Transcriptome Analysis Reveals the Complex Regulatory Pathway of Background Color in Juvenile Plectropomus leopardus Skin Color Variation
Abstract
:1. Introduction
2. Results
2.1. The Types of Chromatophores in P. leopardus under Different Backgrounds
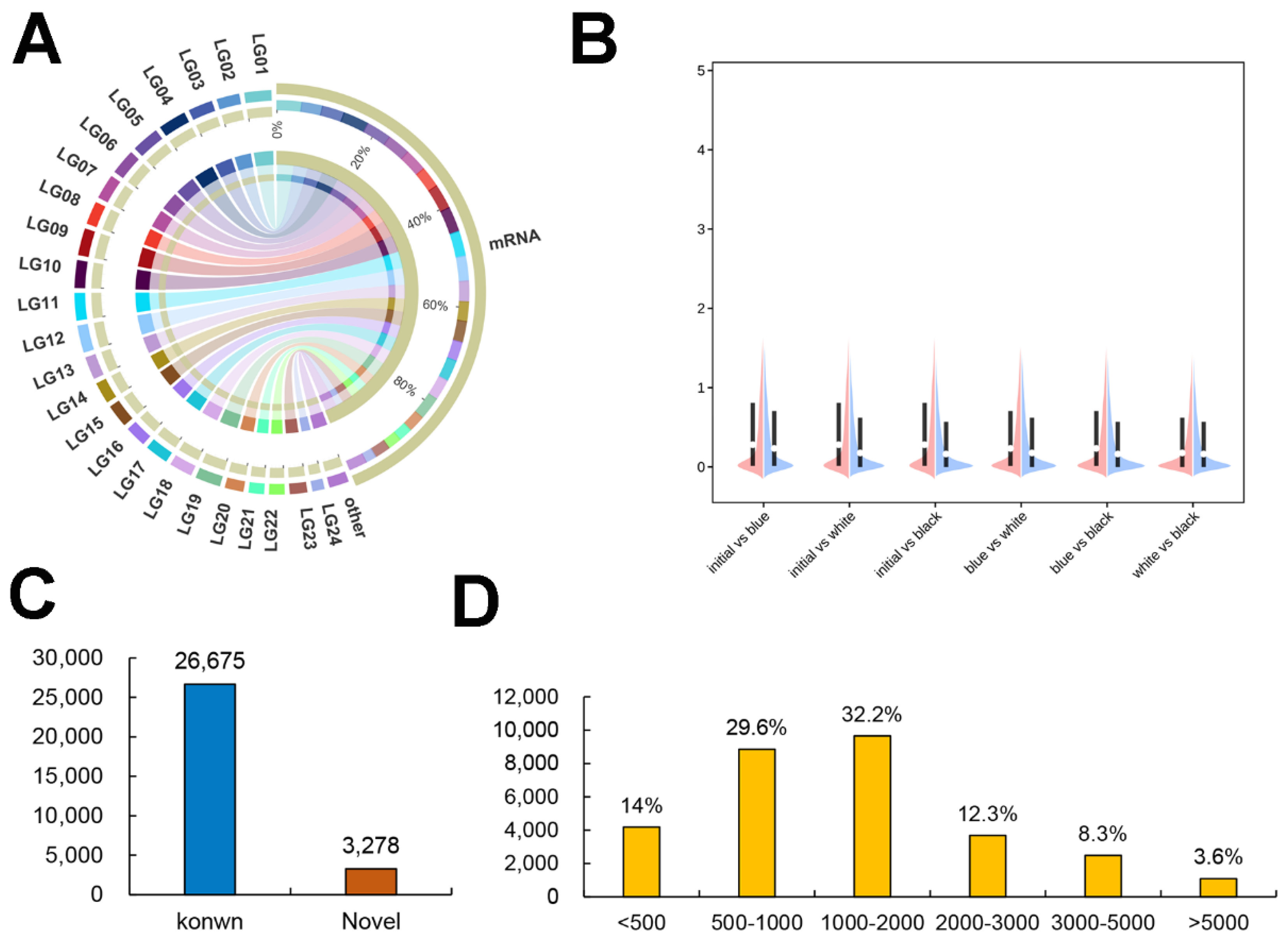
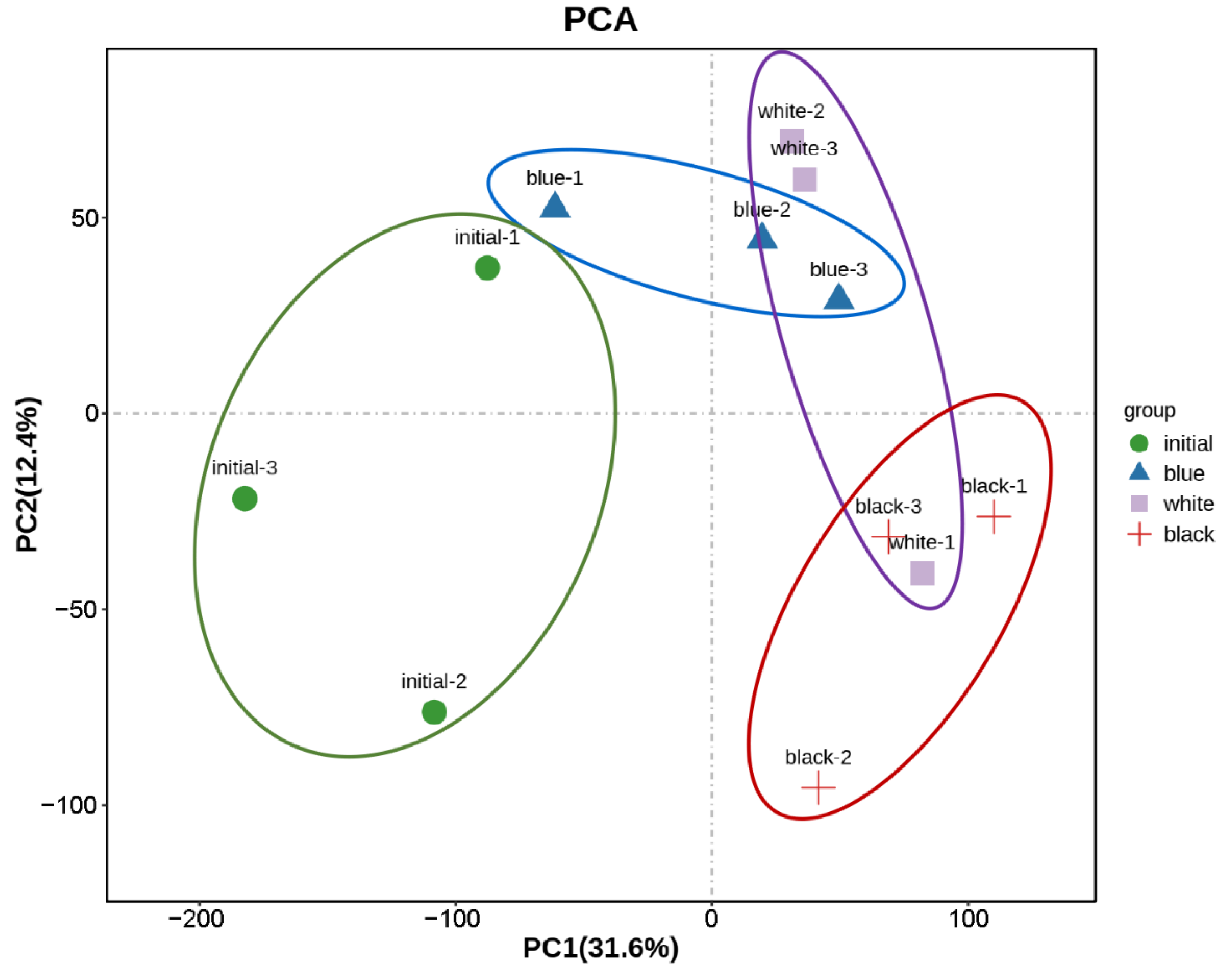
2.2. Overview of RNA Sequencing Data
2.3. Characterization of the Skin mRNAs
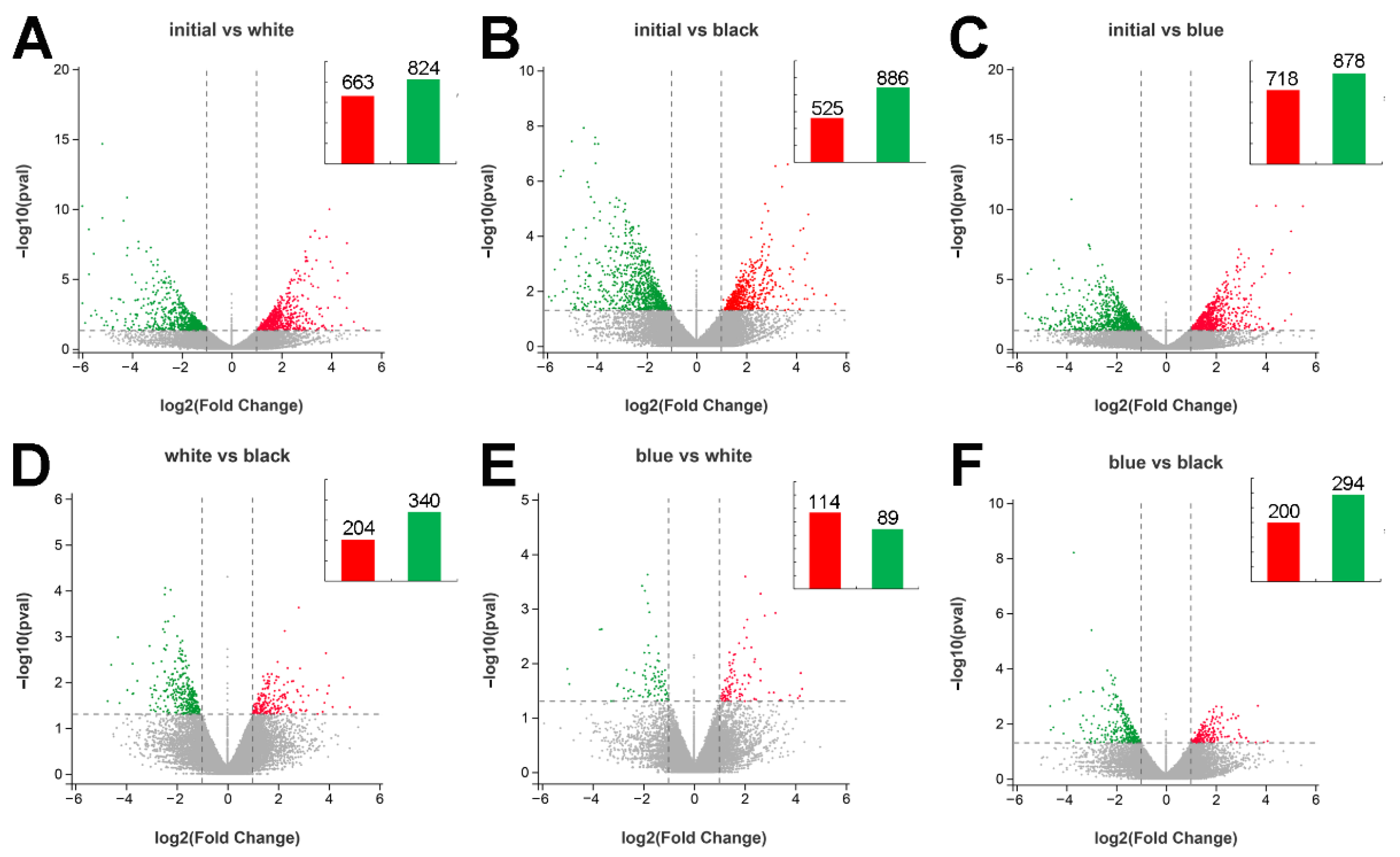
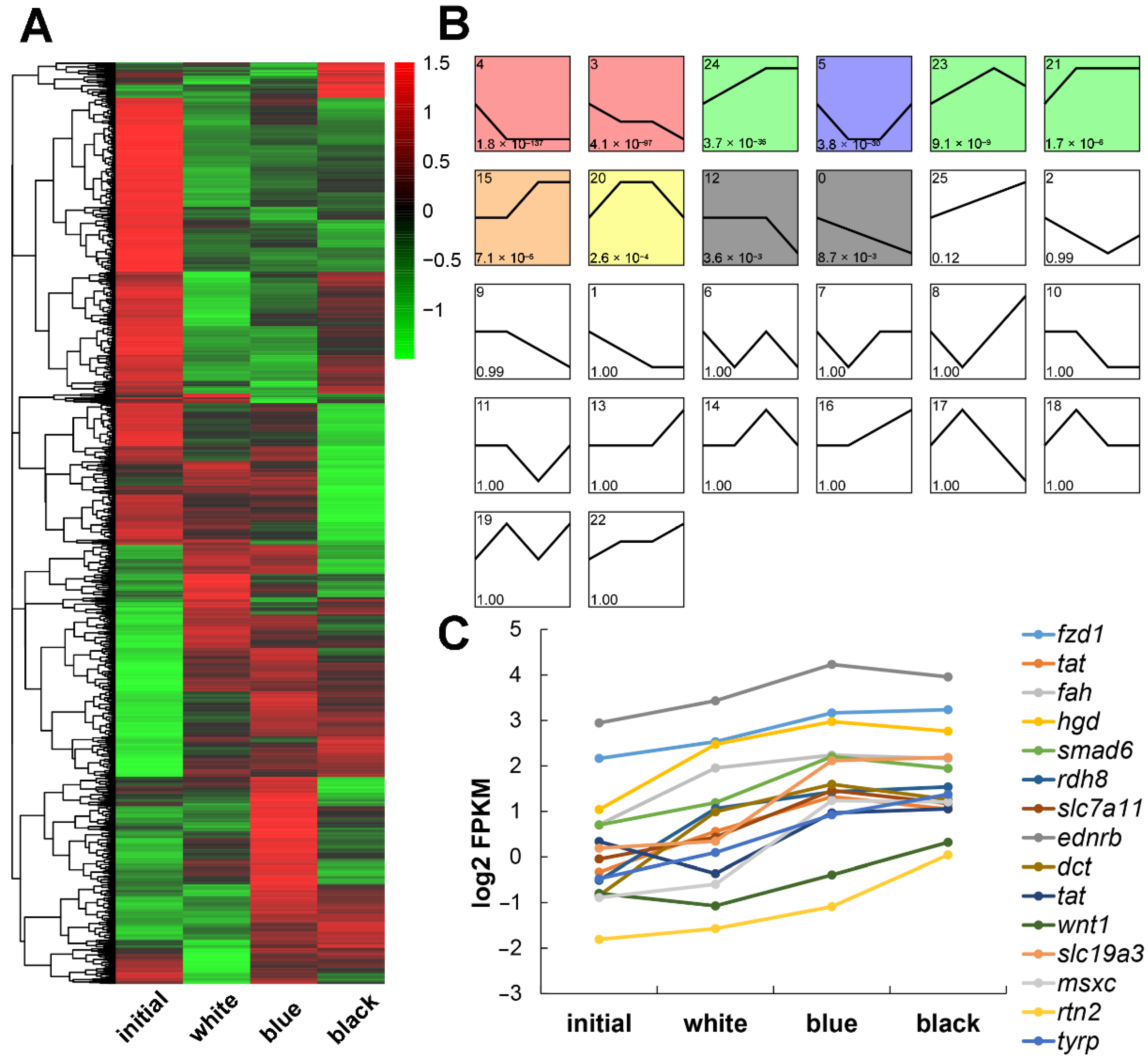
2.4. DEG Analysis among the Four Groups
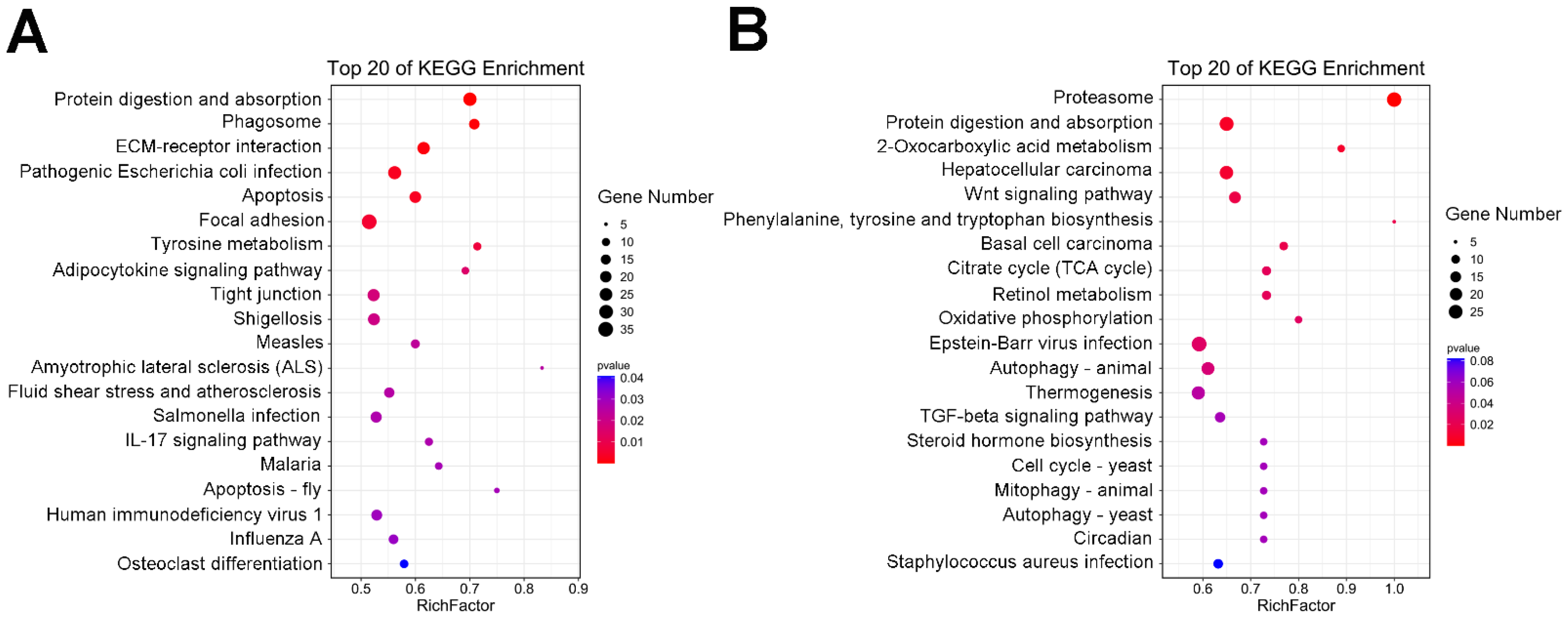
2.5. GO and KEGG Enrichment Analyses of the DEGs
2.6. qRT-PCR Verification
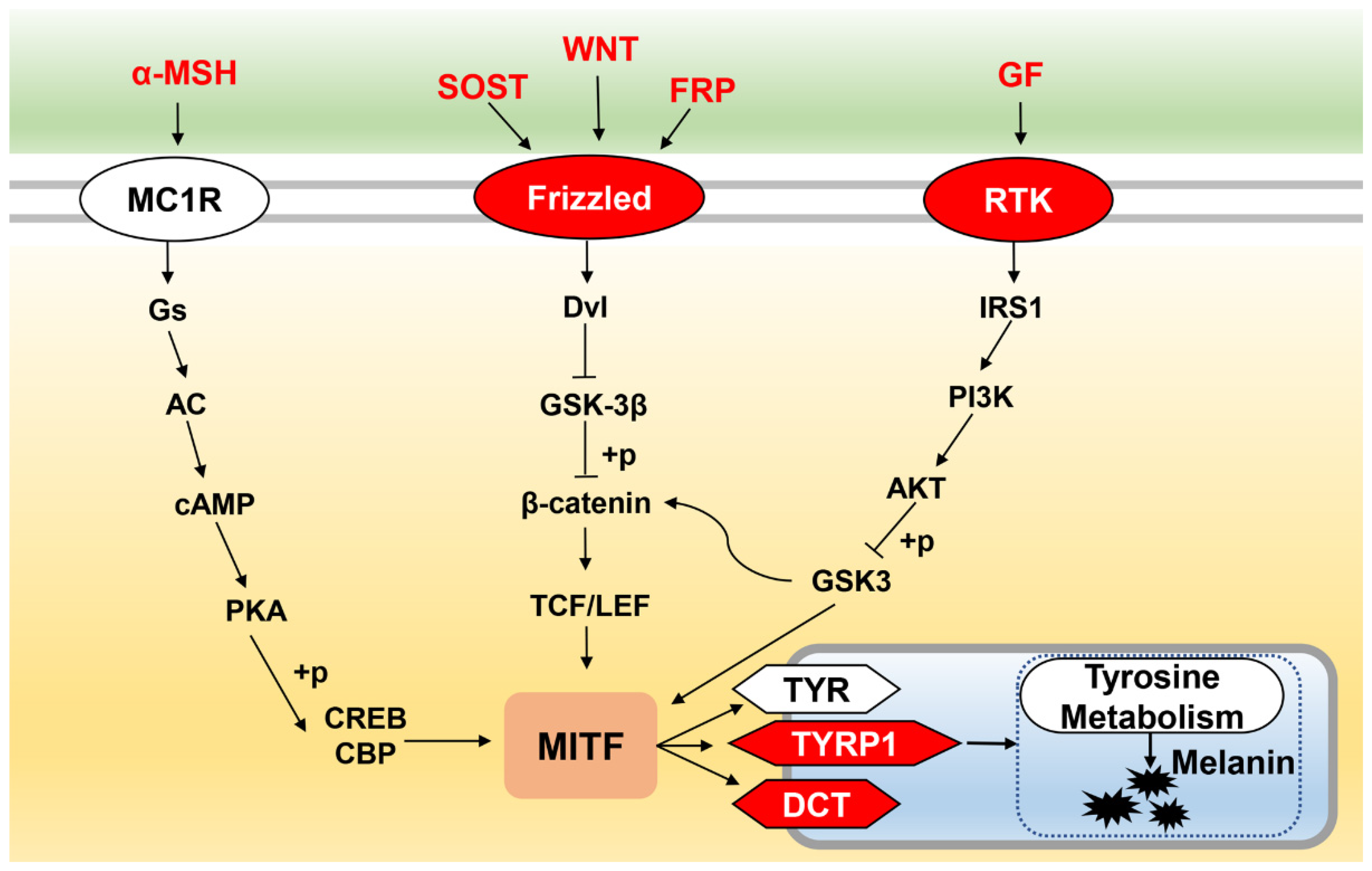
3. Discussion
4. Materials and Methods
4.1. Experiment, Fish Sampling, and Tissue Preparation
4.2. Total RNA Extraction and Qualification
4.3. cDNA Library Construction and Sequencing
4.4. Quality Control, Mapping, and Assembly
4.5. Differently Expressed Genes (DEGs) Analysis
4.6. Functional Annotation and Pathway Enrichment
4.7. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Validation
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, S.L.; Africa, D.; Walker, C.; Weston, J.A. Genetic control of adult pigment stripe development in zebrafish. Dev. Biol. 1995, 167, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, J.K.; Uy, J.A.C.; Hauber, M.E.; Hoekstra, H.E.; Safran, R.J. Vertebrate pigmentation: From underlying genes to adaptive function. Trends Genet. 