Effects of RAGE Deletion on the Cardiac Transcriptome during Aging
Abstract
1. Introduction
2. Results
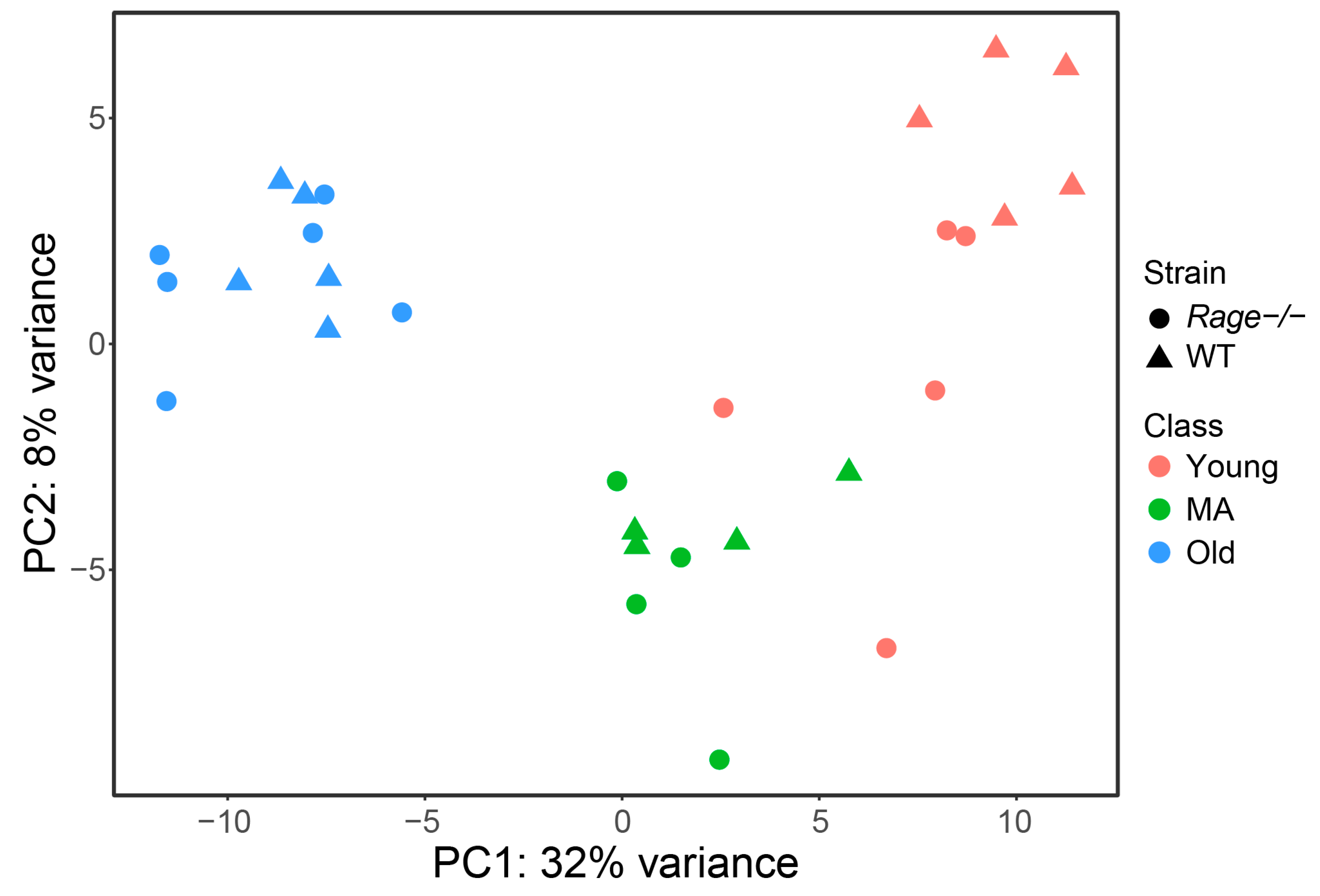
2.1. Gene Expression Dataset
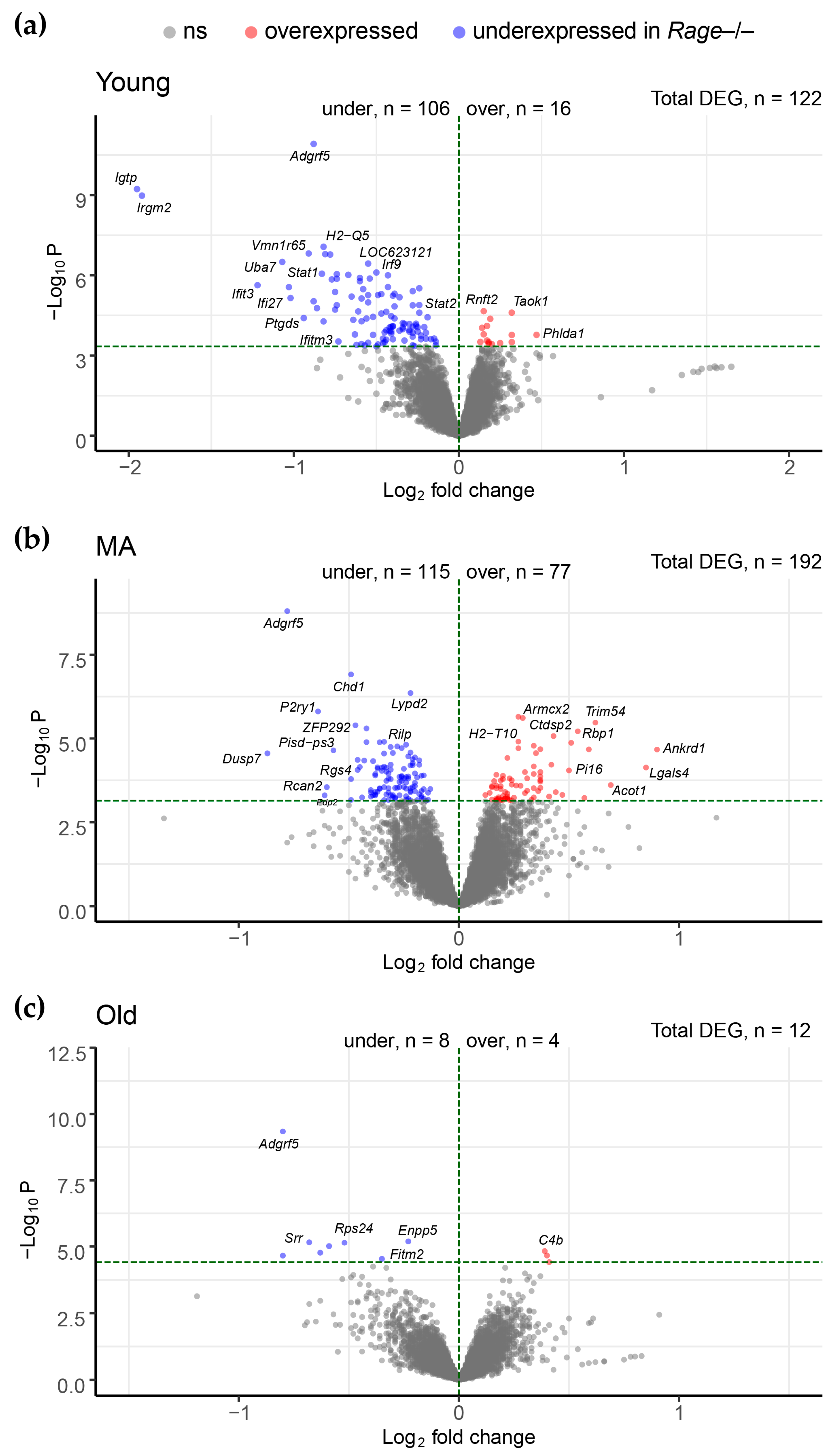
2.2. Differential Expression Analysis between Rage−/− and WT Mice
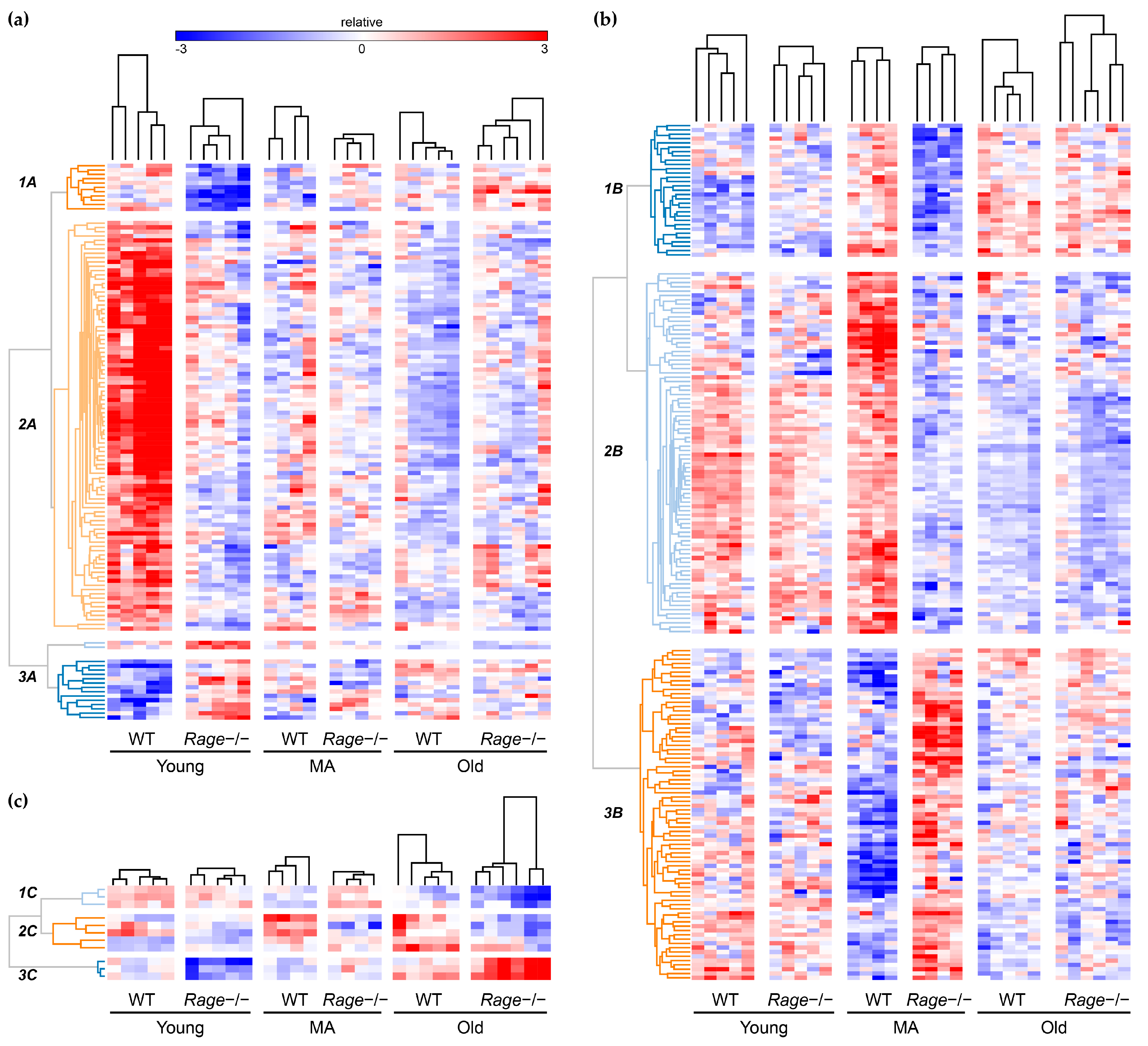
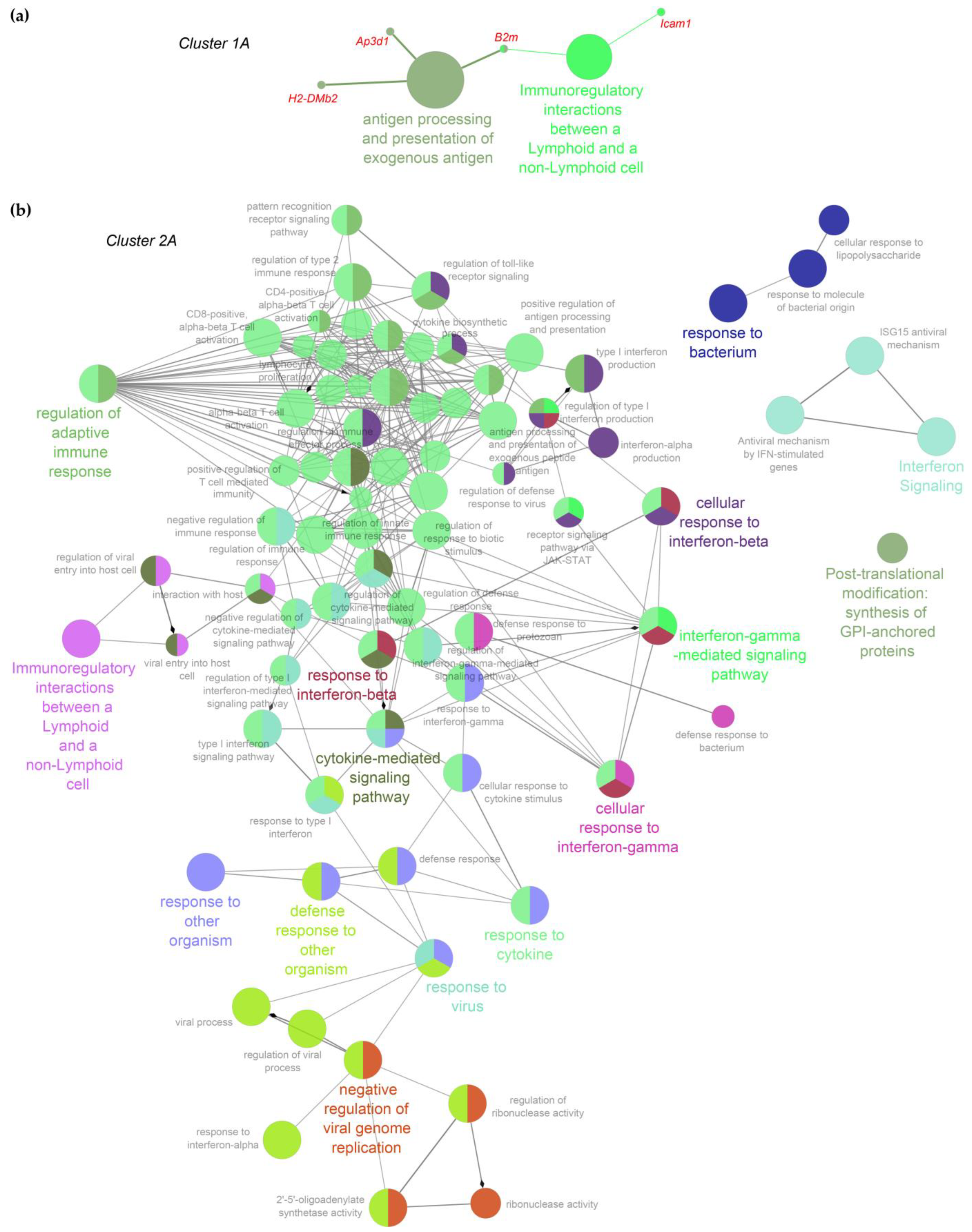
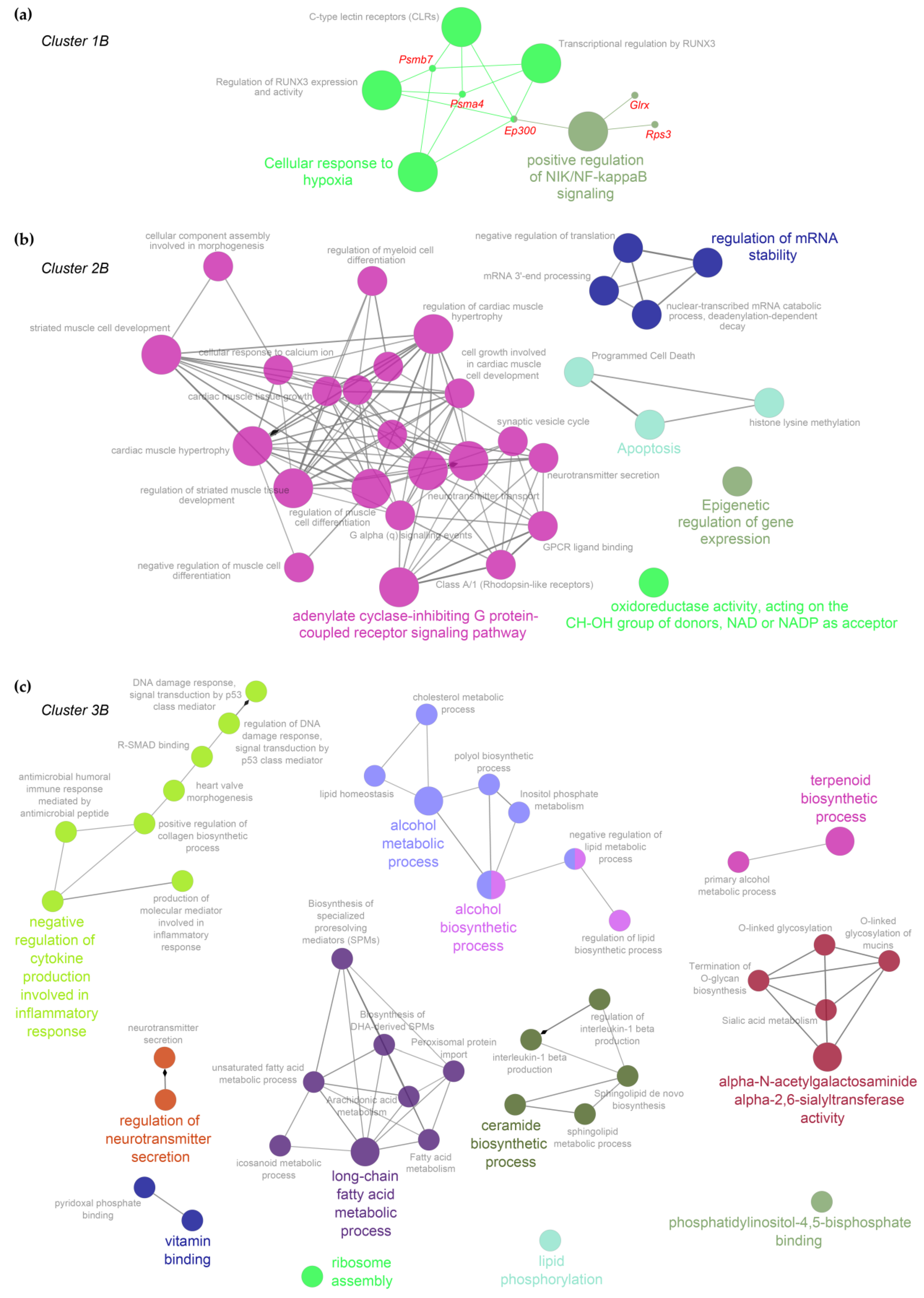
2.3. Functional Inferences from Genome-Wide Differential Expression Analysis
3. Discussion
4. Material and Methods
4.1. Animals
4.2. Microarray Gene Expression Analysis
4.3. Reverse-Transcription Quantitative PCR (RT-qPCR)
4.4. Data Processing
4.5. Statistical Analysis
4.6. Hierarchical Clustering of Gene Expression Data
4.7. Functional Inferences on Gene Expression Profiles
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiao, Y.A.; Ramirez, T.A.; Zamilpa, R.; Okoronkwo, S.M.; Dai, Q.; Zhang, J.; Jin, Y.F.; Lindsey, M.L. Matrix metalloproteinase-9 deletion attenuates myocardial fibrosis and diastolic dysfunction in ageing mice. Cardiovasc. Res. 2012, 96, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Lopez, E.F.; Jin, Y.; Van Remmen, H.; Bauch, T.; Han, H.C.; Lindsey, M.L. Age-related cardiac muscle sarcopenia: Combining experimental and mathematical modeling to identify mechanisms. Exp. Gerontol. 2008, 43, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Steenman, M.; Lande, G. Cardiac aging and heart disease in humans. Biophys. Rev. 2017, 9, 131–137. [Google Scholar] [CrossRef]
- Antonelli, A.; Di Maggio, S.; Rejman, J.; Sanvito, F.; Rossi, A.; Catucci, A.; Gorzanelli, A.; Bragonzi, A.; Bianchi, M.E.; Raucci, A. The shedding-derived soluble receptor for advanced glycation endproducts sustains inflammation during acute Pseudomonas aeruginosa lung infection. Biochim. Biophys. Acta 2017, 1861, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Demling, N.; Ehrhardt, C.; Kasper, M.; Laue, M.; Knels, L.; Rieber, E.P. Promotion of cell adherence and spreading: A novel function of RAGE, the highly selective differentiation marker of human alveolar epithelial type I cells. Cell Tissue Res. 2006, 323, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Queisser, M.A.; Kouri, F.M.; Konigshoff, M.; Wygrecka, M.; Schubert, U.; Eickelberg, O.; Preissner, K.T. Loss of RAGE in pulmonary fibrosis: Molecular relations to functional changes in pulmonary cell types. Am. J. Respir. Cell Mol. Biol. 2008, 39, 337–345. [Google Scholar] [CrossRef]
