Later Growth Cessation and Increased Freezing Tolerance Potentially Result in Later Dormancy in Evergreen Iris Compared with Deciduous Iris
Abstract
:1. Introduction
2. Results
2.1. Dormancy Status Determination
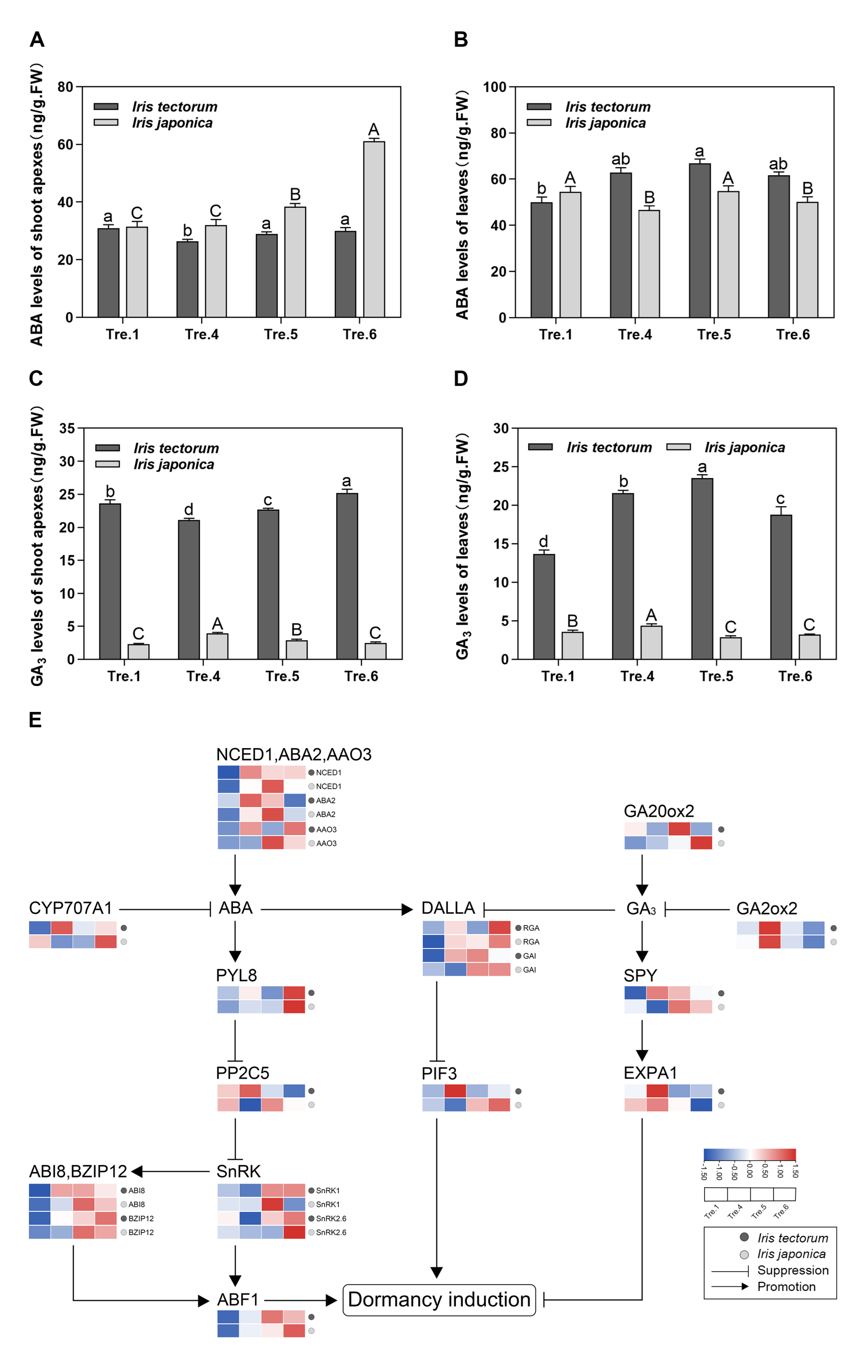
2.2. Changes in ABA and GA3 Levels and Related Gene Expression
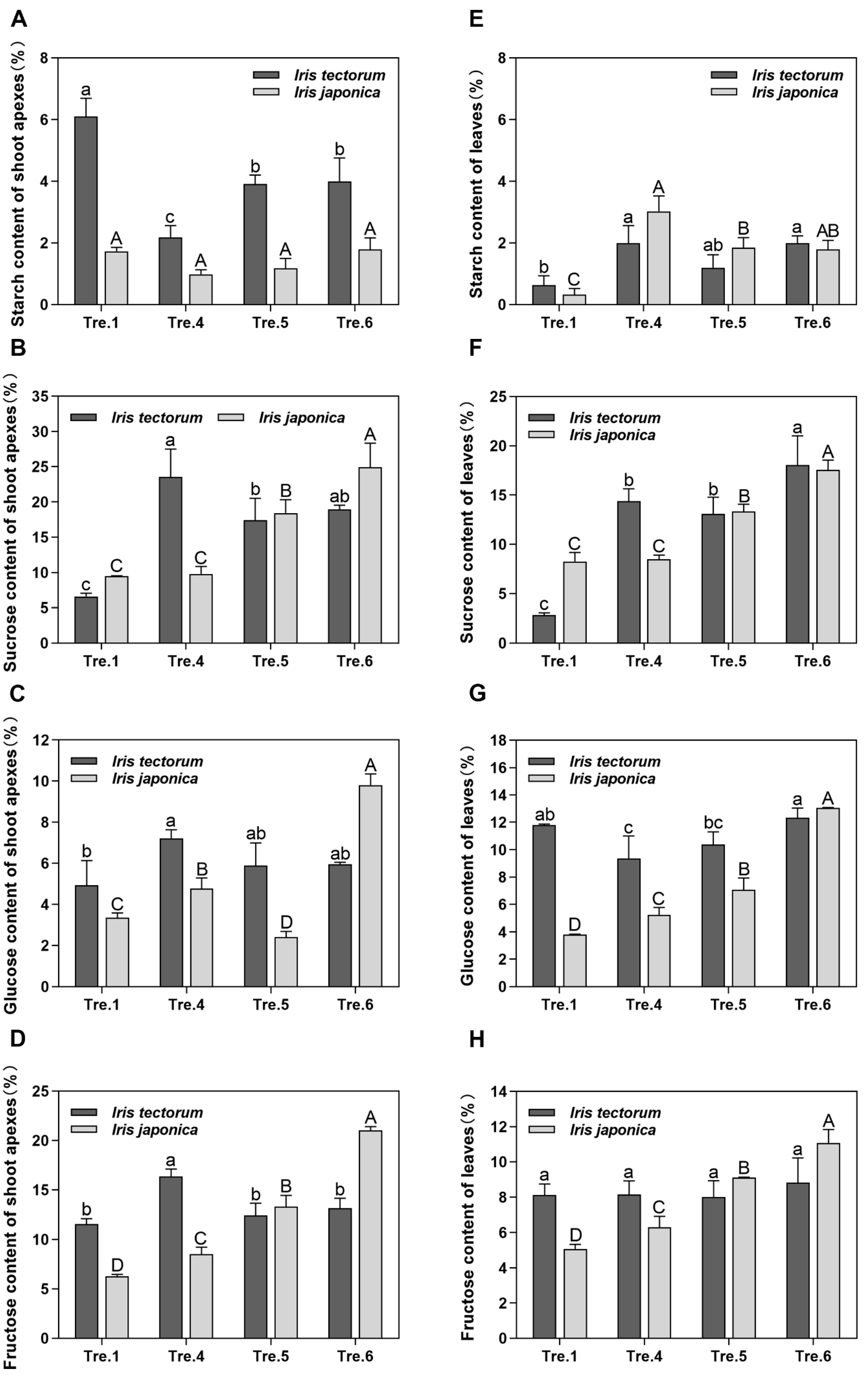
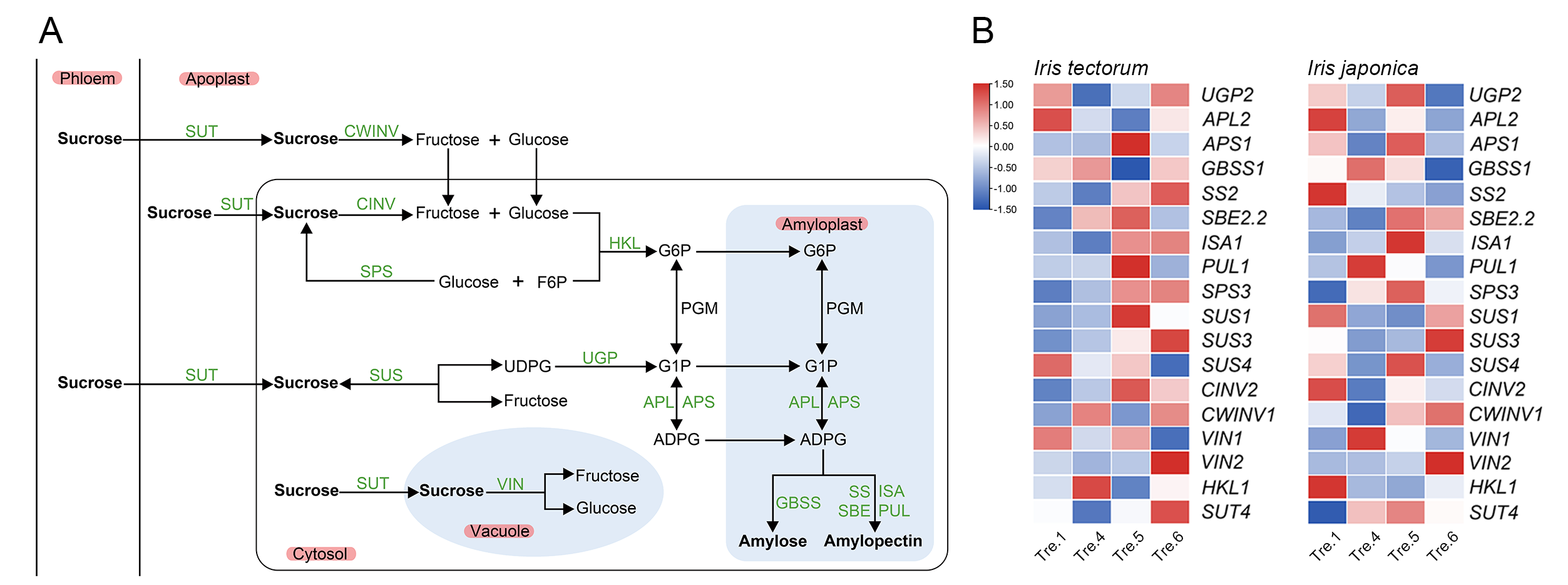
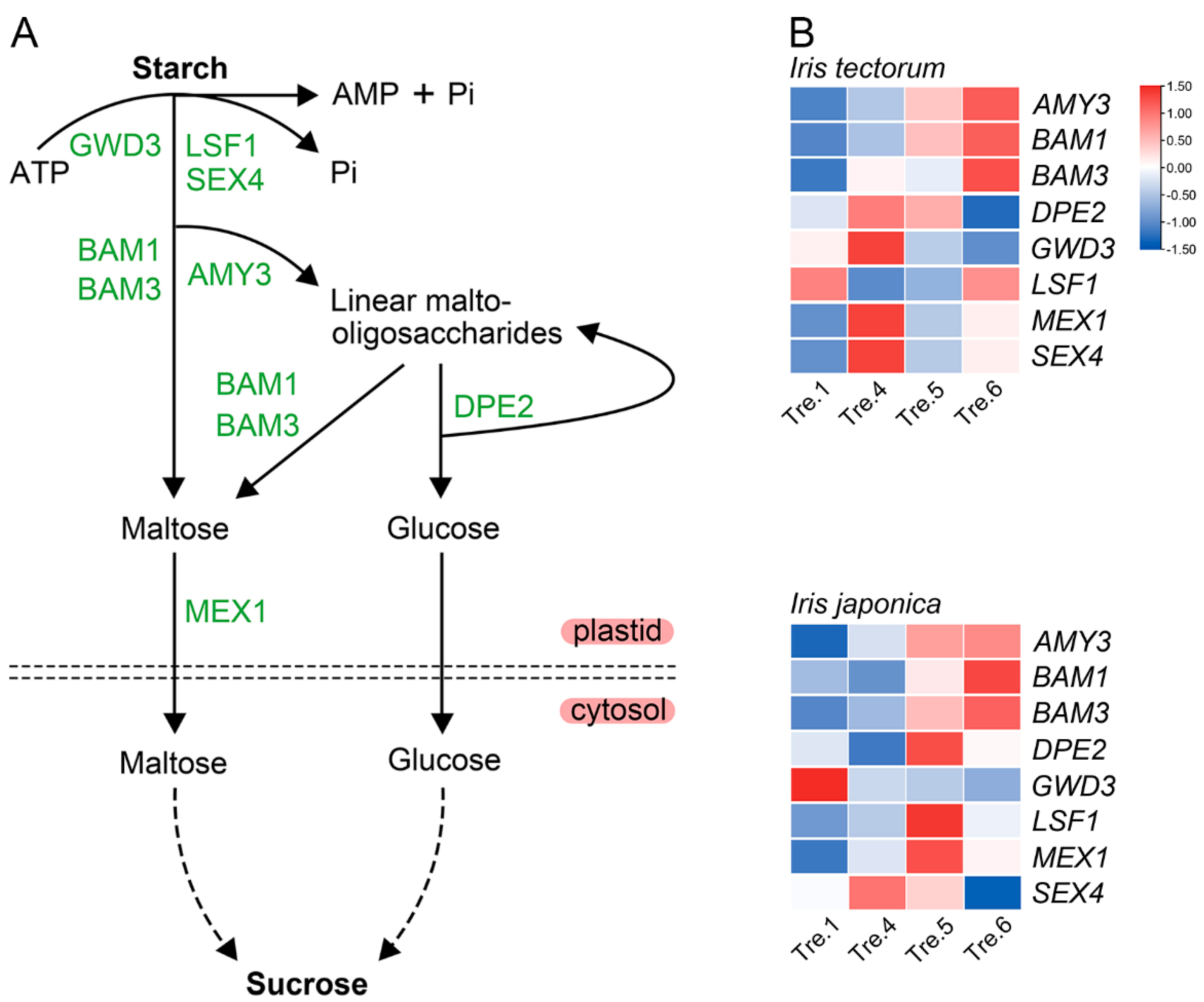
2.3. Changes in Carbohydrate Content and Related Gene Expression
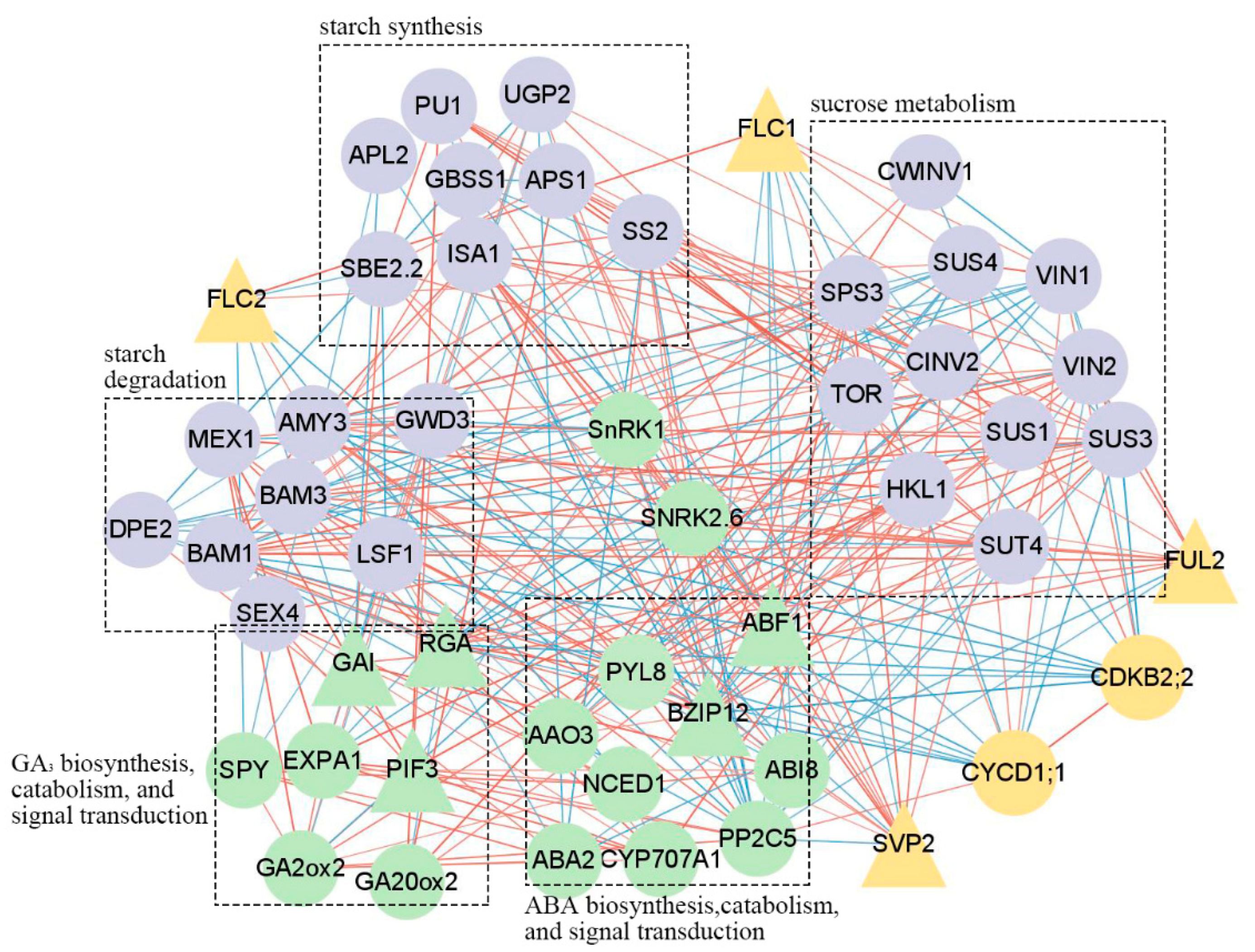
2.4. Correlation of Gene Expression Analysis
3. Discussion
3.1. Early Dormancy in Deciduous Iris under Artificial Dormancy Induction Conditions
3.2. ABA and GA3 Coordinate Growth and Stress Response during the Dormancy of Irises
3.3. Carbohydrates Actively Participate in the Dormancy Transition and Stress Responses in Irises
3.4. Dormancy Related MADS-box Genes and SnRKs Potentially Serve as the Bridge between Carbohydrate and Phytohormone Interaction during Iris Dormancy
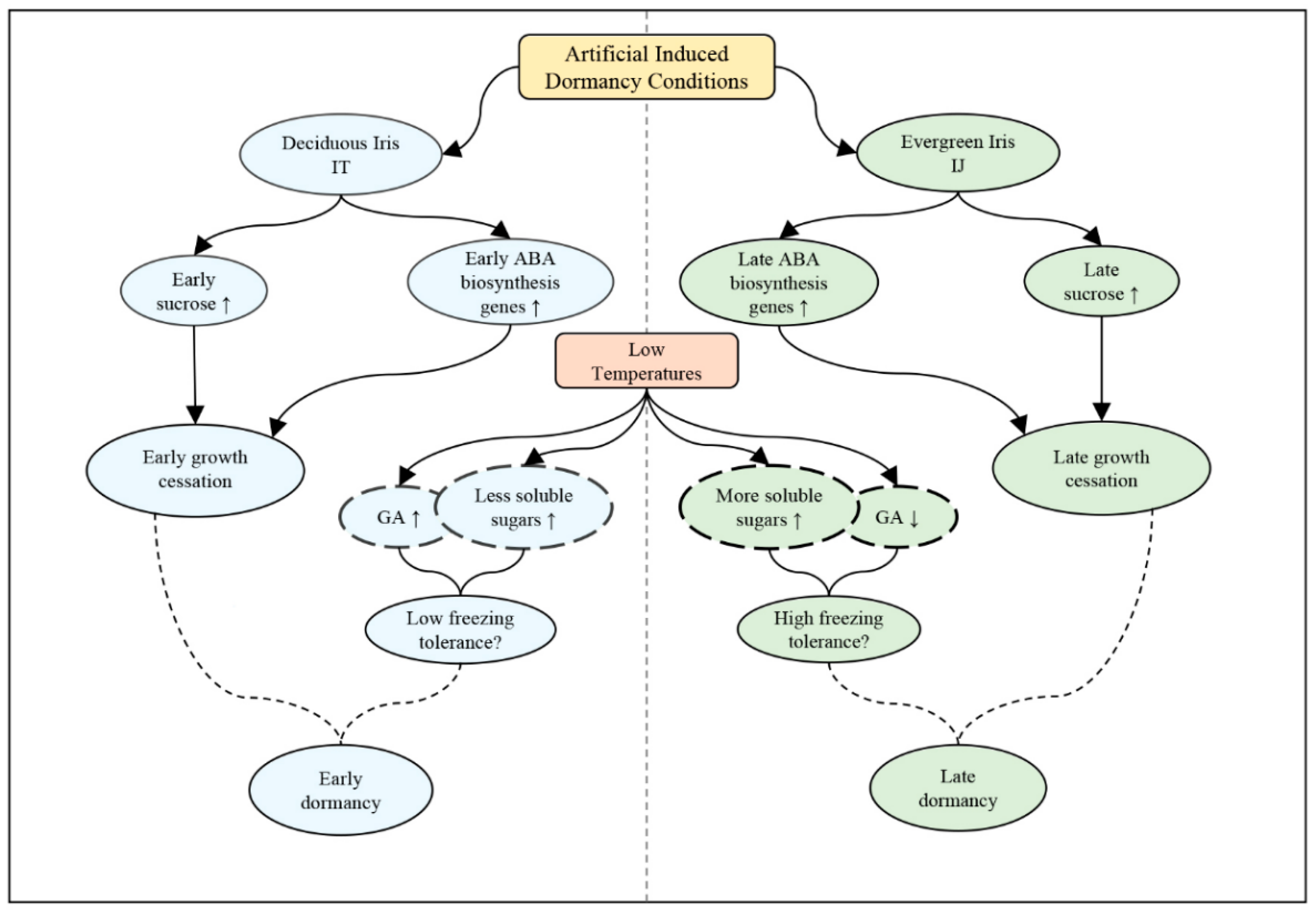
3.5. Later Growth Cessation and Higher Freezing Tolerance Potentially Result in Later Dormancy in Evergreen Iris Compared with Deciduous Iris
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Determination of the Dormancy of Plants
4.3. Determination of Carbohydrate Contents
4.4. Measurement of Endogenous Phytohormone Levels
4.5. Quantitative Real-Time PCR
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Horvath, D.P.; Anderson, J.V.; Chao, W.S.; Foley, M.E. Knowing when to grow: Signals regulating bud dormancy. Trends Plant Sci. 2003, 8, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Lang, G.A.; Early, J.D.; Martin, G.C.; Darnell, R.L. Endodormancy, paradormancy, and ecodormancy—Physiological terminology and classification for dormancy research. Hortscience 1987, 22, 371–377. [Google Scholar]
- Fu, Y.H.; Piao, S.; Vitasse, Y.; Zhao, H.; De Boeck, H.J.; Liu, Q.; Yang, H.; Weber, U.; Hanninen, H.; Janssens, I.A. Increased heat requirement for leaf flushing in temperate woody species over 1980–2012: Effects of chilling, precipitation and insolation. Glob. Chang. Biol. 2015, 21, 2687–2697. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Tomes, S.; Karunairetnam, S.; Tustin, S.D.; Hellens, R.P.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. SVP-like MADS Box Genes Control Dormancy and Budbreak in Apple. Front. Plant Sci. 2017, 8, 477. [Google Scholar] [CrossRef]