2010, 26, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.R.; Islam, M.A.; Wahab, M.A.; Hoq, M.E.; Rahman, M.M.; Azim, M.E. Evaluation of production performance and profitability of hybrid red tilapia and genetically improved farmed tilapia (GIFT) strains in the carbon/nitrogen controlled periphyton-based (C/N-CP) on-farm prawn culture system in Bangladesh. Aquac. Rep. 2016, 4, 101–111. [Google Scholar] [CrossRef]
- Nüsslein-Volhard, C.; Singh, A.P. How fish color their skin: A paradigm for development and evolution of adult patterns: Multipotency, plasticity, and cell competition regulate proliferation and spreading of pigment cells in Zebrafish coloration. BioEssays 2017, 39, 1–11. [Google Scholar] [CrossRef]
- Cal, L.; Suarez-Bregua, P.; Cerdá-Reverter, J.M.; Braasch, I.; Rotllant, J. Fish pigmentation and the melanocortin system. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2017, 211, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.Y.; Huang, J.R.; Li, S.Z.; Lu, J.G. Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis. Aquaculture 2022, 547, 737429. [Google Scholar] [CrossRef]
- Sato, S.; Yamamoto, H. Development of pigment cells in the brain of ascidian tadpole larvae: Insights into the origins of vertebrate pigment cells. Pigment Cell Res. 2001, 14, 428–436. [Google Scholar] [CrossRef]
- McLean, E. Background color and cultured invertebrates-A review. Aquaculture 2021, 537, 736523. [Google Scholar] [CrossRef]
- McLean, E. Fish tank color: An overview. Aquaculture 2021, 530, 735750. [Google Scholar] [CrossRef]
- Fujii, R. The regulation of motile activity of fish chromatophores. Pigment Cell Res. 2000, 13, 300–319. [Google Scholar] [CrossRef]
- Sugimoto, M. Morphological color changes in fish: Regulation of pigment cell density and morphology. Microsc. Res. Tech. 2002, 58, 496–503. [Google Scholar] [CrossRef]
- Bertolesi, G.E.; Mcfarlane, S. Seeing the light to change colour: An evolutionary perspective on the role of melanopsin in neuroendocrine circuits regulating light-mediated skin pigmentation. Pigment Cell Melanoma Res. 2017, 31, 354–373. [Google Scholar] [CrossRef]
- Takahashi, A.; Mizusawa, K.; Amano, M. Multifunctional roles of melanocyte-stimulating hormone and melanin-concentrating hormone in fish: Evolution from classical body color change. Aqua-BioSci. Mono. 2014, 7, 1–46. [Google Scholar] [CrossRef]