- Scavello, F.; Zeni, F.; Milano, G.; Macri, F.; Castiglione, S.; Zuccolo, E.; Scopece, A.; Pezone, G.; Tedesco, C.C.; Nigro, P.; et al. Soluble Receptor for Advanced Glycation End-products regulates age-associated Cardiac Fibrosis. Int. J. Biol. Sci. 2021, 17, 2399–2416. [Google Scholar] [CrossRef]
- Fritz, G. RAGE: A single receptor fits multiple ligands. Trends Biochem. Sci. 2011, 36, 625–632. [Google Scholar] [CrossRef]
- Orlova, V.V.; Choi, E.Y.; Xie, C.; Chavakis, E.; Bierhaus, A.; Ihanus, E.; Ballantyne, C.M.; Gahmberg, C.G.; Bianchi, M.E.; Nawroth, P.P.; et al. A novel pathway of HMGB1-mediated inflammatory cell recruitment that requires Mac-1-integrin. EMBO J. 2007, 26, 1129–1139. [Google Scholar] [CrossRef]
- Sourris, K.C.; Morley, A.L.; Koitka, A.; Samuel, P.; Coughlan, M.T.; Penfold, S.A.; Thomas, M.C.; Bierhaus, A.; Nawroth, P.P.; Yamamoto, H.; et al. Receptor for AGEs (RAGE) blockade may exert its renoprotective effects in patients with diabetic nephropathy via induction of the angiotensin II type 2 (AT2) receptor. Diabetologia 2010, 53, 2442–2451. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, K.; Penn, M.S.; Marso, S.P.; Lauer, M.A.; Forudi, F.; Zhou, X.; Qu, W.; Lu, Y.; Stern, D.M.; et al. Receptor for AGE (RAGE) mediates neointimal formation in response to arterial injury. Circulation 2003, 107, 2238–2243. [Google Scholar] [CrossRef] [PubMed]
- Iannitti, R.G.; Casagrande, A.; De Luca, A.; Cunha, C.; Sorci, G.; Riuzzi, F.; Borghi, M.; Galosi, C.; Massi-Benedetti, C.; Oury, T.D.; et al. Hypoxia promotes danger-mediated inflammation via receptor for advanced glycation end products in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2013, 188, 1338–1350. [Google Scholar] [CrossRef]
- Mahajan, N.; Dhawan, V. Receptor for advanced glycation end products (RAGE) in vascular and inflammatory diseases. Int. J. Cardiol. 2013, 168, 1788–1794. [Google Scholar] [CrossRef]
- Volz, H.C.; Laohachewin, D.; Seidel, C.; Lasitschka, F.; Keilbach, K.; Wienbrandt, A.R.; Andrassy, J.; Bierhaus, A.; Kaya, Z.; Katus, H.A.; et al. S100A8/A9 aggravates post-ischemic heart failure through activation of RAGE-dependent NF-kappaB signaling. Basic Res. Cardiol. 2012, 107, 250. [Google Scholar] [CrossRef]
- Penzo, M.; Molteni, R.; Suda, T.; Samaniego, S.; Raucci, A.; Habiel, D.M.; Miller, F.; Jiang, H.P.; Li, J.; Pardi, R.; et al. Inhibitor of NF-kappa B kinases alpha and beta are both essential for high mobility group box 1-mediated chemotaxis [corrected]. J. Immunol. 2010, 184, 4497–4509. [Google Scholar] [CrossRef] [PubMed]