- Beauvieux, R.; Wenden, B.; Dirlewanger, E. Bud Dormancy in Perennial Fruit Tree Species: A Pivotal Role for Oxidative Cues. Front. Plant Sci. 2018, 9, 657. [Google Scholar] [CrossRef]
- Wu, J.; Jin, Y.; Liu, C.; Vonapartis, E.; Liang, J.; Wu, W.; Gazzarrini, S.; He, J.; Yi, M. GhNAC83 inhibits corm dormancy release by regulating ABA signaling and cytokinin biosynthesis in Gladiolus hybridus. J. Exp. Bot. 2019, 70, 1221–1237. [Google Scholar] [CrossRef]
- Kamenetsky, R.; Okubo, H. Ornamental Geophytes from Basic Science to Sustainable Production Introduction; CRC Press: Boca Raton, FL, USA, 2013; pp. 233–259. [Google Scholar]
- Alamar, M.C.; Anastasiadi, M.; Lopez-Cobollo, R.; Bennett, M.H.; Thompson, A.J.; Turnbull, C.G.N.; Mohareb, F.; Terry, L.A. Transcriptome and phytohormone changes associated with ethylene-induced onion bulb dormancy. Postharvest Biol. Technol. 2020, 168, 111267. [Google Scholar] [CrossRef]
- Zhang, H.Q.; Liu, X.W.; Nie, B.H.; Song, B.H.; Du, P.; Liu, S.X.; Li, L.; Zhao, Z.Q. Nitrogen management can inhibit or induce the sprouting of potato tubers: Consequences of regulation tuberization. Postharvest Biol. Technol. 2022, 183, 111722. [Google Scholar] [CrossRef]
- Jewaria, P.K.; Hanninen, H.; Li, X.; Bhalerao, R.P.; Zhang, R. A hundred years after: Endodormancy and the chilling requirement in subtropical trees. New Phytol. 2021, 23, 565–570. [Google Scholar] [CrossRef]
- Zheng, C.; Kwame Acheampong, A.; Shi, Z.; Halaly, T.; Kamiya, Y.; Ophir, R.; Galbraith, D.W.; Or, E. Distinct gibberellin functions during and after grapevine bud dormancy release. J. Exp. Bot. 2018, 69, 1635–1648. [Google Scholar] [CrossRef]
- Yang, Q.S.; Yang, B.; Li, J.Z.; Wang, Y.; Tao, R.Y.; Yang, F.; Wu, X.Y.; Yan, X.H.; Ahmad, M.; Shen, J.Q.; et al. ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ. 2020, 43, 1360–1375. [Google Scholar] [CrossRef] [PubMed]
- Azeez, A.; Zhao, Y.C.; Singh, R.K.; Yordanov, Y.S.; Dash, M.; Miskolczi, P.; Stojkovic, K.; Strauss, S.H.; Bhalerao, R.P.; Busov, V.B. EARLY BUD-BREAK 1 and EARLY BUD-BREAK 3 control resumption of poplar growth after winter dormancy. Nat. Commun. 2021, 12, 1123. [Google Scholar] [CrossRef] [PubMed]