- Mizusawa, K.; Kobayashi, Y.; Yamanome, T.; Saito, Y.; Takahashi, A. Interrelation between melanocyte-stimulating hormone and melanin-concentrating hormone in physiological body color change: Roles emerging from barfin flounder Verasper moseri. Gen. Comp. Endocrinol. 2013, 181, 229–234. [Google Scholar] [CrossRef]
- Mizusawa, K.; Kobayashi, Y.; Sunuma, T.; Asahida, T.; Saito, Y.; Takahashi, A. Inhibiting roles of melanin-concentrating hormone for skin pigment dispersion in barfin flounder, Verasper moseri. Gen. Comp. Endocrinol. 2011, 171, 75–81. [Google Scholar] [CrossRef]
- Cerdá-Reverter, J.M.; Canosa, L.F.; Peter, R.E. Regulation of the hypothalamic melanin-concentrating hormone neurons by sex steroids in the goldfish: Possible role in the modulation of luteinizing hormone secretion. Neuroendocrinology 2006, 84, 364–377. [Google Scholar] [CrossRef]
- Yang, T.; Kasagi, S.; Takahashi, A.; Mizusawa, K. Effects of background color and feeding status on the expression of genes associated with body color regulation in the goldfish Carassius auratus. Gen. Comp. Endocrinol. 2021, 312, 113860. [Google Scholar] [CrossRef]
- Kasagi, S.; Miura, M.; Okazaki, T.; Mizusawa, K.; Takahashi, A. Effects of tank color brightness on the body color, somatic growth, and endocrine systems of rainbow trout Oncorhynchus mykiss. Gen. Comp. Endocrinol. 2020, 298, 113581. [Google Scholar] [CrossRef]
