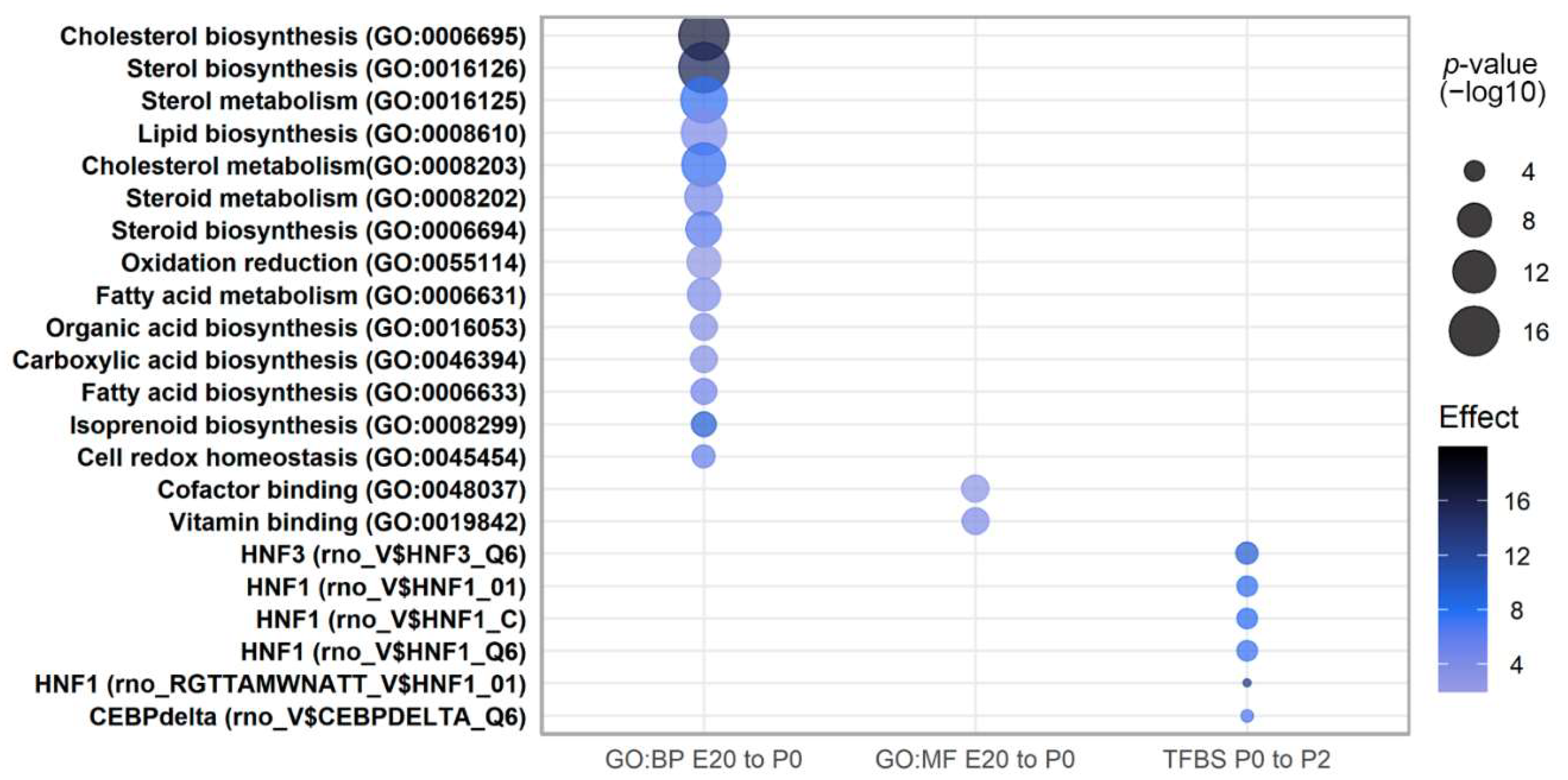
2.3. Clustering and Functional Annotation of Profiles
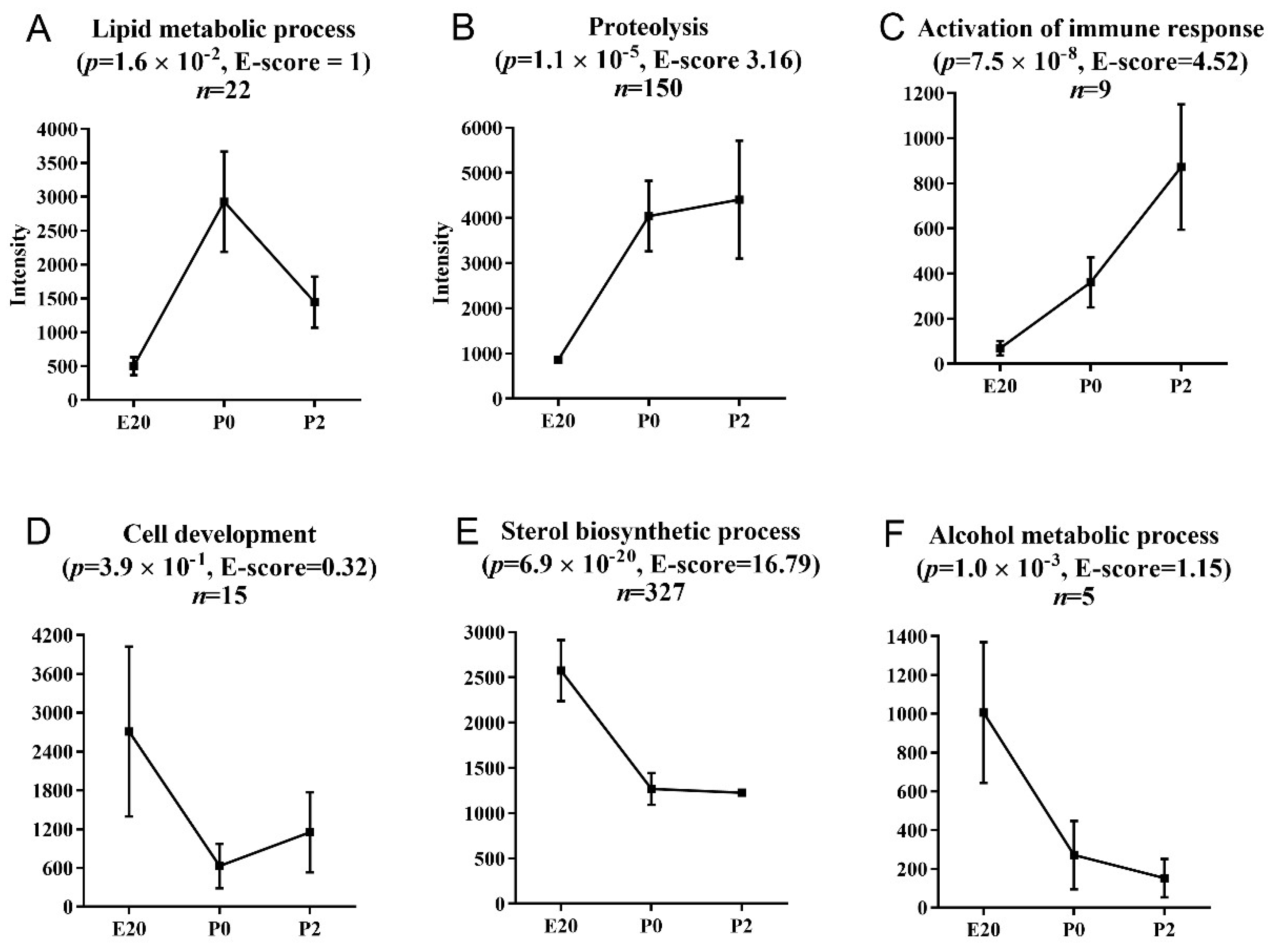
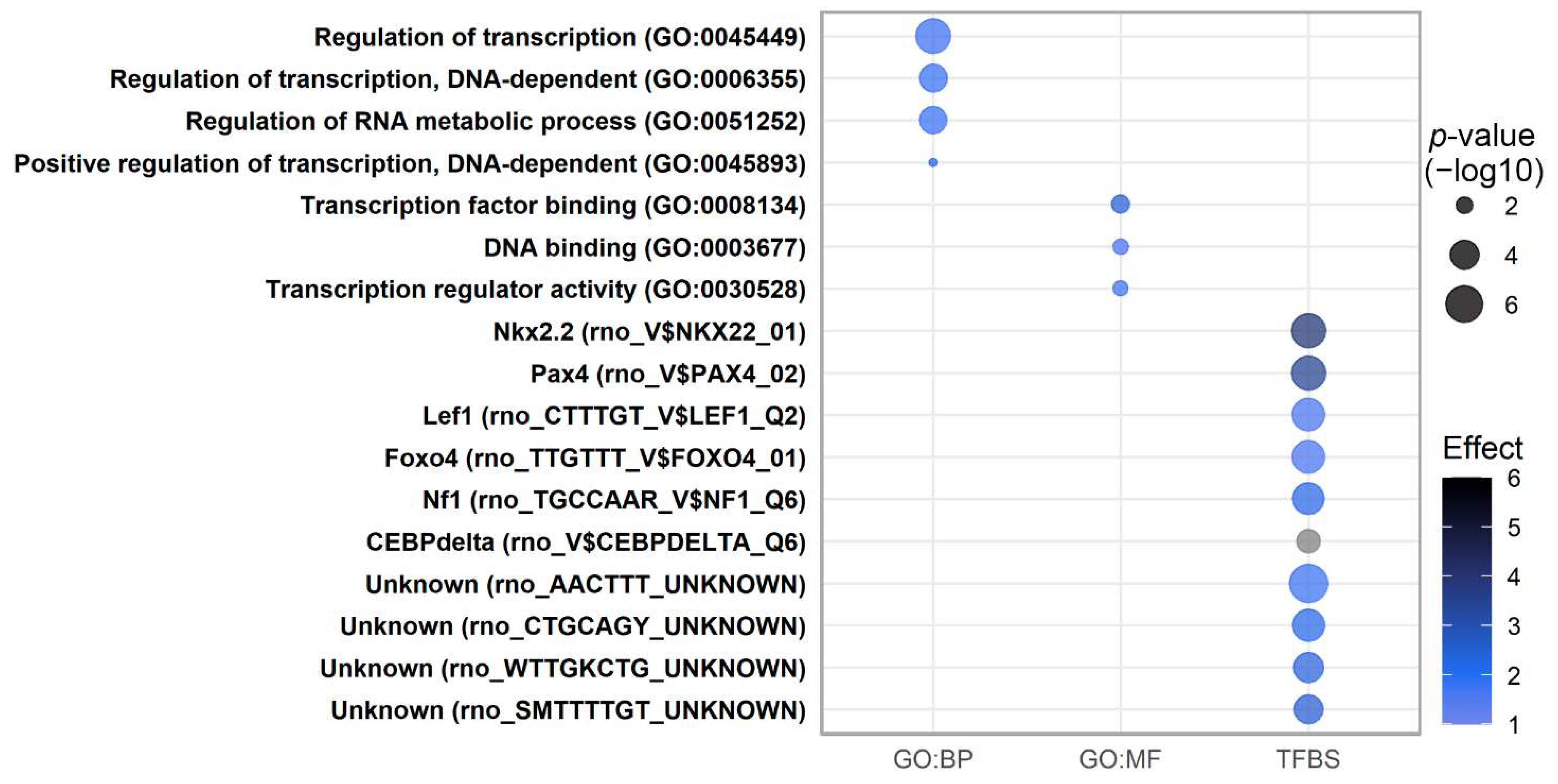
2.4. Pathway Analysis of Perinatally Regulated Transcripts
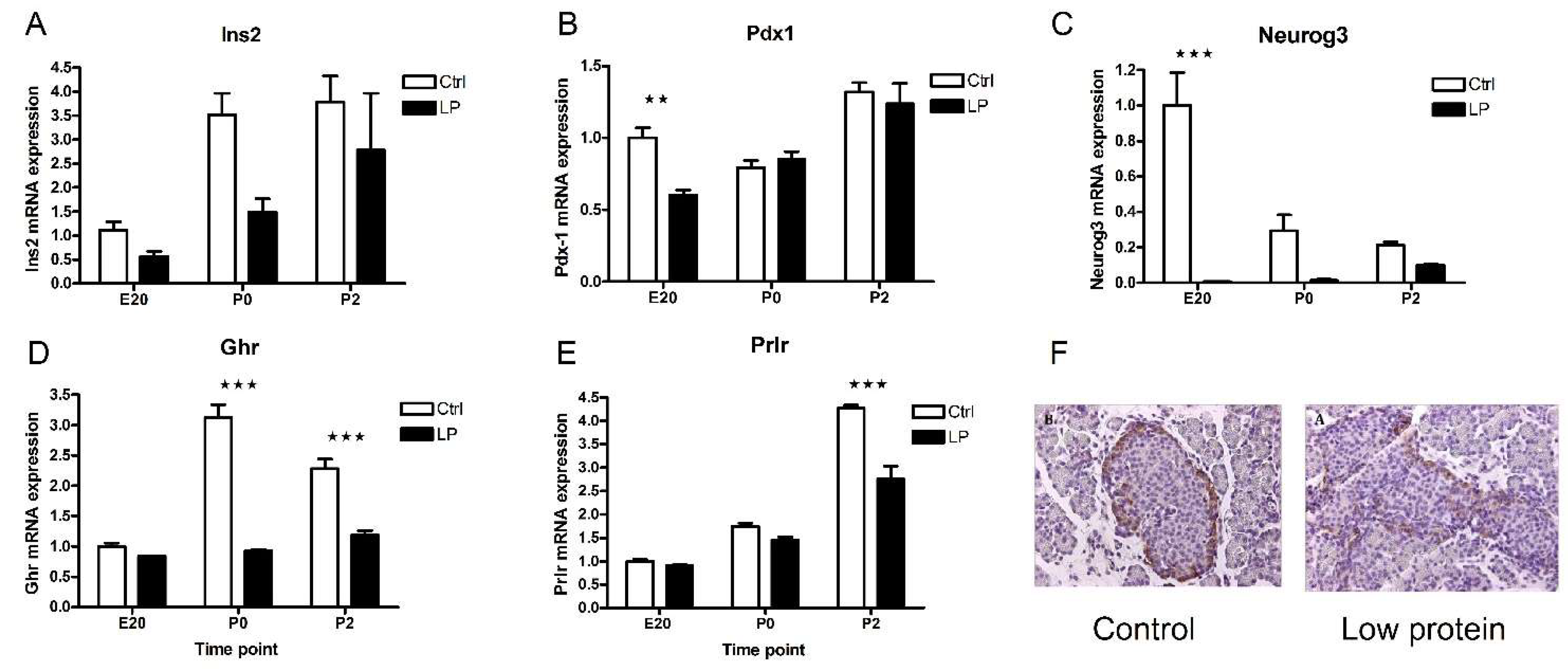
2.5. Characterization of the Low-Protein Malnutrition Model
2.6. Transcripts Regulated by Maternal Low-Protein Diet
2.7. Pathway Analysis of Transcripts Regulated by Low-Protein Diet
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bouret, S.; Levin, B.E.; Ozanne, S.E. Gene-Environment Interactions Controlling Energy and Glucose Homeostasis and the Developmental Origins of Obesity. Physiol. Rev. 2015, 95, 47–82. [Google Scholar] [CrossRef] [PubMed]
- Weir, G.C.; Gaglia, J.; Bonner-Weir, S. Inadequate β-cell mass is essential for the pathogenesis of type 2 diabetes. Lancet Diabetes Endocrinol. 2020, 8, 249–256. [Google Scholar] [CrossRef]
- Nielsen, J.H.; Haase, T.N.; Jaksch, C.; Nalla, A.; Søstrup, B.; Nalla, A.A.; Larsen, L.; Rasmussen, M.; Dalgaard, L.; Gaarn, L.W.; et al. Impact of fetal and neonatal environment on beta cell function and development of diabetes. Acta Obstet. Gynecol. Scand. 2014, 93, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Gradwohl, G.; Dierich, A.; LeMeur, M.; Guillemot, F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. USA 2000, 97, 1607–1611. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, M.C.; Ahnfelt-Ronne, J.; Hald, J.; Madsen, O.D.; Serup, P.; Hecksher-Sorensen, J. An Illustrated Review of Early Pancreas Development in the Mouse. Endocr. Rev. 2007, 28, 685–705. [Google Scholar] [CrossRef]
- Horn, S.; Kobberup, S.; Jørgensen, M.C.; Kalisz, M.; Klein, T.; Kageyama, R.; Gegg, M.; Lickert, H.; Lindner, J.; Magnuson, M.A.; et al. Mind bomb 1 is required for pancreatic β-cell formation. Proc. Natl. Acad. Sci. USA 2012, 109, 7356–7361. [Google Scholar] [CrossRef]
- Bouwens, L.; Rooman, I. Regulation of pancreatic beta-cell mass. Physiol. Rev. 2005, 85, 1255–1270. [Google Scholar] [CrossRef]
- Dhawan, S.; Georgia, S.; Bhushan, A. Formation and regeneration of the endocrine pancreas. Curr. Opin. Cell Biol. 2007, 19, 634–645. [Google Scholar] [CrossRef]
- Dor, Y.; Brown, J.; Martinez, O.I.; Melton, D.A. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature 2004, 429, 41–46. [Google Scholar] [CrossRef]
- Foa, P.P.; Blázquez, E.; Sodoyez, J.C.; Sodoyez-Goffaux, F. The Ontogeny of Mammalian Insular Function. In The Evolution of Pancreatic Islets; Pergamon Press: London, UK, 1976. [Google Scholar]
- Aye, T.; Toschi, E.; Sharma, A.; Sgroi, D.; Bonner-Weir, S. Identification of markers for newly formed beta-cells in the perinatal period: A time of recognized beta-cell immaturity. J. Histochem. Cytochem. 2010, 58, 369–376. [Google Scholar] [CrossRef]
- Hellerstrom, C. The life story of the pancreatic B cell. Diabetologia 1984, 26, 393–400. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, R.C.; Madson, K.L. Pancreatic Insulin-, Glucagon-, and Somatostatin-Positive Islet Cell Populations during the Perinatal Development of the Rat. II. Changes in hormone content and concentration. Biol. Neonatol. 1980, 38, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Hellerstrom, C.; Swenne, I. Functional maturation and proliferation of fetal pancreatic beta-cells. Diabetes 1991, 40 (Suppl. 2), 89–93. [Google Scholar] [CrossRef] [PubMed]
