Relations between Structure and Zn(II) Binding Affinity Shed Light on the Mechanisms of Rad50 Hook Domain Functioning and Its Phosphorylation
Abstract
1. Introduction
2. Results
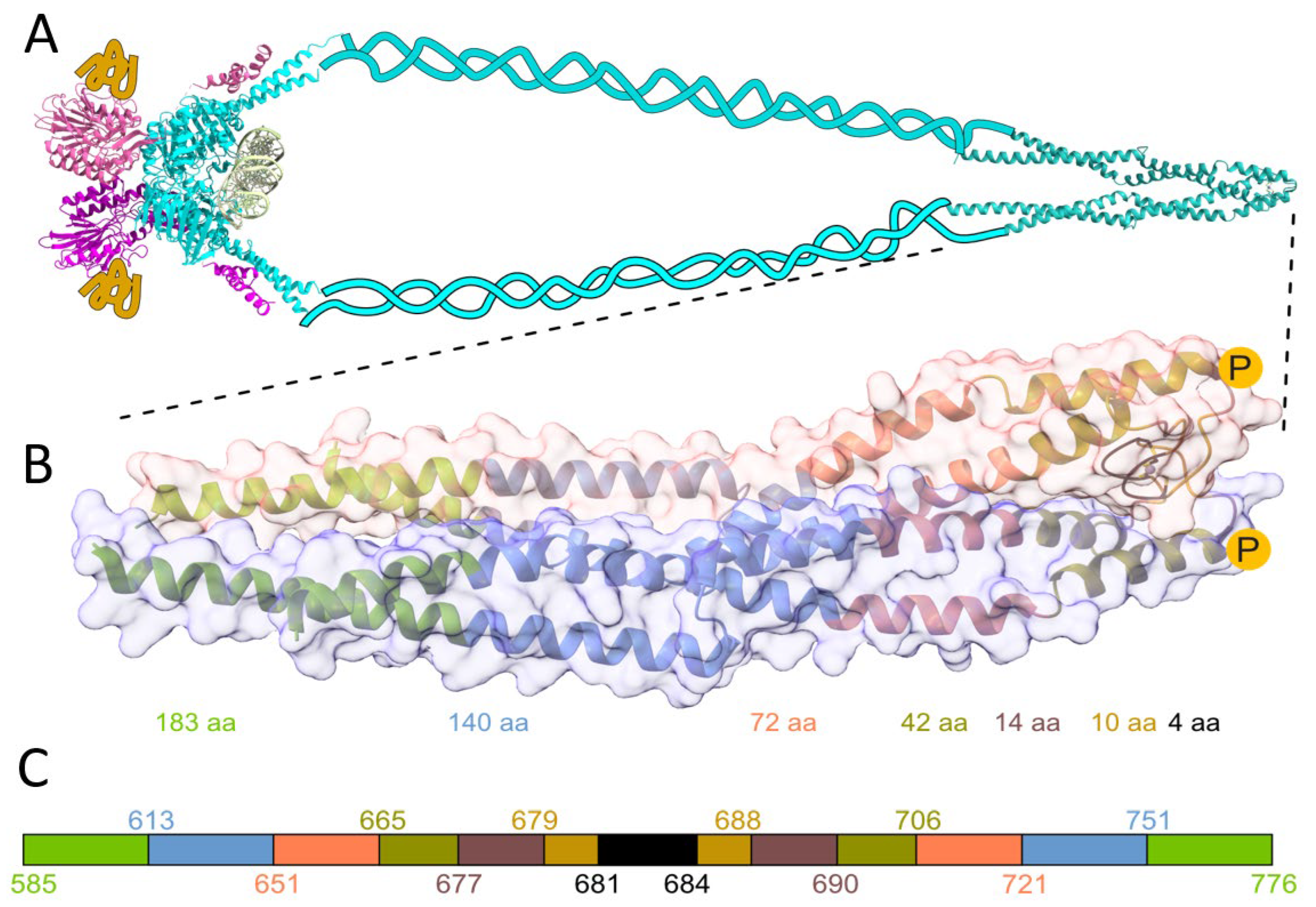
2.1. Folding of the Hook Domain Induced by Zn(II)
2.2. Stability of the Zinc Hook Domain
2.3. Small Angle X-ray Scattering (SAXS) Confirms Rod-Shaped Phosphomimetic Zinc Hook
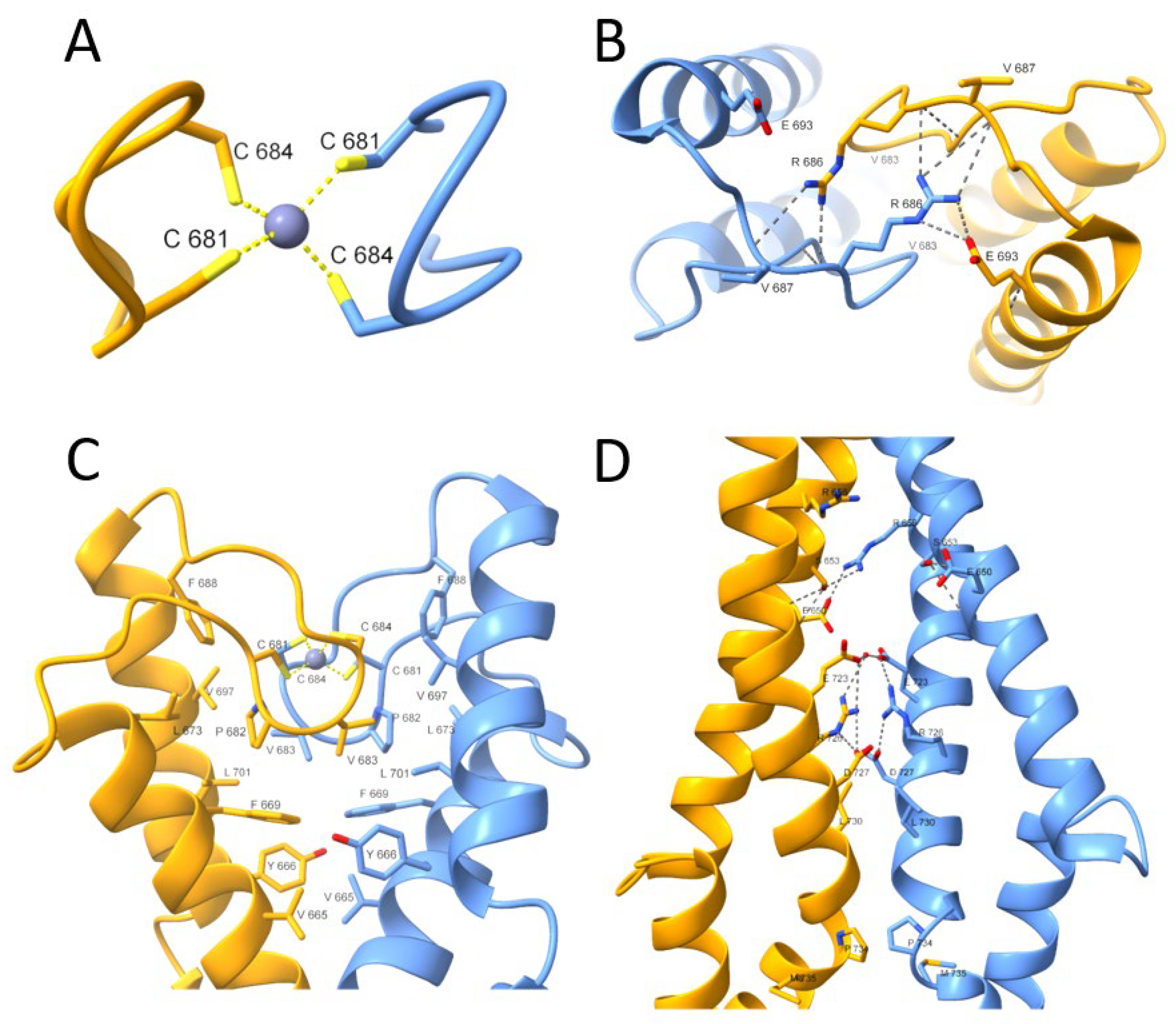
2.4. X-ray Absorption Spectrum in the Near-Edge Structure and Extended X-ray Absorption Fine Structure Show Tetrathiolate Zn(II) in HsHk183
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Peptide Synthesis
4.3. Expression and Purification of Metal-Free HsHk183
4.4. XANES and EXAFS
4.5. SAXS
4.6. Spectroscopic Studies
4.6.1. Electron Absorption Spectroscopy in UV Range
4.6.2. Spectropolarimetric Titrations with Zn(II)
4.7. Competitive Titrations
4.8. Potentiometry
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATM | ataxia-telangiectasia mutated kinase |
| CD | circular dichroism |
| CI | competitivity index |
| EXAFS | extended X-ray absorption fine structure |
| HsHk | zinc hook from H. sapiens Rad50 |
| MRN | Mre11, Rad50, Nbs1 complex |
| NHEJ | non-homologous end joining |
| PDB | Protein Data Bank |
| PfHk | zinc hook from P. furiosus Rad50 |
| SAXS | Small angle X-ray scattering |
| XANES | X-ray absorption near-edge structure |
References
- Hopfner, K.; Craig, L.; Moncalian, G.; Zinkel, R.A.; Usui, T.; Owen, B.A.L.; Karcher, A.; Henderson, B.; Bodmer, J.-L.; McMurray, C.T.; et al. The Rad50 Zinc-Hook is a Structure Joining Mre11 Complexes in DNA Recombination and Repair. Nature 2002, 418, 562–566. [Google Scholar] [CrossRef] [PubMed]
- de Jager, M.; van Noort, J.; van Gent, D.C.; Dekker, C.; Kanaar, R.; Wyman, C. Human Rad50/Mre11 Is a Flexible Complex That Can Tether DNA Ends. Mol. Cell 2001, 8, 1129–1135. [Google Scholar] [CrossRef]
- Andreini, C.; Bertini, I.; Rosato, A. Metalloproteomes: A Bioinformatic Approach. Acc. Chem. Res. 2009, 42, 1471–1479. [Google Scholar] [CrossRef]
- Andreini, C.; Cavallaro, G.; Lorenzini, S.; Rosato, A. MetalPDB: A Database of Metal Sites in Biological Macromolecular Structures. Nucleic Acids Res. 2013, 41, D312–D319. [Google Scholar] [CrossRef]
- Hagedoorn, P.-L. Microbial Metalloproteomics. Proteomes 2015, 3, 424–439. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Banci, L.; Bertini, I.; Rosato, A. Counting the Zinc-Proteins Encoded in the Human Genome. J. Proteome Res. 2006, 5, 196–201. [Google Scholar] [CrossRef]