- Sessa, L.; Gatti, E.; Zeni, F.; Antonelli, A.; Catucci, A.; Koch, M.; Pompilio, G.; Fritz, G.; Raucci, A.; Bianchi, M.E. The receptor for advanced glycation end-products (RAGE) is only present in mammals, and belongs to a family of cell adhesion molecules (CAMs). PLoS ONE 2014, 9, e86903. [Google Scholar] [CrossRef]
- Volz, H.C.; Seidel, C.; Laohachewin, D.; Kaya, Z.; Muller, O.J.; Pleger, S.T.; Lasitschka, F.; Bianchi, M.E.; Remppis, A.; Bierhaus, A.; et al. HMGB1: The missing link between diabetes mellitus and heart failure. Basic Res. Cardiol. 2010, 105, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Raucci, A.; Cugusi, S.; Antonelli, A.; Barabino, S.M.; Monti, L.; Bierhaus, A.; Reiss, K.; Saftig, P.; Bianchi, M.E. A soluble form of the receptor for advanced glycation endproducts (RAGE) is produced by proteolytic cleavage of the membrane-bound form by the sheddase a disintegrin and metalloprotease 10 (ADAM10). FASEB J. 2008, 22, 3716–3727. [Google Scholar] [CrossRef]
- Sabbatinelli, J.; Castiglione, S.; Macri, F.; Giuliani, A.; Ramini, D.; Vinci, M.C.; Tortato, E.; Bonfigli, A.R.; Olivieri, F.; Raucci, A. Circulating levels of AGEs and soluble RAGE isoforms are associated with all-cause mortality and development of cardiovascular complications in type 2 diabetes: A retrospective cohort study. Cardiovasc. Diabetol. 2022, 21, 95. [Google Scholar] [CrossRef]
- Scavello, F.; Tedesco, C.C.; Castiglione, S.; Maciag, A.; Sangalli, E.; Veglia, F.; Spinetti, G.; Puca, A.A.; Raucci, A. Modulation of soluble receptor for advanced glycation end products isoforms and advanced glycation end products in long-living individuals. Biomark. Med. 2021, 15, 785–796. [Google Scholar] [CrossRef]
- Scavello, F.; Zeni, F.; Tedesco, C.C.; Mensa, E.; Veglia, F.; Procopio, A.D.; Bonfigli, A.R.; Olivieri, F.; Raucci, A. Modulation of soluble receptor for advanced glycation end-products (RAGE) isoforms and their ligands in healthy aging. Aging 2019, 11, 1648–1663. [Google Scholar] [CrossRef] [PubMed]
- Park, L.; Raman, K.G.; Lee, K.J.; Lu, Y.; Ferran, L.J., Jr.; Chow, W.S.; Stern, D.; Schmidt, A.M. Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat. Med. 1998, 4, 1025–1031. [Google Scholar] [CrossRef] [PubMed]
- Pickering, R.J.; Tikellis, C.; Rosado, C.J.; Tsorotes, D.; Dimitropoulos, A.; Smith, M.; Huet, O.; Seeber, R.M.; Abhayawardana, R.; Johnstone, E.K.; et al. Transactivation of RAGE mediates angiotensin-induced inflammation and atherogenesis. J. Clin. Investig. 2019, 129, 406–421. [Google Scholar] [CrossRef]