- Martens, G.; Motte, E.; Kramer, G.; Stange, G.; Gaarn, L.W.; Hellemans, K.; Nielsen, J.H.; Aerts, J.; Ling, Z.; Pipeleers, D. Functional characteristics of neonatal rat β cells with distinct markers. J. Mol. Endocrinol. 2013, 52, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, O.; Pages, G.; Gimond, C. The dual-specificity MAP kinase phosphatases: Critical roles in development and cancer. Am. J. Physiol. Cell Physiol. 2010, 299, C189–C202. [Google Scholar] [CrossRef]
- Yamada, T.; Ozaki, N.; Kato, Y.; Miura, Y.; Oiso, Y. Insulin downregulates angiopoietin-like protein 4 mRNA in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2006, 347, 1138–1144. [Google Scholar] [CrossRef]
- Yi, P.; Park, J.-S.; Melton, D.A. RETRACTED: Betatrophin: A Hormone that Controls Pancreatic β Cell Proliferation. Cell 2013, 153, 747–758. [Google Scholar] [CrossRef]
- Møldrup, A.; Petersen, E.D.; Nielsen, J.H. Effects of sex and pregnancy hormones on growth hormone and prolactin receptor gene expression in insulin-producing cells. Endocrinology 1993, 133, 1165–1172. [Google Scholar] [CrossRef]
- Dolva, L.; Nielsen, J.H.; Welinder, B.S.; Hanssen, K.F. Biosynthesis and release of thyrotropin-releasing hormone immunoreactivity in rat pancreatic islets in organ culture. Effects of age, glucose, and streptozotocin. J. Clin. Investig. 1983, 72, 1867–1873. [Google Scholar] [CrossRef]
- Štrbák, V. Pancreatic Thyrotropin Releasing Hormone and Mechanism of Insulin Secretion. Cell. Physiol. Biochem. 2018, 50, 378–384. [Google Scholar] [CrossRef]
- Demarchi, F.; Verardo, R.; Varnum, B.; Brancolini, C.; Schneider, C. Gas6 anti-apoptotic signaling requires NF-kappa B activation. J. Biol. Chem. 2001, 276, 31738–31744. [Google Scholar] [CrossRef] [PubMed]
- Couchie, D.; Lafdil, F.; Martin-Garcia, N.; Laperche, Y.; Zafrani, E.S.; Mavier, P. Expression and role of Gas6 protein and of its receptor Axl in hepatic regeneration from oval cells in the rat. Gastroenterology 2005, 129, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Araki, H.; Sugimoto, T.; Takeuchi, K.; Yamane, T.; Maeda, T.; Yamamoto, Y.; Nishi, K.; Asano, M.; Shirahama-Noda, K.; et al. Legumain/asparaginyl endopeptidase controls extracellular matrix remodeling through the degradation of fibronectin in mouse renal proximal tubular cells. FEBS Lett. 2007, 581, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Sun, C.; Huang, H.; Janda, K.; Edgington, T. Overexpression of legumain in tumors is significant for invasion/metastasis and a candidate enzymatic target for prodrug therapy. Cancer Res. 2003, 63, 2957–2964. [Google Scholar] [PubMed]
- Choi, J.-H.; Lee, M.-Y.; Kim, Y.; Shim, J.-Y.; Han, S.-M.; Lee, K.-A.; Choi, Y.-K.; Jeon, H.-M.; Baek, K.-H. Isolation of genes involved in pancreas regeneration by subtractive hybridization. Biol. Chem. 2010, 391, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Ekman, G.C.; Garcia, T.; Carnes, K.; Zhang, Z.; Murphy, T.; Murphy, K.M.; Hess, R.A.; Cooke, P.S.; Hofmann, M.-C. ETV5 Regulates Sertoli Cell Chemokines Involved in Mouse Stem/Progenitor Spermatogonia Maintenance. Stem Cells 2010, 28, 1882–1892. [Google Scholar] [CrossRef]