- Wang, L.M.; Luo, M.K.; Yin, H.R.; Zhu, W.B.; Fu, J.J.; Dong, Z.J. Effects of background adaptation on the skin color of Malaysian red tilapia. Aquaculture 2020, 521, 735061. [Google Scholar] [CrossRef]
- Luo, M.K.; Lu, G.Q.; Yin, H.R.; Wang, L.M.; Atuganile, M.; Dong, Z.J. Fish pigmentation and coloration: Molecular mechanisms and aquaculture perspectives. Rev. Aquac. 2021, 13, 2395–2412. [Google Scholar] [CrossRef]
- Jiang, Y.L.; Zhang, S.H.; Xu, J.; Feng, J.X.; Mahboob, S.; Al-Ghanim, K.A.; Sun, X.W.; Xu, P. Comparative transcriptome analysis reveals the genetic basis of skin color variation in common carp. PLoS ONE 2014, 9, e108200. [Google Scholar] [CrossRef] [PubMed]
- Maoka, T.; Sato, W.; Nagai, H.; Takahashi, T. Carotenoids of red, brown, and black specimens of Plectropomus leopardus, the coral trout (Suziara in Japanese). J. Oleo Sci. 2017, 66, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Kelsh, R.N.; Inoue, C.; Momoi, A.; Kondoh, H.; Furutani, S.M.; Ozato, K.; Wakamatsu, Y. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech. Dev. 2004, 121, 841–859. [Google Scholar] [CrossRef]