- Howard, C.C.; Folk, R.A.; Beaulieu, J.M.; Cellinese, N. The monocotyledonous underground: Global climatic and phylogenetic patterns of geophyte diversity. Am. J. Bot. 2019, 106, 850–863. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Shao, L.; Zhang, J.; Wang, X.; Zhang, D.; Horvath, D.P.; Zhang, L.; Zhang, J.; Xia, Y. MADS-box transcription factors determine the duration of temporary winter dormancy in closely related evergreen and deciduous Iris spp. J. Exp. Bot. 2022, 73, 1429–1449. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xia, Y.; Lou, J.; Zhang, D.; Wang, X.; Zhang, J. A comparative study between evergreen and deciduous daylily species reveals the potential contributions of winter shoot growth and leaf freezing tolerance to foliar habits. J. Plant Growth Regul. 2020, 39, 1030–1045. [Google Scholar] [CrossRef]
- Shim, D.; Ko, J.-H.; Kim, W.-C.; Wang, Q.; Keathley, D.E.; Han, K.-H. A molecular framework for seasonal growth-dormancy regulation in perennial plants. Hortic. Res. 2014, 1, 14059. [Google Scholar] [CrossRef]
- Niu, Q.F.; Li, J.Z.; Cai, D.Y.; Qian, M.J.; Jia, H.M.; Bai, S.L.; Hussain, S.; Liu, G.Q.; Teng, Y.W.; Zheng, X.Y. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef]
- Singab, A.N.B.; Ayoub, I.M.; El-Shazly, M.; Korinek, M.; Wu, T.Y.; Cheng, Y.B.; Chang, F.R.; Wu, Y.C. Shedding the light on Iridaceae: Ethnobotany, phytochemistry and biological activity. Ind. Crops Prod. 2016, 92, 308–335. [Google Scholar] [CrossRef]
- Luan, Z.J.; Li, P.P.; Li, D.; Meng, X.P.; Sun, J. Optimization of supercritical-CO2 extraction of Iris lactea seed oil: Component analysis and antioxidant activity of the oil. Ind. Crops Prod. 2020, 152, 112553. [Google Scholar] [CrossRef]
- Kemper, T.W. The American iris society. Mo. Bot. Gard. 2012. [Google Scholar]
- Jozghasemi, S.; Rabiei, V.; Soleymani, A.; Khalighi, A. Evaluation of the pigments concentration in the Iris species native to Iran. J. Biodivers. Environ. Sci. 2015, 6, 557–561. [Google Scholar]
- Li, D.; Zhang, J.; Zhang, J.; Li, K.; Xia, Y. Green Period Characteristics and Foliar Cold Tolerance in 12 Iris Species and Cultivars in the Yangtze Delta, China. Horttechnology 2017, 27, 399–407. [Google Scholar] [CrossRef]
- Li, W.; Jiang, D.H. Design study on artificial climate chamber for plant based on computer control system. Build. Technol. Dev. 2016, 43, 56–58. [Google Scholar]
- Zheng, D.C.; Wang, H.; Fan, J.; Zhang, Y.; Xiong, L. Application comparison between shelf-type and box-group-type artificial climate chamber. Lab. Sci. 2021, 24, 22–26. [Google Scholar]
- Zhang, J. Effects of Different Temperature and Light Conditions on Leaf Bud Dormancy and Growth of Evergreen and Deciduous Iris in Jiangnan Region; Greystone Books Ltd.: Vancouver, BC, Canada, 2019. [Google Scholar]