- Kobberup, S.; Nyeng, P.; Juhl, K.; Hutton, J.; Jensen, J. ETS-family genes in pancreatic development. Dev. Dyn. 2007, 236, 3100–3110. [Google Scholar] [CrossRef]
- Haase, T.N.; Rasmussen, M.; Jaksch, C.A.M.; Gaarn, L.W.; Petersen, C.K.; Billestrup, N.; Nielsen, J.H. Growth arrest specific protein (GAS) 6: A role in the regulation of proliferation and functional capacity of the perinatal rat beta cell. Diabetologia 2013, 56, 763–773. [Google Scholar] [CrossRef]
- Winkel, L.; Bagge, A.; Larsen, L.; Haase, T.N.; Rasmussen, M.; Lykke, J.; Holmgaard, D.B.; Thim, L.; Nielsen, J.H.; Dalgaard, L.T. Trefoil factor 3 in perinatal pancreas is increased by gestational low protein diet and associated with accelerated β-cell maturation. Islets 2018, 10, e1472186-25. [Google Scholar] [CrossRef]
- Jackerott, M.; Lee, Y.C.; Møllgård, K.; Kofod, H.; Jensen, J.; Rohleder, S.; Neubauer, N.; Gaarn, L.W.; Lykke, J.; Dodge, R.; et al. Trefoil Factors Are Expressed in Human and Rat Endocrine Pancreas: Differential Regulation by Growth Hormone. Endocrinology 2006, 147, 5752–5759. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Lin, Z.; Lin, Q.; Bei, W.; Guo, J. Pathological and therapeutic roles of bioactive peptide trefoil factor 3 in diverse diseases: Recent progress and perspective. Cell Death Dis. 2022, 13, 62. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Fang, Z.; Yu, X.; Zhang, M. Transcription factors involved in glucose-stimulated insulin secretion of pancreatic beta cells. Biochem. Biophys. Res. Commun. 2009, 384, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Iwasaki, Y.; Yatoh, S.; Toyonori, K.; Shin, K.; Noriyuki, I.; Takashi, Y.; Takashi, M.; Yoshimi, N.; Naoya, Y.; et al. Cholesterol accumulation and diabetes in pancreatic beta-cell-specific SREBP-2 transgenic mice: A new model for lipotoxicity. J. Lipid Res. 2008, 49, 2524–2534. [Google Scholar] [CrossRef]
- Reusens, B.; Theys, N.; Dumortier, O.; Goosse, K.; Remacle, C. Maternal malnutrition programs the endocrine pancreas in progeny. Am. J. Clin. Nutr. 2011, 94 (Suppl. 6), 1824S–1829S. [Google Scholar] [CrossRef]
- Søstrup, B.; Gaarn, L.W.; Nalla, A.; Billestrup, N.; Nielsen, J.H. Co-ordinated regulation of neurogenin-3 expression in the maternal and fetal pancreas during pregnancy. Acta Obstet. Gynecol. Scand. 2014, 93, 1190–1197. [Google Scholar] [CrossRef]
- Golden, T.N.; Simmons, R.A. Immune dysfunction in developmental programming of type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2021, 17, 235–245. [Google Scholar] [CrossRef]
- Bruun, C.; Christensen, G.L.; Jacobsen, M.L.B.; Kanstrup, M.B.; Jensen, P.R.; Fjordvang, H.; Mandrup-Poulsen, T.; Billestrup, N. Inhibition of beta cell growth and function by bone morphogenetic proteins. Diabetologia 2014, 57, 2546–2554. [Google Scholar] [CrossRef]
- Dumartin, L.; Whiteman, H.J.; Weeks, M.E.; Hariharan, D.; Dmitrovic, B.; Iacobuzio-Donahue, C.A.; Brentnall, T.A.; Bronner, M.P.; Feakins, R.M.; Timms, J.F.; et al. AGR2 Is a Novel Surface Antigen That Promotes the Dissemination of Pancreatic Cancer Cells through Regulation of Cathepsins B and D. Cancer Res. 2011, 71, 7091–7102. [Google Scholar] [CrossRef]