- Parichy, D.M. Evolution of danio pigment pattern development. Heredity 2006, 97, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Maan, M.; Sefc, K.M. Colour variation in cichlid fish: Developmental mechanisms, selective pressures and evolutionary consequences. Semin Cell Dev. Biol. 2013, 24, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.H.; Ma, B.H.; Guo, X.L.; Wang, H.H.; Ma, A.J.; Sun, Z.B.; Wang, Q.M. Comparative transcriptome analysis of the molecular mechanism underlying the golden red colour in mutant Taiwanese loach. Aquaculture 2021, 543, 736979. [Google Scholar] [CrossRef]
- Hao, R.J.; Zhu, X.W.; Tian, C.X.; Jiang, M.Y.; Huang, Y.; Zhu, C.H. Integrated analysis of the role of miRNA-mRNA in determining different body colors of leopard coral grouper (Plectropomus leopardus). Aquaculture 2022, 548, 737575. [Google Scholar] [CrossRef]
- Mizusawa, K.; Kasagi, S.; Takahashi, K. Melanin-concentrating hormone is a major substance mediating light wavelength-dependent skin color change in larval zebrafish. Gen. Comp. Endocrinol. 2018, 269, 141–148. [Google Scholar] [CrossRef]
- Marmol, V.D.; Beermann, F. Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett. 1996, 381, 165–168. [Google Scholar] [CrossRef]
- Koga, A.; Inagaki, H.; Bessho, Y.; Hori, H. Insertion of a novel transposable element in the tyrosinase gene is responsible for an albino mutation in the medaka fish, Oryzias latipes. Mol. Gen. Genet. 1995, 249, 400–405. [Google Scholar] [CrossRef]