- Volz, H.C.; Kaya, Z.; Katus, H.A.; Andrassy, M. The role of HMGB1/RAGE in inflammatory cardiomyopathy. Semin. Thromb. Hemost. 2010, 36, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Englert, J.M.; Kliment, C.R.; Ramsgaard, L.; Milutinovic, P.S.; Crum, L.; Tobolewski, J.M.; Oury, T.D. Paradoxical function for the receptor for advanced glycation end products in mouse models of pulmonary fibrosis. Int. J. Clin. Exp. Pathol. 2011, 4, 241–254. [Google Scholar]
- Kumar, V.; Fleming, T.; Terjung, S.; Gorzelanny, C.; Gebhardt, C.; Agrawal, R.; Mall, M.A.; Ranzinger, J.; Zeier, M.; Madhusudhan, T.; et al. Homeostatic nuclear RAGE-ATM interaction is essential for efficient DNA repair. Nucleic Acids Res. 2017, 45, 10595–10613. [Google Scholar] [CrossRef]
- Bui, T.M.; Wiesolek, H.L.; Sumagin, R. ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J. Leukoc. Biol. 2020, 108, 787–799. [Google Scholar] [CrossRef]
- Hofmann, U.; Beyersdorf, N.; Weirather, J.; Podolskaya, A.; Bauersachs, J.; Ertl, G.; Kerkau, T.; Frantz, S. Activation of CD4+ T lymphocytes improves wound healing and survival after experimental myocardial infarction in mice. Circulation 2012, 125, 1652–1663. [Google Scholar] [CrossRef]
- Ramos, G.C.; van den Berg, A.; Nunes-Silva, V.; Weirather, J.; Peters, L.; Burkard, M.; Friedrich, M.; Pinnecker, J.; Abesser, M.; Heinze, K.G.; et al. Myocardial aging as a T-cell-mediated phenomenon. Proc. Natl. Acad. Sci. USA 2017, 114, E2420–E2429. [Google Scholar] [CrossRef]
- Weirather, J.; Hofmann, U.D.; Beyersdorf, N.; Ramos, G.C.; Vogel, B.; Frey, A.; Ertl, G.; Kerkau, T.; Frantz, S. Foxp3+ CD4+ T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ. Res. 2014, 115, 55–67. [Google Scholar] [CrossRef]
- Ramos, G.; Hofmann, U.; Frantz, S. Myocardial fibrosis seen through the lenses of T-cell biology. J. Mol. Cell. Cardiol. 2016, 92, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Laroumanie, F.; Douin-Echinard, V.; Pozzo, J.; Lairez, O.; Tortosa, F.; Vinel, C.; Delage, C.; Calise, D.; Dutaur, M.; Parini, A.; et al. CD4+ T cells promote the transition from hypertrophy to heart failure during chronic pressure overload. Circulation 2014, 129, 2111–2124. [Google Scholar] [CrossRef]
- Skorska, A.; von Haehling, S.; Ludwig, M.; Lux, C.A.; Gaebel, R.; Kleiner, G.; Klopsch, C.; Dong, J.; Curato, C.; Altarche-Xifro, W.; et al. The CD4(+) AT2R(+) T cell subpopulation improves post-infarction remodelling and restores cardiac function. J. Cell. Mol. Med. 2015, 19, 1975–1985. [Google Scholar] [CrossRef]