- Park, S.-W.; Zhen, G.; Verhaeghe, C.; Nakagami, Y.; Nguyenvu, L.T.; Barczak, A.J.; Killeen, N.; Erle, D.J. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. USA 2009, 106, 6950–6955. [Google Scholar] [CrossRef]
- Kinsey, C.; Balakrishnan, V.; O’Dell, M.R.; Huang, J.L.; Newman, L.; Whitney-Miller, C.L.; Hezel, A.F.; Land, H. Plac8 Links Oncogenic Mutations to Regulation of Autophagy and Is Critical to Pancreatic Cancer Progression. Cell Rep. 2014, 7, 1143–1155. [Google Scholar] [CrossRef]
- Conrad, E.; Stein, R.; Hunter, C.S. Revealing transcription factors during human pancreatic β cell development. Trends Endocrinol. Metab. 2014, 25, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Swenne, I.; Eriksson, U. Diabetes in pregnancy: Islet cell proliferation in the fetal rat pancreas. Diabetologia 1982, 23, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Diraison, F.; Ravier, M.A.; Richards, S.K.; Smith, R.M.; Shimano, H.; Rutter, G.A. SREBP1 is required for the induction by glucose of pancreatic beta-cell genes involved in glucose sensing. J. Lipid Res. 2008, 49, 814–822. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.; Rosenstierne, M.W.; Gaarn, L.W.; Bagge, A.; Pedersen, L.; Dahmcke, C.M.; Nielsen, J.H.; Dalgaard, L.T. Expression and Localization of microRNAs in Perinatal Rat Pancreas: Role of miR-21 in Regulation of Cholesterol Metabolism. PLoS ONE 2011, 6, e25997. [Google Scholar] [CrossRef] [PubMed]
- Freie, H.M.; Pasma, A.; Bouman, P.R. Quantitative analysis of pancreatic islet development and insulin storage in the foetal and newborn rat. Acta Endocrinol. 1975, 80, 657–666. [Google Scholar] [CrossRef]
- Chen, Z.; Downing, S.; Tzanakakis, E.S. Four Decades After the Discovery of Regenerating Islet-Derived (Reg) Proteins: Current Understanding and Challenges. Front. Cell Dev. Biol. 2019, 7, 235. [Google Scholar] [CrossRef]
- Okamoto, H. The Reg gene family and Reg proteins: With special attention to the regeneration of pancreatic beta-cells. J. Hepatobiliary-Pancreat. Surg. 1999, 6, 254–262. [Google Scholar] [CrossRef]
- Sala, P.; Torrinhas, R.S.; Fonseca, D.C.; Heymsfield, S.; Giannella-Neto, D.; Waitzberg, D.L. Type 2 Diabetes Remission After Roux-en-Y Gastric Bypass: Evidence for Increased Expression of Jejunal Genes Encoding Regenerating Pancreatic Islet-Derived Proteins as a Potential Mechanism. Obes. Surg. 2017, 27, 1123–1127. [Google Scholar] [CrossRef]
- Rezania, A.; Bruin, J.E.; Arora, P.; Rubin, A.; Batushansky, I.; Asadi, A.; O’Dwyer, S.; Quiskamp, N.; Mojibian, M.; Albrecht, T.; et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 1121–1133. [Google Scholar] [CrossRef]
- Zhang, L.; Theise, N.; Chua, M.; Reid, L.M. The stem cell niche of human livers: Symmetry between development and regeneration. Hepatology 2008, 48, 1598–1607. [Google Scholar] [CrossRef]
- Jelnes, P.; Santoni-Rugiu, E.; Rasmussen, M.; Friis, S.L.; Nielsen, J.H.; Tygstrup, N.; Bisgaard, H.C. Remarkable heterogeneity displayed by oval cells in rat and mouse models of stem cell–mediated liver regeneration. Hepatology 2007, 45, 1462–1470. [Google Scholar] [CrossRef]