- Boonanuntanasarn, S.; Yoshizaki, G.; Iwai, K.; Takeuchi, T. Molecular cloning, gene expression in albino mutants and gene knockdown studies of tyrosinase mRNA in rainbow trout. Pigment Cell Res. 2004, 17, 413–421. [Google Scholar] [CrossRef]
- Yamanome, T.; Chiba, H.; Takahashi, A. Melanocyte-stimulating hormone facilitates hypermelanosis on the non-eyed side of the barfin flounder, a pleuronectiform fish. Aquaculture 2007, 270, 505–511. [Google Scholar] [CrossRef]
- Sato, H.; Tamba, M.; Ishii, T.; Bannai, S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 1999, 274, 11455–11458. [Google Scholar] [CrossRef]
- Verrey, F.; Closs, E.I.; Wagner, C.A.; Palacin, M.; Endou, H.; Kanai, Y. CATs and HATs: The SLC7 family of amino acid transporters. Pflugers Arch-Eur. J. Physiol. 2004, 447, 532–542. [Google Scholar] [CrossRef]
- Cerdá-Reverter, J.M.; Haitina, T.; Schiöth, H.B.; Peter, R.E. Gene structure of the goldfish agouti-signaling protein: A putative role in the dorsal-ventral pigment pattern of fish. Endocrinology 2005, 146, 1597–1610. [Google Scholar] [CrossRef]
- Wang, L.M.; Jiang, B.J.; Zhu, W.B.; Fu, J.J.; Luo, M.K.; Liu, W.; Dong, Z.J. The role of melanocortin 1 receptor on melanogenesis pathway in skin color differentiation of red tilapia. Aquac. Rep. 2022, 22, 100946. [Google Scholar] [CrossRef]