- Maret, W.; Li, Y. Coordination Dynamics of Zinc in Proteins. Chem. Rev. 2009, 109, 4682–4707. [Google Scholar] [CrossRef]
- Maret, W. Zinc Biochemistry: From a Single Zinc Enzyme to a Key Element of Life. Adv. Nutr. 2013, 4, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Krężel, A.; Maret, W. The Biological Inorganic Chemistry of Zinc Ions. Arch. Biochem. Biophys. 2016, 611, 3–19. [Google Scholar] [CrossRef]
- Kochańczyk, T.; Drozd, A.; Krężel, A. Relationship between the Architecture of Zinc Coordination and Zinc Binding Affinity in Proteins—Insights into Zinc Regulation. Metallomics 2015, 7, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S. Zinc Coordination Sphere in Biochemical Zinc Sites. Biometals 2001, 14, 271–313. [Google Scholar] [CrossRef]
- Padjasek, M.; Kocyła, A.; Kluska, K.; Kerber, O.; Tran, J.B.; Krężel, A. Structural Zinc Binding Sites Shaped for Greater Works: Structure-Function Relations in Classical Zinc Finger, Hook and Clasp Domains. J. Inorg. Biochem. 2020, 204, 110955. [Google Scholar] [CrossRef] [PubMed]
- Kocyła, A.; Tran, J.B.; Krężel, A. Galvanization of Protein—Protein Interactions in a Dynamic Zinc Interactome. Trends Biochem. Sci. 2021, 46, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Tran, J.B.; Krężel, A. InterMetalDB: A Database and Browser of Intermolecular Metal Binding Sites in Macromolecules with Structural Information. J. Proteome Res. 2021, 20, 1889–1901. [Google Scholar] [CrossRef]
- Kochańczyk, T.; Nowakowski, M.; Wojewska, D.; Kocyła, A.; Ejchart, A.; Koźmiński, W.; Krężel, A. Metal-Coupled Folding as the Driving Force for the Extreme Stability of Rad50 Zinc Hook Dimer Assembly. Sci. Rep. 2016, 6, 36346. [Google Scholar] [CrossRef]
- Stracker, T.H.; Petrini, J.H.J. The MRE11 Complex: Starting from the Ends. Nat. Rev. Mol. Cell Biol. 2011, 12, 90–103. [Google Scholar] [CrossRef]
- Syed, A.; Tainer, J.A. The MRE11–RAD50–NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu. Rev. Biochem. 2018, 87, 263–294. [Google Scholar] [CrossRef]
- Käshammer, L.; Saathoff, J.H.; Lammens, K.; Gut, F.; Bartho, J.; Alt, A.; Kessler, B.; Hopfner, K.P. Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex. Mol. Cell 2019, 76, 382–394.e6. [Google Scholar] [CrossRef]
- Park, Y.B.; Hohl, M.; Padjasek, M.; Jeong, E.; Jin, K.S.; Krężel, A.; Petrini, J.H.J.; Cho, Y. Eukaryotic Rad50 Functions as a Rod-Shaped Dimer. Nat. Struct. Mol. Biol. 2017, 24, 248–257. [Google Scholar] [CrossRef]