- Finkelstein, R. Abscisic Acid synthesis and response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Gao, Z.; Du, P.; Xiao, W.; Tan, Q.; Chen, X.; Li, L.; Gao, D. Expression of ABA metabolism-related genes suggests similarities and differences between seed dormancy and bud dormancy of peach (Prunus persica). Front. Plant Sci. 2015, 6, 1248. [Google Scholar] [CrossRef]
- Yue, C.; Cao, H.; Hao, X.; Zeng, J.; Qian, W.; Guo, Y.; Ye, N.; Yang, Y.; Wang, X. Differential expression of gibberellin- and abscisic acid-related genes implies their roles in the bud activity-dormancy transition of tea plants. Plant Cell Rep. 2018, 37, 425–441. [Google Scholar] [CrossRef]
- Hernández, J.A.; Díaz-Vivancos, P.; Acosta-Motos, J.R.; Alburquerque, N.; Martínez, D.; Carrera, E.; García-Bruntón, J.; Barba-Espín, G. Interplay among antioxidant system, hormone profile and carbohydrate metabolism during bud dormancy breaking in a high-chill peach variety. Antioxidants 2021, 10, 560. [Google Scholar] [CrossRef]
- Sapkota, S.; Liu, J.; Islam, M.T.; Sherif, S.M. Changes in reactive oxygen species, antioxidants and carbohydrate metabolism in relation to dormancy transition and bud break in apple (Malus × domestica Borkh) Cultivars. Antioxidants 2021, 10, 1549. [Google Scholar] [CrossRef]
- Díaz-Riquelme, J.; Lijavetzky, D.; Martínez-Zapater, J.M.; Carmona, M.J. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol. 2009, 149, 354–369. [Google Scholar] [CrossRef]
- Takahashi, H.; Nishihara, M.; Yoshida, C.; Itoh, K. Gentian FLOWERING LOCUS T orthologs regulate phase transitions: Floral induction and endodormancy release. Plant Physiol. 2022, 188, 1887–1899. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Miskolczi, P.; Maurya, J.P.; Bhalerao, R.P. A Tree Ortholog of SHORT VEGETATIVE PHASE Floral Repressor Mediates Photoperiodic Control of Bud Dormancy. Curr. Biol. 2019, 29, 128–133.e2. [Google Scholar] [CrossRef] [PubMed]
- Andres, F.; Porri, A.; Torti, S.; Mateos, J.; Romera-Branchat, M.; Luis Garcia-Martinez, J.; Fornara, F.; Gregis, V.; Kater, M.M.; Coupland, G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 2014, 111, E2760–E2769. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Maurya, J.P.; Azeez, A.; Miskolczi, P.; Tylewicz, S.; Stojkovič, K.; Delhomme, N.; Busov, V.; Bhalerao, R.P. A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 2018, 9, 4173. [Google Scholar] [CrossRef]