- Delgobo, M.; Heinrichs, M.; Hapke, N.; Ashour, D.; Appel, M.; Srivastava, M.; Heckel, T.; Spyridopoulos, I.; Hofmann, U.; Frantz, S.; et al. Terminally Differentiated CD4(+) T Cells Promote Myocardial Inflammaging. Front. Immunol. 2021, 12, 584538. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, S.; Espitia-Corredor, J.A.; Olivares-Silva, F.; Valenzuela, P.; Humeres, C.; Anfossi, R.; Castro, E.; Vivar, R.; Salas-Hernandez, A.; Pardo-Jimenez, V.; et al. In cardiac fibroblasts, interferon-beta attenuates differentiation, collagen synthesis, and TGF-beta1-induced collagen gel contraction. Cytokine 2021, 138, 155359. [Google Scholar] [CrossRef] [PubMed]
- Levick, S.P.; Goldspink, P.H. Could interferon-gamma be a therapeutic target for treating heart failure? Heart Fail. Rev. 2014, 19, 227–236. [Google Scholar] [CrossRef]
- Bartlett, E.J.; Lenzo, J.C.; Sivamoorthy, S.; Mansfield, J.P.; Cull, V.S.; James, C.M. Type I IFN-beta gene therapy suppresses cardiac CD8+ T-cell infiltration during autoimmune myocarditis. Immunol. Cell Biol. 2004, 82, 119–126. [Google Scholar] [CrossRef]
- Deonarain, R.; Cerullo, D.; Fuse, K.; Liu, P.P.; Fish, E.N. Protective role for interferon-beta in coxsackievirus B3 infection. Circulation 2004, 110, 3540–3543. [Google Scholar] [CrossRef]
- Kimura, A.; Ishida, Y.; Furuta, M.; Nosaka, M.; Kuninaka, Y.; Taruya, A.; Mukaida, N.; Kondo, T. Protective Roles of Interferon-gamma in Cardiac Hypertrophy Induced by Sustained Pressure Overload. J. Am. Heart Assoc. 2018, 7, e008145. [Google Scholar] [CrossRef]
- Lee, J.W.; Oh, J.E.; Rhee, K.J.; Yoo, B.S.; Eom, Y.W.; Park, S.W.; Lee, J.H.; Son, J.W.; Youn, Y.J.; Ahn, M.S.; et al. Co-treatment with interferon-gamma and 1-methyl tryptophan ameliorates cardiac fibrosis through cardiac myofibroblasts apoptosis. Mol. Cell. Biochem. 2019, 458, 197–205. [Google Scholar] [CrossRef]
- Frommhold, D.; Kamphues, A.; Hepper, I.; Pruenster, M.; Lukic, I.K.; Socher, I.; Zablotskaya, V.; Buschmann, K.; Lange-Sperandio, B.; Schymeinsky, J.; et al. RAGE and ICAM-1 cooperate in mediating leukocyte recruitment during acute inflammation in vivo. Blood 2010, 116, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, T.; Horstkotte, M.; Lange, P.; Ng, J.; Bongiovanni, D.; Hinkel, R.; Laugwitz, K.L.; Sperandio, M.; Horstkotte, J.; Kupatt, C. Endothelial RAGE exacerbates acute postischaemic cardiac inflammation. Thromb. Haemost. 2016, 116, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Akirav, E.M.; Chen, W.; Henegariu, O.; Moser, B.; Desai, D.; Shen, J.M.; Webster, J.C.; Andrews, R.C.; Mjalli, A.M.; et al. RAGE ligation affects T cell activation and controls T cell differentiation. J. Immunol. 2008, 181, 4272–4278. [Google Scholar] [CrossRef] [PubMed]