- Liu, Y.; Sung, S.; Kim, Y.; Li, F.; Gwon, G.; Jo, A.; Kim, A.; Kim, T.; Song, O.; Lee, S.E.; et al. ATP-dependent DNA Binding, Unwinding, and Resection by the Mre11/Rad50 Complex. EMBO J. 2016, 35, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Soh, Y.; Basquin, J.; Gruber, S. A Rod Conformation of the Pyrococcus Furiosus Rad50 Coiled Coil. Proteins Struct. Funct. Bioinforma. 2021, 89, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Kochańczyk, T.; Jakimowicz, P.; Krężel, A. Femtomolar Zn(II) Affinity of Minimal Zinc Hook Peptides—A Promising Small Tag for Protein Engineering. Chem. Commun. 2013, 49, 1312–1314. [Google Scholar] [CrossRef] [PubMed]
- Herdendorf, T.J.; Albrecht, D.W.; Benkovic, S.J.; Nelson, S.W. Biochemical Characterization of Bacteriophage T4 Mre11-Rad50 Complex. J. Biol. Chem. 2011, 286, 2382–2392. [Google Scholar] [CrossRef]
- Hohl, M.; Kochańczyk, T.; Tous, C.; Aguilera, A.; Krężel, A.; Petrini, J.H.J. Interdependence of the Rad50 Hook and Globular Domain Functions. Mol. Cell 2015, 57, 479–491. [Google Scholar] [CrossRef]
- Hohl, M.; Kwon, Y.; Galván, S.M.; Xue, X.; Tous, C.; Aguilera, A.; Sung, P.; Petrini, J.H.J. The Rad50 Coiled-Coil Domain Is Indispensable for Mre11 Complex Functions. Nat. Struct. Mol. Biol. 2011, 18, 1124–1131. [Google Scholar] [CrossRef]
- Lafrance-Vanasse, J.; Williams, G.J.; Tainer, J.A. Envisioning the Dynamics and Flexibility of Mre11-Rad50-Nbs1 Complex to Decipher Its Roles in DNA Replication and Repair. Prog. Biophys. Mol. Biol. 2015, 117, 182–193. [Google Scholar] [CrossRef]
- Padjasek, M.; Maciejczyk, M.; Nowakowski, M.; Kerber, O.; Pyrka, M.; Koźmiński, W.; Krężel, A. Metal Exchange in the Interprotein Zn(II)-Binding Site of the Rad50 Hook Domain: Structural Insights into Cd(II)-Induced DNA-Repair Inhibition. Chem. A Eur. J. 2020, 26, 3297–3313. [Google Scholar] [CrossRef]
- Dephoure, N.; Zhou, C.; Villén, J.; Beausoleil, S.A.; Bakalarski, C.E.; Elledge, S.J.; Gygi, S.P. A Quantitative Atlas of Mitotic Phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 10762–10767. [Google Scholar] [CrossRef]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A.; et al. Quantitative Phosphoproteomics Reveals Widespread Full Phosphorylation Site Occupancy During Mitosis. Sci. Signal. 2010, 3, ra3. [Google Scholar] [CrossRef]
- Zhou, H.; Di Palma, S.; Preisinger, C.; Peng, M.; Polat, A.N.; Heck, A.J.R.; Mohammed, S. Toward a Comprehensive Characterization of a Human Cancer Cell Phosphoproteome. J. Proteome Res. 2013, 12, 260–271. [Google Scholar] [CrossRef]