- Wu, R.; Wang, T.; Warren, B.A.W.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 2017, 68, 1071–1082. [Google Scholar] [CrossRef]
- Chao, W.S.; Serpe, M.D. Changes in the expression of carbohydrate metabolism genes during three phases of bud dormancy in leafy spurge. Plant Mol. Biol. 2010, 73, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Rubio, S.; Donoso, A.; Pérez, F.J. The dormancy-breaking stimuli “chilling, hypoxia and cyanamide exposure” up-regulate the expression of α-amylase genes in grapevine buds. J. Plant Physiol. 2014, 171, 373–381. [Google Scholar] [CrossRef]
- Park, J.Y.; Canam, T.; Kang, K.Y.; Unda, F.; Mansfield, S.D. Sucrose phosphate synthase expression influences poplar phenology. Tree physiol. 2009, 29, 937–946. [Google Scholar] [CrossRef]
- Perry, T.O. Dormancy of trees in winter. Science 1971, 171, 29–36. [Google Scholar] [CrossRef]
- Yang, Q.; Gao, Y.; Wu, X.; Moriguchi, T.; Bai, S.; Teng, Y. Bud endodormancy in deciduous fruit trees: Advances and prospects. Hortic. Res. 2021, 8, 139. [Google Scholar] [CrossRef]
- Gillespie, L.M.; Volaire, F.A. Are winter and summer dormancy symmetrical seasonal adaptive strategies? The case of temperate herbaceous perennials. Ann. Bot. 2017, 119, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Walton, E.F.; Wu, R.-M.; Richardson, A.C.; Davy, M.; Hellens, R.P.; Thodey, K.; Janssen, B.J.; Gleave, A.P.; Rae, G.M.; Wood, M.; et al. A rapid transcriptional activation is induced by the dormancy-breaking chemical hydrogen cyanamide in kiwifruit (Actinidia deliciosa) buds. J. Exp. Bot. 2009, 60, 3835–3848. [Google Scholar] [CrossRef]
- Li, Z.; Reighard, G.L.; Abbott, A.G.; Bielenberg, D.G. Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 2009, 60, 3521–3530. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, R.; Yamane, H.; Ooka, T.; Jotatsu, H.; Kitamura, Y.; Akagi, T.; Tao, R. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 2011, 157, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yang, X.Y.; Su, X.X.; Gao, K.; Rao, P.A.; An, X.M. A comprehensive gene network for fine tuning floral development in poplar. Genes Genom. 2017, 39, 793–803. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Li, Z.M.; Mei, L.; Yao, J.L.; Hu, C.G. PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuation in expression level. Planta 2009, 229, 847–859. [Google Scholar] [CrossRef]