- Moser, B.; Desai, D.D.; Downie, M.P.; Chen, Y.; Yan, S.F.; Herold, K.; Schmidt, A.M.; Clynes, R. Receptor for advanced glycation end products expression on T cells contributes to antigen-specific cellular expansion in vivo. J. Immunol. 2007, 179, 8051–8058. [Google Scholar] [CrossRef]
- Dang, M.; Zeng, X.; Chen, B.; Wang, H.; Li, H.; Du, F.; Guo, C. Interferon-gamma mediates the protective effects of soluble receptor for advanced glycation end-product in myocardial ischemia/reperfusion. Lab. Investig. 2019, 99, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cao, X.; Dang, M.; Wang, H.; Chen, B.; Du, F.; Li, H.; Zeng, X.; Guo, C. Soluble receptor for advanced glycation end-products enhanced the production of IFN-gamma through the NF-kappaB pathway in macrophages recruited by ischemia/reperfusion. Int. J. Mol. Med. 2019, 43, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Godec, J.; Ben-Aissa, K.; Cui, K.; Zhao, K.; Pucsek, A.B.; Lee, Y.K.; Weaver, C.T.; Yagi, R.; Lazarevic, V. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity 2014, 40, 355–366. [Google Scholar] [CrossRef]
- Wong, W.F.; Kohu, K.; Chiba, T.; Sato, T.; Satake, M. Interplay of transcription factors in T-cell differentiation and function: The role of Runx. Immunology 2011, 132, 157–164. [Google Scholar] [CrossRef]
- Ghosh, A.K. p300 in Cardiac Development and Accelerated Cardiac Aging. Aging Dis. 2020, 11, 916–926. [Google Scholar] [CrossRef]
- Kawamura, T.; Hasegawa, K.; Morimoto, T.; Iwai-Kanai, E.; Miyamoto, S.; Kawase, Y.; Ono, K.; Wada, H.; Akao, M.; Kita, T. Expression of p300 protects cardiac myocytes from apoptosis in vivo. Biochem. Biophys. Res. Commun. 2004, 315, 733–738. [Google Scholar] [CrossRef]
- Kyrychenko, V.O.; Nagibin, V.S.; Tumanovska, L.V.; Pashevin, D.O.; Gurianova, V.L.; Moibenko, A.A.; Dosenko, V.E.; Klionsky, D.J. Knockdown of PSMB7 induces autophagy in cardiomyocyte cultures: Possible role in endoplasmic reticulum stress. Pathobiology 2014, 81, 8–14. [Google Scholar] [CrossRef]
- Sithara, T.; Drosatos, K. Metabolic Complications in Cardiac Aging. Front. Physiol. 2021, 12, 669497. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.H.; Tatum, S.M.; Symons, J.D.; Summers, S.A.; Holland, W.L. Ceramides and other sphingolipids as drivers of cardiovascular disease. Nat. Rev. Cardiol. 