- Nishi, H.; Hashimoto, K.; Panchenko, A.R. Phosphorylation in Protein-Protein Binding: Effect on Stability and Function. Structure 2011, 19, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- Krężel, A.; Latajka, R.; Bujacz, G.D.; Bal, W. Coordination Properties of Tris(2-Carboxyethyl)Phosphine, a Newly Introduced Thiol Reductant, and Its Oxide. Inorg. Chem. 2003, 42, 1994–2003. [Google Scholar] [CrossRef] [PubMed]
- Rambo, R.P.; Tainer, J.A. Characterizing Flexible and Intrinsically Unstructured Biological Macromolecules by SAS Using the Porod-Debye Law. Biopolymers 2011, 95, 559–571. [Google Scholar] [CrossRef]
- Svergun, D.I.; Koch, M.H.J. Small-Angle Scattering Studies of Biological Macromolecules in Solution. Rep. Prog. Phys. 2003, 66, 1735–1782. [Google Scholar] [CrossRef]
- Grant, T.D. Ab Initio Electron Density Determination Directly from Solution Scattering Data. Nat. Methods 2018, 15, 191–193. [Google Scholar] [CrossRef]
- Bobyr, E.; Lassila, J.K.; Wiersma-Koch, H.I.; Fenn, T.D.; Lee, J.J.; Nikolic-Hughes, I.; Hodgson, K.O.; Rees, D.C.; Hedman, B.; Herschlag, D. High-Resolution Analysis of Zn2+ Coordination in the Alkaline Phosphatase Superfamily by EXAFS and X-ray Crystallography. J. Mol. Biol. 2012, 415, 102–117. [Google Scholar] [CrossRef]
- Clark-Baldwin, K.; Tierney, D.L.; Govindaswamy, N.; Gruff, E.S.; Kim, C.; Berg, J.; Koch, S.A.; Penner-Hahn, J.E. The Limitations of X-ray Absorption Spectroscopy for Determining the Structure of Zinc Sites in Proteins. When is a Tetrathiolate Not a Tetrathiolate? J. Am. Chem. Soc. 1998, 120, 8401–8409. [Google Scholar] [CrossRef]
- Heinz, U.; Kiefer, M.; Tholey, A.; Adolph, H.-W. On the Competition for Available Zinc. J. Biol. Chem. 2005, 280, 3197–3207. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Thornton, J.M. Principles of Protein-Protein Interactions. Proc. Natl. Acad. Sci. USA 1996, 93, 13–20. [Google Scholar] [CrossRef]
- Nooren, I.M.A. Diversity of Protein–Protein Interactions. EMBO J. 2003, 22, 3486–3492. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S. Structural Zinc Sites. In Handbook of Metalloproteins; John Wiley & Sons, Ltd.: Chichester, UK, 2006; pp. 1–13. ISBN 9780470028636. [Google Scholar]
- Sontz, P.A.; Song, W.J.; Tezcan, F.A. Interfacial Metal Coordination in Engineered Protein and Peptide Assemblies. Curr. Opin. Chem. Biol. 2014, 19, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Kocyła, A.; Adamczyk, J.; Krężel, A. Interdependence of Free Zinc Changes and Protein Complex Assembly—Insights into Zinc Signal Regulation. Metallomics 2018, 10, 120–131. [Google Scholar] [CrossRef]