- Burgoyne, T.; O’Connor, M.N.; Seabra, M.C.; Cutler, D.F.; Futter, C.E. Regulation of melanosome number, shape and movement in the zebrafish retinal pigment epithelium by OA1 and PMEL. J. Cell Sci. 2015, 128, 1400–1407. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Livak, J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, F.; Wang, L.; Yang, Z.; Shi, L.; Zheng, D.; Zhang, K.; Sun, J.; Luo, J. Transcriptome Analysis Reveals the Complex Regulatory Pathway of Background Color in Juvenile Plectropomus leopardus Skin Color Variation. Int. J. Mol. Sci. 2022, 23, 11186. https://doi.org/10.3390/ijms231911186
Song F, Wang L, Yang Z, Shi L, Zheng D, Zhang K, Sun J, Luo J. Transcriptome Analysis Reveals the Complex Regulatory Pathway of Background Color in Juvenile Plectropomus leopardus Skin Color Variation. International Journal of Molecular Sciences. 2022; 23(19):11186. https://doi.org/10.3390/ijms231911186
Chicago/Turabian StyleSong, Feibiao, Lei Wang, Zihang Yang, Liping Shi, Da Zheng, Kaixi Zhang, Junlong Sun, and Jian Luo. 2022. "Transcriptome Analysis Reveals the Complex Regulatory Pathway of Background Color in Juvenile Plectropomus leopardus Skin Color Variation" International Journal of Molecular Sciences 23, no. 19: 11186. https://doi.org/10.3390/ijms231911186