- Mizejewski, G.J. Biological roles of alpha-fetoprotein during pregnancy and perinatal development. Exp. Biol. Med. 2004, 229, 439–463. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Guo, J.; Yuan, L.; Cheng, M.; Cao, L.; Shi, H.; Tong, H.; Wang, N.; De, W. Alpha-fetoprotein is dynamically expressed in rat pancreas during development. Dev. Growth Differ. 2007, 49, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, G.; Jabs, N.; Konstantinova, I.; Domogatskaya, A.; Tryggvason, K.; Sorokin, L.; Fässler, R.; Gu, G.; Gerber, H.-P.; Ferrara, N.; et al. The vascular basement membrane: A niche for insulin gene expression and Beta cell proliferation. Dev. Cell 2006, 10, 397–405. [Google Scholar] [CrossRef]
- Eberhard, D.; Kragl, M.; Lammert, E. ‘Giving and taking’: Endothelial and beta-cells in the islets of Langerhans. Trends Endocrinol. Metab. 2010, 21, 457–463. [Google Scholar] [CrossRef]
- Schumacher, A.; Costa, S.-D.; Zenclussen, A.C. Endocrine Factors Modulating Immune Responses in Pregnancy. Front. Immunol. 2014, 5, 196. [Google Scholar] [CrossRef]
- Tian, L.; Zhou, J.; Casimiro, M.C.; Pestell, R.G.; Wang, C. Activating peroxisome proliferator-activated receptor gamma mutant promotes tumor growth in vivo by enhancing angiogenesis. Cancer Res. 2009, 69, 9236–9244. [Google Scholar] [CrossRef]
- Chen, J.M.; Fortunato, M.; Stevens, R.A.; Barrett, A.J. Activation of progelatinase A by mammalian legumain, a recently discovered cysteine proteinase. Biol. Chem. 2001, 382, 777–783. [Google Scholar] [CrossRef]
- Gutierrez-Aguilar, R.; Kim, D.-H.; Casimir, M.; Dai, X.-Q.; Pfluger, P.; Park, J.; Haller, A.; Donelan, E.; Park, J.; D’Alessio, D.; et al. The role of the transcription factor ETV5 in insulin exocytosis. Diabetologia 2013, 57, 383–391. [Google Scholar] [CrossRef]
- Planaguma, J.; Liljestrom, M.; Alameda, F.; Bützow, R.; Virtanen, I.; Reventós, J.; Hukkanen, M. Matrix metalloproteinase-2 and matrix metalloproteinase-9 codistribute with transcription factors RUNX1/AML1 and ETV5/ERM at the invasive front of endometrial and ovarian carcinoma. Hum. Pathol. 2011, 42, 57–67. [Google Scholar] [CrossRef]
- Miralles, F.; Battelino, T.; Czernichow, P.; Scharfmann, R. TGF-beta plays a key role in morphogenesis of the pancreatic islets of Langerhans by controling the activity of the matrix metalloproteinase MMP-2. J. Cell Biol. 1998, 143, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, P.; Huotari, M.; Koivisto, T.; Ustinov, J.; Palgi, J.; Rasilainen, S.; Lehtonen, E.; Keski-Oja, J.; Otonkoski, T. Impaired migration and delayed differentiation of pancreatic islet cells in mice lacking EGF-receptors. Development 2000, 127, 2617–2627. [Google Scholar] [CrossRef] [PubMed]
- Stetler-Stevenson, W.G.; Aznavoorian, S.; Liotta, L.A. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu. Rev. Cell Biol. 1993, 9, 541–573. [Google Scholar] [CrossRef] [PubMed]
- Liotta, L.A.; Tryggvason, K.; Garbisa, S.; Hart, I.; Foltz, C.M.; Shafie, S. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature 1980, 284, 67–68. [Google Scholar] [CrossRef]
- Miller, K.; Kim, A.; Kilimnik, G.; Jo, J.; Moka, U.; Periwal, V.; Hara, M. Islet Formation during the Neonatal Development in Mice. PLoS ONE 2009, 4, e7739. [Google Scholar] [CrossRef]