- Kluska, K.; Adamczyk, J.; Krężel, A. Metal Binding Properties, Stability and Reactivity of Zinc Fingers. Coord. Chem. Rev. 2018, 367, 18–64. [Google Scholar] [CrossRef]
- Koohi-Moghadam, M.; Wang, H.; Wang, Y.; Yang, X.; Li, H.; Wang, J.; Sun, H. Predicting Disease-Associated Mutation of Metal-Binding Sites in Proteins Using a Deep Learning Approach. Nat. Mach. Intell. 2019, 1, 561–567. [Google Scholar] [CrossRef]
- Reddi, A.R.; Gibney, B.R. Role of Protons in the Thermodynamic Contribution of a Zn(II)-Cys4 Site toward Metalloprotein Stability. Biochemistry 2007, 46, 3745–3758. [Google Scholar] [CrossRef] [PubMed]
- Reddi, A.R.; Guzman, T.R.; Breece, R.M.; Tierney, D.L.; Gibney, B.R. Deducing the Energetic Cost of Protein Folding in Zinc Finger Proteins Using Designed Metallopeptides. J. Am. Chem. Soc. 2007, 129, 12815–12827. [Google Scholar] [CrossRef]
- Sénèque, O.; Bonnet, E.; Joumas, F.L.; Latour, J.M. Cooperative Metal Binding and Helical Folding in Model Peptides of Treble-Clef Zinc Fingers. Chem. A Eur. J. 2009, 15, 4798–4810. [Google Scholar] [CrossRef]
- Sikorska, M.; Krężel, A.; Otlewski, J. Femtomolar Zn2+ Affinity of LIM Domain of PDLIM1 Protein Uncovers Crucial Contribution of Protein-Protein Interactions to Protein Stability. J. Inorg. Biochem. 2012, 115, 28–35. [Google Scholar] [CrossRef]
- Bombarda, E.; Morellet, N.; Cherradi, H.; Spiess, B.; Bouaziz, S.; Grell, E.; Roques, B.; Mély, Y. Determination of the PKa of the Four Zn2+-Coordinating Residues of the Distal Finger Motif of the HIV-1 Nucleocapsid Protein: Consequences on the Binding of Zn2+. J. Mol. Biol. 2001, 310, 659–672. [Google Scholar] [CrossRef]
- Kim, P.W.; Sun, Z.-Y.J.; Blacklow, S.C.; Wagner, G.; Eck, M.J. A Zinc Clasp Structure Tethers Lck to T Cell Coreceptors CD4 and CD8. Science 2003, 301, 1725–1728. [Google Scholar] [CrossRef] [PubMed]
- Kocyła, A.; Krężel, A. Zinc Clasp-Based Reversible Toolset for Selective Metal-Mediated Protein Heterodimerization. Chem. Commun. 2018, 54, 13539–13542. [Google Scholar] [CrossRef]
- Krężel, A.; Wójcik, J.; Maciejczyk, M.; Bal, W. May GSH and L-His Contribute to Intracellular Binding of Zinc? Thermodynamic and Solution Structural Study of a Ternary Complex. Chem. Commun. 2003, 3, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Blindauer, C.A.; Harvey, I.; Bunyan, K.E.; Stewart, A.J.; Sleep, D.; Harrison, D.J.; Berezenko, S.; Sadler, P.J. Structure, Properties, and Engineering of the Major Zinc Binding Site on Human Albumin. J. Biol. Chem. 2009, 284, 23116–23124. [Google Scholar] [CrossRef] [PubMed]