- Singh, R.K.; Svystun, T.; AlDahmash, B.; Jönsson, A.M.; Bhalerao, R.P. Photoperiod- and temperature-mediated control of phenology in trees—A molecular perspective. New Phytol. 2017, 213, 511–524. [Google Scholar] [CrossRef]
- Xue, X.; Yu, Y.C.; Wu, Y.; Xue, H.; Chen, L.Q. Locally restricted glucose availability in the embryonic hypocotyl determines seed germination under abscisic acid treatment. New Phytol. 2021, 231, 1832–1844. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Kaur, H.; Ozga, J.A.; Reinecke, D.M. Balancing of hormonal biosynthesis and catabolism pathways, a strategy to ameliorate the negative effects of heat stress on reproductive growth. Plant Cell Environ. 2021, 44, 1486–1503. [Google Scholar] [CrossRef]
- Liang, D.; Huang, X.; Shen, Y.; Shen, T.; Zhang, H.; Lin, L.; Wang, J.; Deng, Q.; Lyu, X.; Xia, H. Hydrogen cyanamide induces grape bud endodormancy release through carbohydrate metabolism and plant hormone signaling. BMC Genom. 2019, 20, 1034. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wu, W.; Liang, J.; Jin, Y.; Gazzarrini, S.; He, J.; Yi, M. GhTCP19 transcription factor regulates corm dormancy release by repressing GhNCED expression in gladiolus. Plant Cell Physiol. 2019, 60, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Sudawan, B.; Chang, C.S.; Chao, H.F.; Ku, M.S.; Yen, Y.F. Hydrogen cyanamide breaks grapevine bud dormancy in the summer through transient activation of gene expression and accumulation of reactive oxygen and nitrogen species. BMC Plant Biol. 2016, 16, 202. [Google Scholar] [CrossRef] [Green Version]
- Falavigna, V.D.S.; Guitton, B.; Costes, E.; Andrés, F. I want to (bud) break free: The potential role of DAM and SVP-like genes in regulating dormancy cycle in temperate fruit trees. Front. Plant Sci. 2018, 9, 1990. [Google Scholar] [CrossRef]
- Xue, J.; Li, T.; Wang, S.; Xue, Y.; Hu, F.; Zhang, X. Elucidation of the mechanism of reflowering in tree peony (Paeonia suffruticosa) ‘Zi Luo Lan’ by defoliation and gibberellic acid application. Plant Physiol. Biochem. 2018, 132, 571–578. [Google Scholar] [CrossRef]
- Lange, T.; Krämer, C.; Pimenta Lange, M.J. The Class III Gibberellin 2-Oxidases AtGA2ox9 and AtGA2ox10 Contribute to Cold Stress Tolerance and Fertility. Plant Physiol. 2020, 184, 478–486. [Google Scholar] [CrossRef]