2021, 18, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Gellings Lowe, N.; Swaney, J.S.; Moreno, K.M.; Sabbadini, R.A. Sphingosine-1-phosphate and sphingosine kinase are critical for transforming growth factor-beta-stimulated collagen production by cardiac fibroblasts. Cardiovasc. Res. 2009, 82, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Pchejetski, D.; Foussal, C.; Alfarano, C.; Lairez, O.; Calise, D.; Guilbeau-Frugier, C.; Schaak, S.; Seguelas, M.H.; Wanecq, E.; Valet, P.; et al. Apelin prevents cardiac fibroblast activation and collagen production through inhibition of sphingosine kinase 1. Eur. Heart J. 2012, 33, 2360–2369. [Google Scholar] [CrossRef] [PubMed]
- Imig, J.D.; Cervenka, L.; Neckar, J. Epoxylipids and soluble epoxide hydrolase in heart diseases. Biochem. Pharmacol. 2022, 195, 114866. [Google Scholar] [CrossRef]
- Gaens, K.H.; Goossens, G.H.; Niessen, P.M.; van Greevenbroek, M.M.; van der Kallen, C.J.; Niessen, H.W.; Rensen, S.S.; Buurman, W.A.; Greve, J.W.; Blaak, E.E.; et al. Nepsilon-(carboxymethyl)lysine-receptor for advanced glycation end product axis is a key modulator of obesity-induced dysregulation of adipokine expression and insulin resistance. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1199–1208. [Google Scholar] [CrossRef]
- Mastrocola, R.; Collino, M.; Nigro, D.; Chiazza, F.; D’Antona, G.; Aragno, M.; Minetto, M.A. Accumulation of advanced glycation end-products and activation of the SCAP/SREBP Lipogenetic pathway occur in diet-induced obese mouse skeletal muscle. PLoS ONE 2015, 10, e0119587. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, M.Y.; Zheng, H.; Zhang, H.; Jiang, Y. Pentoxifylline alleviates high-fat diet-induced non-alcoholic steatohepatitis and early atherosclerosis in rats by inhibiting AGE and RAGE expression. Acta Pharmacol. Sin. 2010, 31, 1367–1375. [Google Scholar] [CrossRef]
- Chen, M.; Li, H.; Wang, G.; Shen, X.; Zhao, S.; Su, W. Atorvastatin prevents advanced glycation end products (AGEs)-induced cardiac fibrosis via activating peroxisome proliferator-activated receptor gamma (PPAR-gamma). Metabolism 2016, 65, 441–453. [Google Scholar] [CrossRef]
- Fu, M.X.; Requena, J.R.; Jenkins, A.J.; Lyons, T.J.; Baynes, J.W.; Thorpe, S.R. The advanced glycation end product, Nepsilon-(carboxymethyl)lysine, is a product of both lipid peroxidation and glycoxidation reactions. J. Biol. Chem. 1996, 271, 9982–9986. [Google Scholar] [CrossRef] [PubMed]
- Burch, M.L.; Zheng, W.; Little, P.J. Smad linker region phosphorylation in the regulation of extracellular matrix synthesis. Cell. Mol. Life Sci. 2011, 68, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Euler-Taimor, G.; Heger, J. The complex pattern of SMAD signaling in the cardiovascular system. Cardiovasc. Res. 2006, 69, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.G.; Rodriguez-Pascual, F.; Magan-Marchal, N.; Reguero, J.R.; Alonso-Montes, C.; Moris, C.; Alvarez, V.; Lamas, S.; Coto, E. Screening of the endothelin1 gene (EDN1) in a cohort of patients with essential left ventricular hypertrophy. Ann. Hum. Genet. 2007, 71, 601–610. [Google Scholar] [CrossRef]