- Kleifeld, O. The Conserved Glu-60 Residue in Thermoanaerobacter Brockii Alcohol Dehydrogenase is Not Essential for Catalysis. Protein Sci. 2003, 12, 468–479. [Google Scholar] [CrossRef]
- Feiters, M.C.; Eijkelenboom, A.P.A.M.; Nolting, H.-F.; Krebs, B.; van den Ent, F.M.I.; Plasterk, R.H.A.; Kaptein, R.; Boelens, R. X-ray Absorption Spectroscopic Studies of Zinc in the N-Terminal Domain of HIV-2 Integrase and Model Compounds. J. Synchrotron Radiat. 2003, 10, 86–95. [Google Scholar] [CrossRef]
- Zhang, K.; Auld, D.S. Structure of Binary and Ternary Complexes of Zinc and Cobalt Carboxypeptidase A as Determined by X-ray Absorption Fine Structure. Biochemistry 1995, 34, 16306–16312. [Google Scholar] [CrossRef] [PubMed]
- Tobin, D.A.; Pickett, J.S.; Hartman, H.L.; Fierke, C.A.; Penner-Hahn, J.E. Structural Characterization of the Zinc Site in Protein Farnesyltransferase. J. Am. Chem. Soc. 2003, 125, 9962–9969. [Google Scholar] [CrossRef] [PubMed]
- Akabayov, B.; Lee, S.-J.; Akabayov, S.R.; Rekhi, S.; Zhu, B.; Richardson, C.C. DNA Recognition by the DNA Primase of Bacteriophage T7: A Structure−Function Study of the Zinc-Binding Domain. Biochemistry 2009, 48, 1763–1773. [Google Scholar] [CrossRef]
- Krizek, B.A.; Merkle, D.L.; Berg, J.M. Ligand Variation and Metal Ion Binding Specificity in Zinc Finger Peptides. Inorg. Chem. 1993, 32, 937–940. [Google Scholar] [CrossRef]
- Miłoch, A.; Krężel, A. Metal Binding Properties of the Zinc Finger Metallome—Insights into Variations in Stability. Metallomics 2014, 6, 2015–2024. [Google Scholar] [CrossRef]
- Guerrerio, A.L.; Berg, J.M. Metal Ion Affinities of the Zinc Finger Domains of the Metal Responsive Element-Binding Transcription Factor-1 (MTF1). Biochemistry 2004, 43, 5437–5444. [Google Scholar] [CrossRef] [PubMed]
- Gatei, M.; Jakob, B.; Chen, P.; Kijas, A.W.; Becherel, O.J.; Gueven, N.; Birrell, G.; Lee, J.; Paull, T.T.; Lerenthal, Y.; et al. ATM Protein-Dependent Phosphorylation of Rad50 Protein Regulates DNA Repair and Cell Cycle Control. J. Biol. Chem. 2011, 286, 31542–31556. [Google Scholar] [CrossRef]
- Moreno-Herrero, F.; de Jager, M.; Dekker, N.H.; Kanaar, R.; Wyman, C.; Dekker, C. Mesoscale Conformational Changes in the DNA-Repair Complex Rad50/Mre11/Nbs1 upon Binding DNA. Nature 2005, 437, 440–443. [Google Scholar] [CrossRef]
- Zabolotnaya, E.; Mela, I.; Williamson, M.J.; Bray, S.M.; Yau, S.K.; Papatziamou, D.; Edwardson, J.M.; Robinson, N.P.; Henderson, R.M. Modes of Action of the Archaeal Mre11/Rad50 DNA-Repair Complex Revealed by Fast-Scan Atomic Force Microscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 14936–14947. [Google Scholar] [CrossRef]