- Piskurewicz, U.; Jikumaru, Y.; Kinoshita, N.; Nambara, E.; Kamiya, Y.; Lopez-Molina, L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 2008, 20, 2729–2745. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Lv, L.; Huo, X.; Wen, L.; Gao, Z.; Khalil-Ur-Rehman, M. Isolation and Role of PmRGL2 in GA-mediated Floral Bud Dormancy Release in Japanese Apricot (Prunus mume Siebold et Zucc.). Front. Plant Sci. 2018, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Rubio, S.; Noriega, X.; Pérez, F.J. ABA promotes starch synthesis and storage metabolism in dormant grapevine buds. J. Plant Physiol. 2019, 234–235, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.; Moyle, R.; Bhalerao, R.; Hertzberg, M.; Lundeberg, J.; Nilsson, P.; Bhalerao, R.P. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 2004, 40, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Wanner, L.A.; Junttila, O. Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 1999, 120, 391–399. [Google Scholar] [CrossRef]
- Tarkowski, L.P.; Van den Ende, W. Cold tolerance triggered by soluble sugars: A multifaceted countermeasure. Front. Plant Sci. 2015, 6, 203. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yang, Y.; Liu, X.; Huang, J.; Wang, Q.; Gu, J.; Lu, Y. Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genom. 2014, 15, 203. [Google Scholar] [CrossRef]
- Yuan, L.Y.; Xie, S.L.; Nie, L.B.; Zheng, Y.S.; Wang, J.; Huang, J.; Zhao, M.R.; Zhu, S.D.; Hou, J.F.; Chen, G.H.; et al. Comparative proteomics reveals cold acclimation machinery through enhanced carbohydrate and amino acid metabolism in Wucai (Brassica Campestris L.). Plants 2019, 8, 474. [Google Scholar] [CrossRef]
- Rohde, A.; Bhalerao, R.P. Plant dormancy in the perennial context. Trends Plant Sci. 2007, 12, 217–223. [Google Scholar] [CrossRef]
- Li, X.; Wang, C.; Cheng, J.; Zhang, J.; da Silva, J.A.; Liu, X.; Duan, X.; Li, T.; Sun, H. Transcriptome analysis of carbohydrate metabolism during bulblet formation and development in Lilium davidii var. unicolor. BMC Plant Biol. 2014, 14, 358. [Google Scholar] [CrossRef]
- Sonnewald, S.; Sonnewald, U. Regulation of potato tuber sprouting. Planta 2014, 239, 27–38. [Google Scholar] [CrossRef]
- Pan, W.; Liang, J.; Sui, J.; Li, J.; Liu, C.; Xin, Y.; Zhang, Y.; Wang, S.; Zhao, Y.; Zhang, J.; et al. ABA and bud dormancy in perennials: Current knowledge and future perspective. Genes 2021, 12, 1635. [Google Scholar] [CrossRef] [PubMed]