- Fang, J.; Li, T.; Zhu, X.; Deng, K.Q.; Ji, Y.X.; Fang, C.; Zhang, X.J.; Guo, J.H.; Zhang, P.; Li, H.; et al. Control of Pathological Cardiac Hypertrophy by Transcriptional Corepressor IRF2BP2 (Interferon Regulatory Factor-2 Binding Protein 2). Hypertension 2017, 70, 515–523. [Google Scholar] [CrossRef]
- Du, P.; Kibbe, W.A.; Lin, S.M. lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008, 24, 1547–1548. [Google Scholar] [CrossRef]
- Du, P.F.G.; Kibbe, W.; Lin, S. lumiMouseIDMapping: Illumina Identifier Mapping for Mouse, R package version 1.10.0; Bioconductor: Boston, MA, USA, 2011. [Google Scholar]
- Chiesa, M.; Colombo, G.I.; Piacentini, L. DaMiRseq-an R/Bioconductor package for data mining of RNA-Seq data: Normalization, feature selection and classification. Bioinformatics 2018, 34, 1416–1418. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Leek, J.T.; Storey, J.D. A general framework for multiple testing dependence. Proc. Nat. Acad. Sci. USA 2008, 105, 18718–18723. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scavello, F.; Piacentini, L.; Castiglione, S.; Zeni, F.; Macrì, F.; Casaburo, M.; Vinci, M.C.; Colombo, G.I.; Raucci, A. Effects of RAGE Deletion on the Cardiac Transcriptome during Aging. Int. J. Mol. Sci. 2022, 23, 11130. https://doi.org/10.3390/ijms231911130
Scavello F, Piacentini L, Castiglione S, Zeni F, Macrì F, Casaburo M, Vinci MC, Colombo GI, Raucci A. Effects of RAGE Deletion on the Cardiac Transcriptome during Aging. International Journal of Molecular Sciences. 2022; 23(19):11130. https://doi.org/10.3390/ijms231911130
Chicago/Turabian StyleScavello, Francesco, Luca Piacentini, Stefania Castiglione, Filippo Zeni, Federica Macrì, Manuel Casaburo, Maria Cristina Vinci, Gualtiero I. Colombo, and Angela Raucci. 2022. "Effects of RAGE Deletion on the Cardiac Transcriptome during Aging" International Journal of Molecular Sciences 23, no. 19: 11130. https://doi.org/10.3390/ijms231911130
APA StyleScavello, F., Piacentini, L., Castiglione, S., Zeni, F., Macrì, F., Casaburo, M., Vinci, M. C., Colombo, G. I., & Raucci, A. (2022). Effects of RAGE Deletion on the Cardiac Transcriptome during Aging. International Journal of Molecular Sciences, 23(19), 11130. https://doi.org/10.3390/ijms231911130