- Eyer, P.; Worek, F.; Kiderlen, D.; Sinko, G.; Stuglin, A.; Simeon-Rudolf, V.; Reiner, E. Molar Absorption Coefficients for the Reduced Ellman Reagent: Reassessment. Anal. Biochem. 2003, 312, 224–227. [Google Scholar] [CrossRef]
- Chong, S.; Mersha, F.B.; Comb, D.G.; Scott, M.E.; Landry, D.; Vence, L.M.; Perler, F.B.; Benner, J.; Kucera, R.B.; Hirvonen, C.A.; et al. Single-Column Purification of Free Recombinant Proteins Using a Self-Cleavable Affinity Tag Derived from a Protein Splicing Element. Gene 1997, 192, 271–281. [Google Scholar] [CrossRef]
- Drozd, A.; Wojewska, D.; Peris-Díaz, M.D.; Jakimowicz, P.; Krężel, A. Crosstalk of the Structural and Zinc Buffering Properties of Mammalian Metallothionein-2. Metallomics 2018, 10, 595–613. [Google Scholar] [CrossRef]
- Goulon, J.; Goulon-Ginet, C.; Cortes, R.; Dubois, J.M. On Experimental Attenuation Factors of the Amplitude of the EXAFS Oscillations in Absorption, Reflectivity and Luminescence Measurements. J. Phys. 1982, 43, 539–548. [Google Scholar] [CrossRef]
- Ravel, B.; Newville, M. ATHENA, ARTEMIS, HEPHAESTUS: Data Analysis for X-ray Absorption Spectroscopy Using IFEFFIT. J. Synchrotron Radiat. 2005, 12, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Newville, M. IFEFFIT: Interactive XAFS Analysis and FEFF Fitting. J. Synchrotron Radiat. 2001, 8, 322–324. [Google Scholar] [CrossRef]
- Smith, R.M.; Martell, A.E. Aminocarboxylic Acids. In Critical Stability Constants: Second Supplement; Springer: Boston, MA, USA, 1989; pp. 1–66. ISBN 978-1-4615-6764-6. [Google Scholar]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad Simulation and Speciation (HySS): A Utility Program for the Investigation of Equilibria Involving Soluble and Partially Soluble Species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. SUPERQUAD: An Improved General Program for Computation of Formation Constants from Potentiometric Data. J. Chem. Soc. Dalt. Trans. 1985, 6, 1195. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and Sequence Analysis Tools Services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]






| Zinc Hook Peptide | logK12–Competition | logK12–Potentiometry |
|---|---|---|
| HsHk4 | n.d. a | 14.93 b |
| HsHk6 | n.d. | 15.13 |
| HsHk10 | 16.5 ± 0.4 | 16.15 |
| HsHk14 | 15.8 ± 0.4 | n.d. |
| HsHk42 | 17.26 ± 0.08 | n.d. |
| HsHk42 pT690 | 16.4 ± 0.2 | n.d. |
| HsHk42 T690E | 16.5 ± 0.1 | n.d. |
| HsHk72 | 18.3 ± 0.4 | n.d. |
| HsHk140 | 19.5 ± 0.2 | n.d. |
| HsHk183 | 19.53 ± 0.06 | n.d. |
| HsHk183 T690E | 19.27 ± 0.05 | n.d. |