- Friedrichsen, B.N.; Carlsson, C.; Moldrup, A.; Michelsen, B.; Jensen, C.H.; Teisner, B.; Nielsen, J.H. Expression, biosynthesis and release of preadipocyte factor-1/ delta-like protein/fetal antigen-1 in pancreatic beta-cells: Possible physiological implications. J. Endocrinol. 2003, 176, 257–266. [Google Scholar] [CrossRef]
- Pagliuca, F.W.; Millman, J.R.; Gürtler, M.; Segel, M.; Van Dervort, A.; Ryu, J.H.; Peterson, Q.P.; Greiner, D.; Melton, D.A. Generation of Functional Human Pancreatic β Cells In Vitro. Cell 2014, 159, 428–439. [Google Scholar] [CrossRef]
- Simon, R.; Lam, A.; Li, M.C.; Ngan, M.; Menenzes, S.; Zhao, Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007, 3, 11–17. [Google Scholar] [CrossRef]
- Korn, E.L.; Troendle, J.F.; McShane, L.M.; Simon, R. Controling the number of false discoveries: Application to high-dimensional genomic data. J. Stat. Plan. Inference 2004, 124, 379–398. [Google Scholar] [CrossRef]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef]
- Li, C.; Wong, W.H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 2001, 98, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, 3. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Gerds, T.A.; Nielsen, O.H.; Seidelin, J.B.; Troelsen, J.T.; Olsen, J. pcaGoPromoter—An R Package for Biological and Regulatory Interpretation of Principal Components in Genome-Wide Gene Expression Data. PLoS ONE 2012, 7, e32394. [Google Scholar] [CrossRef]
- Sandelin, A.; Alkema, W.; Engstrom, P.; Wasserman, W.W.; Lenhard, B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucl. Acids Res. 2004, 32, D91–D94. [Google Scholar] [CrossRef]
- Matys, V. TRANSFAC(R) and its module TRANSCompel(R): Transcriptional gene regulation in eukaryotes. Nucl. Acids Res. 2006, 34, D108–D110. [Google Scholar] [CrossRef]

| Increased at P0 vs. E20 | Increased at P2 vs. P0 | ||||||
|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | FC CO | FC LP | Gene Symbol | Gene Name | FC CO | FC LP |
| Reg3a/3b | Regenerating family member 3 alpha/beta | 461/335 | 617/617 | LOC688750 | CD209 antigen | 5.1 | 1.8 |
| Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | 270 | 55 | Prss35 | Protease, serine, 35 | 4.9 | 1.2 |
| LOC286960 | Preprotrypsinogen IV | 155 | 103 | C5 | Complement C5 | 4.8 | −1.3 |
| LOC312273 | Trypsin V-A | 145 | 151 | LOC500183 | NGF-binding Ig light chain | 4.7 | 1.3 |
| Gif | Gastric intrinsic factor | 61 | 61 | C4a | Complement component 4a | 3.6 | 1.6 |
| Alb | Albumin | 58 | 12 | LOC365985 | Adenylate kinase 5 isoform 1 | 3.4 | 1.6 |
| Angptl4 | Angiopoietin-like 4 | 54 | 18 | Tinag | Tubulointerstitial nephritis antigen | 3.1 | −1.5 |
| Apoa1 | Apolipoprotein A1 | 39 | 117 | Spink3 | Serine protease inhibitor, Kazal type 3 | 2.9 | 1.7 |
| Try10 | Pancreatic trypsin 1 | 30 | 15 | LOC686268 | SUMO/sentrin specific protease 5 | 2.9 | 1.4 |
| Ahsg | Alpha-2-HS-glycoprotein | 29 | 19 | Cuzd1 | CUB and zona pellucida-like domains 1 | 2.9 | 1.1 |
| Spink1 | Serine protease inhibitor, Kazal type 1 | 29 | 11 | Egfl6 | EGF-like-domain, multiple 6 | 2.9 | 2.0 |
| Kng1 | Kininogen 1 | 28 | 23 | Ak7 | Adenylate kinase 7 | 2.8 | −1.1 |