- Considine, M.J.; Considine, J.A. On the language and physiology of dormancy and quiescence in plants. J. Exp. Bot. 2016, 67, 3189–3203. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Wang, H.; Li, X.; Sui, X.; Zhang, Z. Down-regulating cucumber sucrose synthase 4 (CsSUS4) suppresses the growth and development of flowers and fruits. Plant Cell Physiol. 2019, 60, 752–764. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, D.; Miao, Q.; Yang, J.; Xuan, Y.; Hu, Y. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef]
- Laurentin, A.; Edwards, C.A. A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal. Biochem. 2003, 315, 143–145. [Google Scholar] [CrossRef]
- Wang, H.; Sui, X.; Guo, J.; Wang, Z.; Cheng, J.; Ma, S.; Li, X.; Zhang, Z. Antisense suppression of cucumber (Cucumis sativus L.) sucrose synthase 3 (CsSUS3) reduces hypoxic stress tolerance. Plant Cell Environ. 2014, 37, 795–810. [Google Scholar] [CrossRef]
- Chen, Y.; Ji, F.; Xie, H.; Liang, J.; Zhang, J. The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol. 2006, 140, 302–310. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
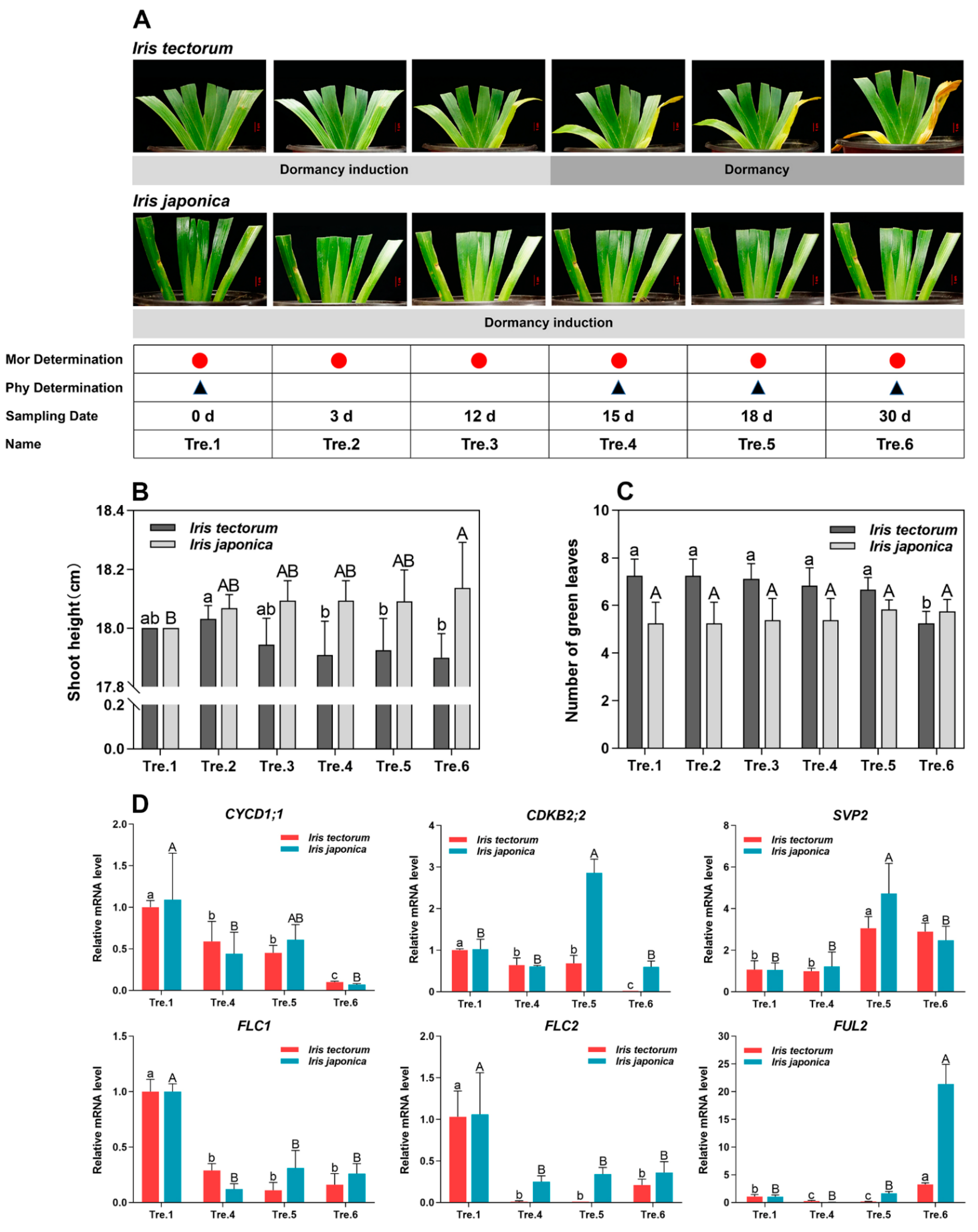
| Temperature (°C) | Processing Time (h) | Time Distribution | Light Intensity (μmol Photons m−2 s−1) |
|---|---|---|---|
| 0 | 1.0 | 0:00–5:00 | 0 |
| 3 | 2.0 | 5:00–7:00 | 0 |
| 3 | 2.0 | 7:00–9:00 | 60 |
| 7 | 5.0 | 9:00–14:00 | 60 |
| 4 | 3.5 | 14:00–17:30 | 60 |
| 4 | 0.5 | 17:30–18:00 | 0 |
| 0 | 1.0 | 18:00–24:00 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, T.; Zhang, J.; Shao, L.; Wang, X.; Zhang, R.; Ji, C.; Xia, Y.; Zhang, L.; Zhang, J.; Li, D. Later Growth Cessation and Increased Freezing Tolerance Potentially Result in Later Dormancy in Evergreen Iris Compared with Deciduous Iris. Int. J. Mol. Sci. 2022, 23, 11123. https://doi.org/10.3390/ijms231911123
Xu T, Zhang J, Shao L, Wang X, Zhang R, Ji C, Xia Y, Zhang L, Zhang J, Li D. Later Growth Cessation and Increased Freezing Tolerance Potentially Result in Later Dormancy in Evergreen Iris Compared with Deciduous Iris. International Journal of Molecular Sciences. 2022; 23(19):11123. https://doi.org/10.3390/ijms231911123
Chicago/Turabian StyleXu, Tong, Jiao Zhang, Lingmei Shao, Xiaobin Wang, Runlong Zhang, Chenxi Ji, Yiping Xia, Liangsheng Zhang, Jiaping Zhang, and Danqing Li. 2022. "Later Growth Cessation and Increased Freezing Tolerance Potentially Result in Later Dormancy in Evergreen Iris Compared with Deciduous Iris" International Journal of Molecular Sciences 23, no. 19: 11123. https://doi.org/10.3390/ijms231911123