| Protein Complex | Residues in Coordination Sphere (Complex Stoichiometry) | Experimental Conditions | logK a | logK Estimated to pH 7.4 b | CI c Estimated to pH 7.4 b | Reference |
|---|---|---|---|---|---|---|
| CD4-Lck kinase complex (H. sapiens) | 2 × CC (ZnL′L″) | 150 mM NaCl, 10 mM Na2SO4, 0.08%, β-ME, 50 mM ZnSO4, 10 mM HEPES, pH 7.0 | 6.4 | 8 | 4.7 | [51] |
| CD4-Lck kinase complex (H. sapiens) | 2 × CC (ZnL′L″) | 20 mM Tris, 10 mM NaClO4, 15 μM TCEP, pH 7.4 | 18.97 | 18.97 | 15.67 | [52] |
| CD8α-Lck kinase complex (H. sapiens) | 2 × CC (ZnL′L″) | 150 mM NaCl, 10 mM Na2SO4, 0.08%, β-ME, 50 mM ZnSO4, 10 mM HEPES, pH 7.0 | 6.05 | 7.65 | 4.35 | [51] |
| Rad50 (P. furiosus) | 2 × CC (ZnL2) | 20 mM Tris-HCl, 100 mM NaClO4, pH 7.4 | 20.82 | 20.82 | 17.52 | [27] |
| Rad50 (H. sapiens) | 2 × CC (ZnL2) | 20 mM Tris-HCl, 100 mM NaClO4, pH 7.4 | 19.52 | 19.52 | 16.22 | this study |
| CP1 | CCCC (ZnL) | 50 mM HEPES, 100 mM NaCl, pH 7.0 | 12.0 | 13.6 | 13.6 | [60] |
| CP1 | CCHH (ZnL) | 50 mM HEPES, 100 mM NaCl, pH 7.0 | 11.2 | 12 | 12 | [60] |
| MTF1-1 (H. sapiens) | CCHH (ZnL) | 50 mM HEPES, pH 7.4, 100 mM NaClO4 | 11.6 | 11.6 | 11.6 | [61] |
| MTF1-1 (H. sapiens) | CCHH (ZnL) | 100 mM HEPES, 50 mM NaCl, pH 7.0 | 10.5 | 11.3 | 11.3 | [62] |
| PDLIM1 (H. sapiens) | CCCC (ZnL) | 50 mM Tris, 100 mM NaCl, pH 7.4 | 14.5 | 14.5 | 14.5 | [49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, J.B.; Padjasek, M.; Krężel, A. Relations between Structure and Zn(II) Binding Affinity Shed Light on the Mechanisms of Rad50 Hook Domain Functioning and Its Phosphorylation. Int. J. Mol. Sci. 2022, 23, 11140. https://doi.org/10.3390/ijms231911140
Tran JB, Padjasek M, Krężel A. Relations between Structure and Zn(II) Binding Affinity Shed Light on the Mechanisms of Rad50 Hook Domain Functioning and Its Phosphorylation. International Journal of Molecular Sciences. 2022; 23(19):11140. https://doi.org/10.3390/ijms231911140
Chicago/Turabian StyleTran, Józef Ba, Michał Padjasek, and Artur Krężel. 2022. "Relations between Structure and Zn(II) Binding Affinity Shed Light on the Mechanisms of Rad50 Hook Domain Functioning and Its Phosphorylation" International Journal of Molecular Sciences 23, no. 19: 11140. https://doi.org/10.3390/ijms231911140
APA StyleTran, J. B., Padjasek, M., & Krężel, A. (2022). Relations between Structure and Zn(II) Binding Affinity Shed Light on the Mechanisms of Rad50 Hook Domain Functioning and Its Phosphorylation. International Journal of Molecular Sciences, 23(19), 11140. https://doi.org/10.3390/ijms231911140