| Gas6 | Growth arrest specific 6 | 28 | 18 | Nradd | Neurotrophin receptor associated death domain | 2.8 | 1.3 |
| Apob | Apolipoprotein B | 25 | 84 | Ret | Ret proto-oncogene | 2.7 | −1.0 |
| Slc18a2 | Solute carrier family 18 (vesicular monoamine), member 2 | 23 | 5.3 | Zcchc12 | Zinc finger, CCHC domain containing 12 | 2.6 | −1.1 |
| Decreased at P0 vs. E20 | Decreased at P2 vs. P0 | ||||||
| Gene Symbol | Gene Name | FC CO | FC LP | Gene Symbol | Gene Name | FC CO | FC LP |
| Serpina6 | Serpin family A member 6 | −47 | −74 | Ahsg | Alpha-2-HS-glycoprotein | −31 | −1.4 |
| Hbe1 | Hemoglobin subunit epsilon 1 | −44 | −30 | Fga | Fibrinogen, alpha polypeptide | −23 | −1.2 |
| Tinag | Tubulointerstitial nephritis antigen | −30 | −7.6 | Fabp1 | Fatty acid binding protein 1 | −23 | −1.3 |
| Tnni | Troponin I, skeletal, slow | −28 | −23 | Kng1 | Kininogen 1 | −21 | −1.6 |
| Hbg1 | Hemoglobin, gamma A | −27 | −19 | Apoc2 | Apolipoprotein C-II | −19 | −4.6 |
| Hdc | Histidine decarboxylase | −24 | −17 | Fgg | Fibrinogen, gamma polypeptide | −17 | 2.0 |
| Ptges | Prostaglandin E synthase | −22 | −14 | Apoc1 | Apolipoprotein C-I | −16 | −1.2 |
| Tm7sf2 | Transmembrane 7 superfamily member 2 | −17 | −4.1 | Apoh | Apolipoprotein H | −15 | −1.0 |
| Pln | Phospholamban | −17 | −4.2 | Fgb | Fibrinogen, beta polypeptide | −13 | −1.1 |
| Nags | N-acetylglutamate synthase | −17 | −3.2 | Apoa1 | Apolipoprotein A-I | −13 | −5.4 |
| Clic3 | Chloride intracellular channel 3 | −13 | −7.1 | Hpx | Hemopexin | −13 | −1.2 |
| Adam2 | A disintegrin and metalloprotease domain 2 | −12 | −9.8 | LOC299282 | Serine protease inhibitor | −12 | −1.3 |
| Fdft1 | Farnesyl diphosphate farnesyl transferase 1 | −12 | −4.4 | Itih3 | Inter-alpha trypsin inhibitor, heavy chain 3 | −10 | −1.2 |
| LOC682690 | Chromodomain helicase DNA binding protein 9 | −11 | 1.2 | Pck1 | Phosphoenolpyruvate carboxykinase 1 | −9.9 | −5.3 |
| Camkk2 | Calcium/calmodulin-dependent protein kinase kinase 2, beta | −11 | −2.4 | Serpina3k | Serine peptidase inhibitor, clade A, member 3K | −9.7 | −1.2 |
| Gene Symbol | Gene Name | E20 | Gene Symbol | Gene Name | P0 | Gene Symbol | Gene Name | P2 |
|---|---|---|---|---|---|---|---|---|
| Increased | LP vs. Control (Fold Regulation) | Increased | LP vs. Control (Fold Regulation) | Increased | LP vs. Control (Fold Regulation) | |||
| Agtr2 | Angiotensin II receptor, type 2 | 11 | Apoa4 | Apolipoprotein A4 | 142 | Lgals4 | Lectin, galactose binding, soluble 4 | 236 |
| LOC686892 | Muscleblind-like 1 isoform d | 8.7 | Pga5 | Pepsinogen 5 | 112 | Pga5 | Pepsinogen 5 | 215 |
| Cav | Caveolin | 8.6 | Rbp2 | Retinol binding protein 2 | 86 | Agr2 | Anterior gradient 2 | 110 |
| Ppp3r1 | Calcineurin B, type I | 8.5 | Lgals4 | Lectin, galactose binding, soluble 4 | 79 | Retnla | Resistin like alpha | 93 |
| Zfp260 | Zinc finger protein 260 | 7.7 | Tff1 | Trefoil factor 1 | 73 | Fabp1 | Fatty acid binding protein 1 | 92 |
| Sept2 | Septin 2 | 7.6 | Clca3 | Chloride channel calcium activated 3 | 55 | Tff1 | Trefoil factor 1 | 87 |
| Ogn | Osteoglycin | 7.4 | Agr2 | Anterior gradient 2 | 48 | Gkn1 | Gastrokine 1 | 82 |
| Mat2a | Methionine adenosyltransferase II, alpha | 6.3 | Fabp2 | Fatty acid binding protein 2 | 46 | LOC56825 | Prochymosin | 66 |
| Il13ra1 | Interleukin 13 receptor, alpha 1 | 5.9 | Clca6 | Chloride channel calcium activated 6 | 45 | Clca3 | Chloride channel calcium activated 3 | 42 |
| LOC498358 | Solute carrier family 30 (zinc transporte), member 9 | 5.6 | Gkn1 | Gastrokine 1 | 42 | Sult1b1 | Sulfotransferase family 1B | 39 |
| Decreased | LP vs. Control (Fold Regulation) | E20 | Decreased | LP vs. Control (Fold Regulation) | P0 | Decreased | LP vs. Control (Fold Regulation) | P2 |
| Fos | FBJ murine osteosarcoma viral oncogene | −14 | Myo5c | Myosin Vc | −5.1 | Atp8b1 | ATPase, Class I, type 8B, member 1 | −11 |
| LOC680231 | Chromodomain helicase DNA binding protein 9 | −12 | Phlda1 | Chromodomain helicase DNA binding protein 9 | −4.3 | Aff4 | AF4/FMR2 family, member 4 | −9.9 |
| Zfhx1b | Zinc finger homeobox 1b (ZEB2) | −6.8 | Gtl2 | GTL2, imprinted maternally expressed untranslated | −4.2 | Foxo1a | Forkhead box O1A | −9.6 |
| Atxn2 | Ataxin 2 | −6.7 | Smoc1 | SPARC-related modular calcium binding protein 1 | −4.1 | Eif2c2 | Eukaryotic translation initiation factor 2C, 2 | −9.4 |
| RGD1561386 | CBL E3 ubiquitin protein ligase | −6.4 | LOC682488 | Ras-related protein Rab-1B | −4.1 | Tns | Tensin | −9.0 |
| Adipor2 | Adiponectin receptor 2 | −6.1 | Lamc1 | Laminin, gamma 1 | −3.9 | Akap9 | A kinase (PRKA) anchor protein 9 | −8.9 |
| Eif2c2 | Eukaryotic translation initiation factor 2C, 2 | −5.8 | Adhfe1 | Alcohol dehydrogenase, iron containing, 1 | −3.9 | Ash1l | Absent, small, or homeotic)-like | −8.8 |
| Mt1a | Metallothionein 1a | −5.8 | Fgfr1 | Fibroblast growth factor receptor 1 | −3.8 | Rck | DEAD box protein rck/p54 | −7.4 |
| Tbl1x | Transducin (beta)-like 1 X-linked | −5.8 | P34 | P34 protein | −3.7 | Tbl1x | Transducin (beta)-like 1 X-linked | −7.4 |
| Atp8b1 | ATPase, Class I, type 8B, member 1 | −5.4 | Ccnl2 | Cyclin L2 | −3.5 | Ubn1 | Ubinuclein 1 | −6.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winkel, L.; Rasmussen, M.; Larsen, L.; Dalgaard, L.T.; Nielsen, J.H. Fetal Programming of the Endocrine Pancreas: Impact of a Maternal Low-Protein Diet on Gene Expression in the Perinatal Rat Pancreas. Int. J. Mol. Sci. 2022, 23, 11057. https://doi.org/10.3390/ijms231911057
Winkel L, Rasmussen M, Larsen L, Dalgaard LT, Nielsen JH. Fetal Programming of the Endocrine Pancreas: Impact of a Maternal Low-Protein Diet on Gene Expression in the Perinatal Rat Pancreas. International Journal of Molecular Sciences. 2022; 23(19):11057. https://doi.org/10.3390/ijms231911057
Chicago/Turabian StyleWinkel, Louise, Morten Rasmussen, Louise Larsen, Louise T. Dalgaard, and Jens H. Nielsen. 2022. "Fetal Programming of the Endocrine Pancreas: Impact of a Maternal Low-Protein Diet on Gene Expression in the Perinatal Rat Pancreas" International Journal of Molecular Sciences 23, no. 19: 11057. https://doi.org/10.3390/ijms231911057
APA StyleWinkel, L., Rasmussen, M., Larsen, L., Dalgaard, L. T., & Nielsen, J. H. (2022). Fetal Programming of the Endocrine Pancreas: Impact of a Maternal Low-Protein Diet on Gene Expression in the Perinatal Rat Pancreas. International Journal of Molecular Sciences, 23(19), 11057. https://doi.org/10.3390/ijms231911057







