Functional and Expressional Analyses Reveal the Distinct Role of Complement Factor I in Regulating Complement System Activation during GCRV Infection in Ctenopharyngodon idella
Abstract
:1. Introduction
2. Results
2.1. Sequence Feature of CiCFI
2.2. The Predicted Domain Architecture and Three-Dimensional Structure Characteristic of CiCFI
2.3. The Phylogenetic Tree of CFIs
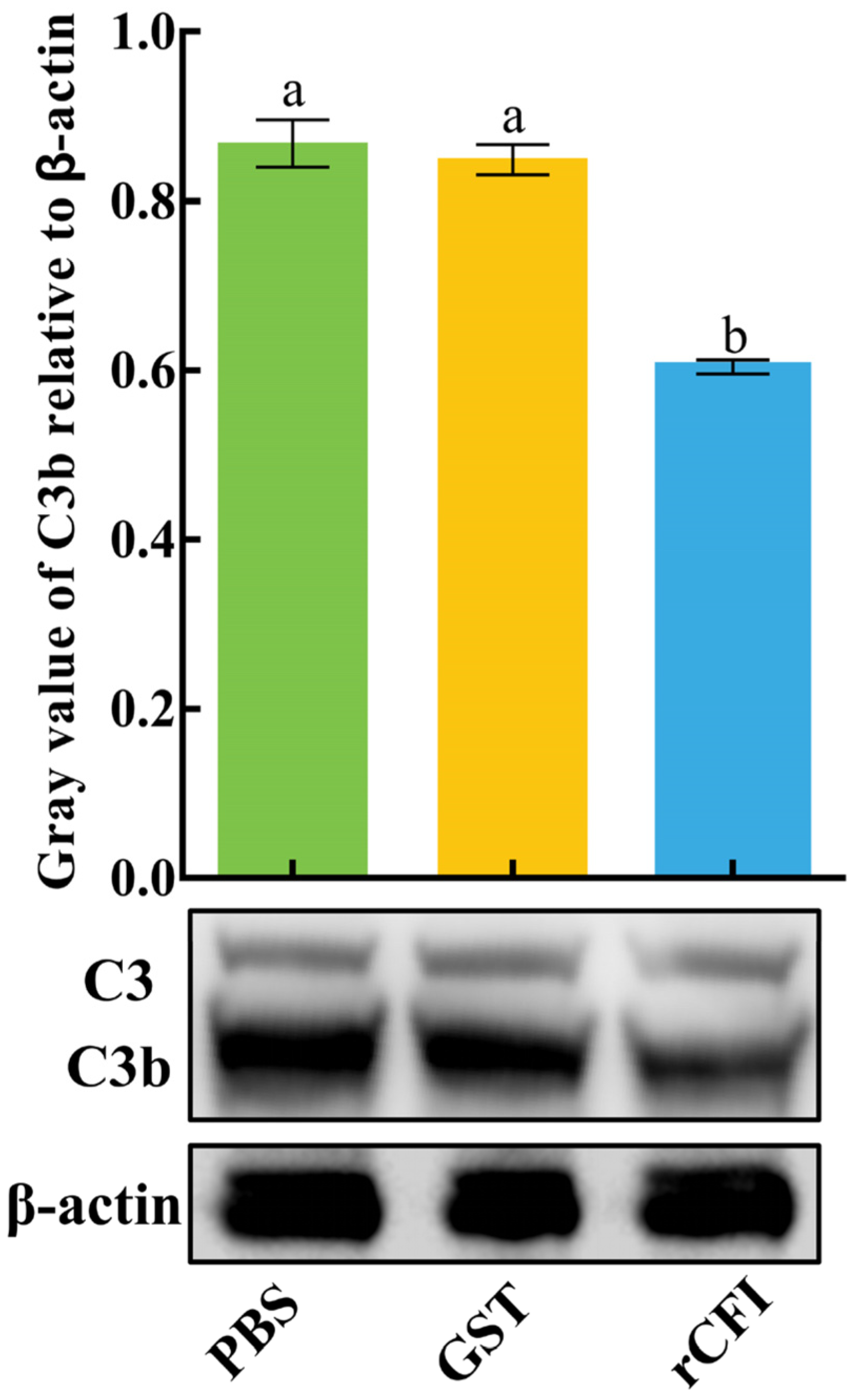
2.4. The Activity of CiCFI to Degrade CiC3b
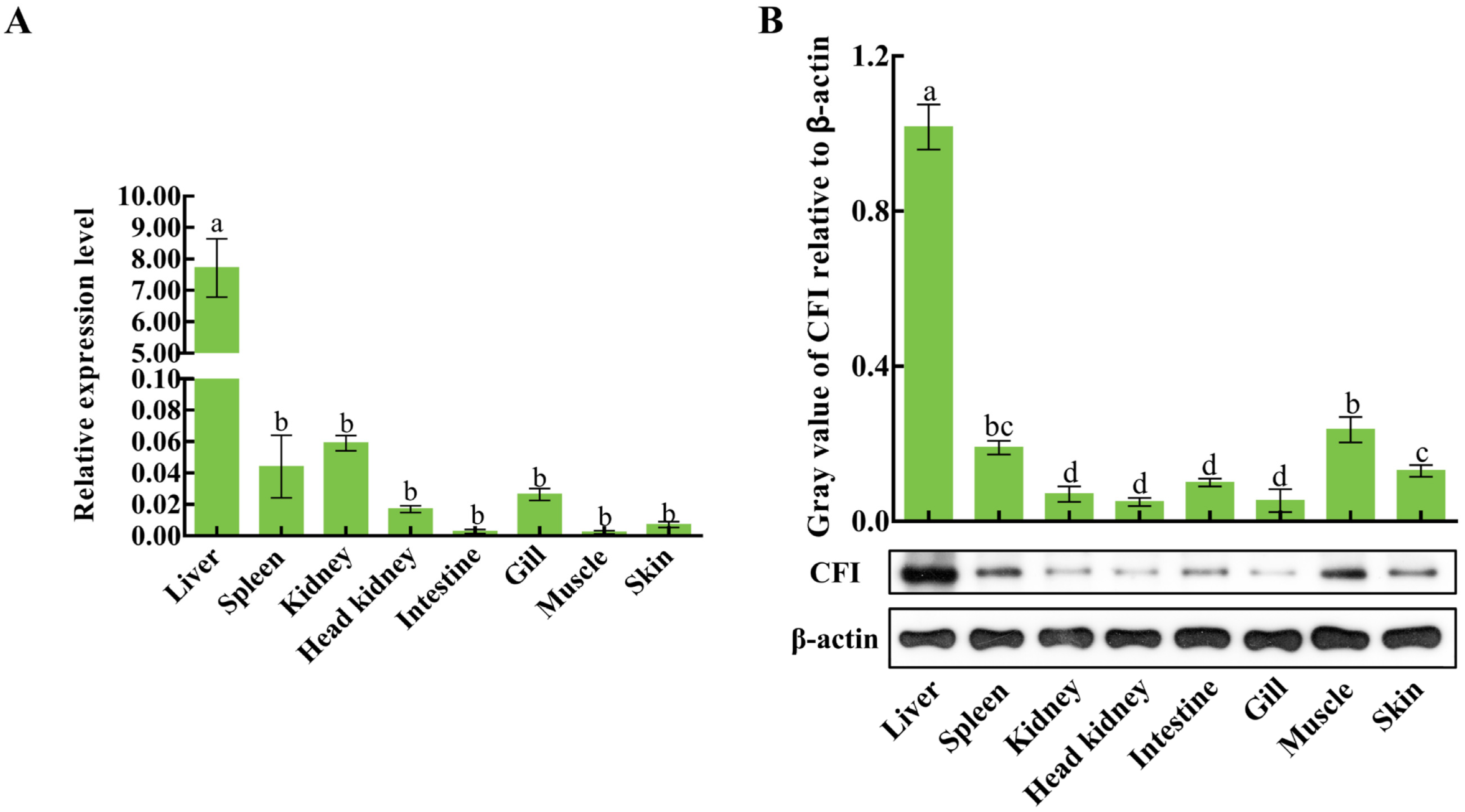
2.5. The Distributions of CiCFI mRNA Transcripts and Proteins in Grass Carp Tissues
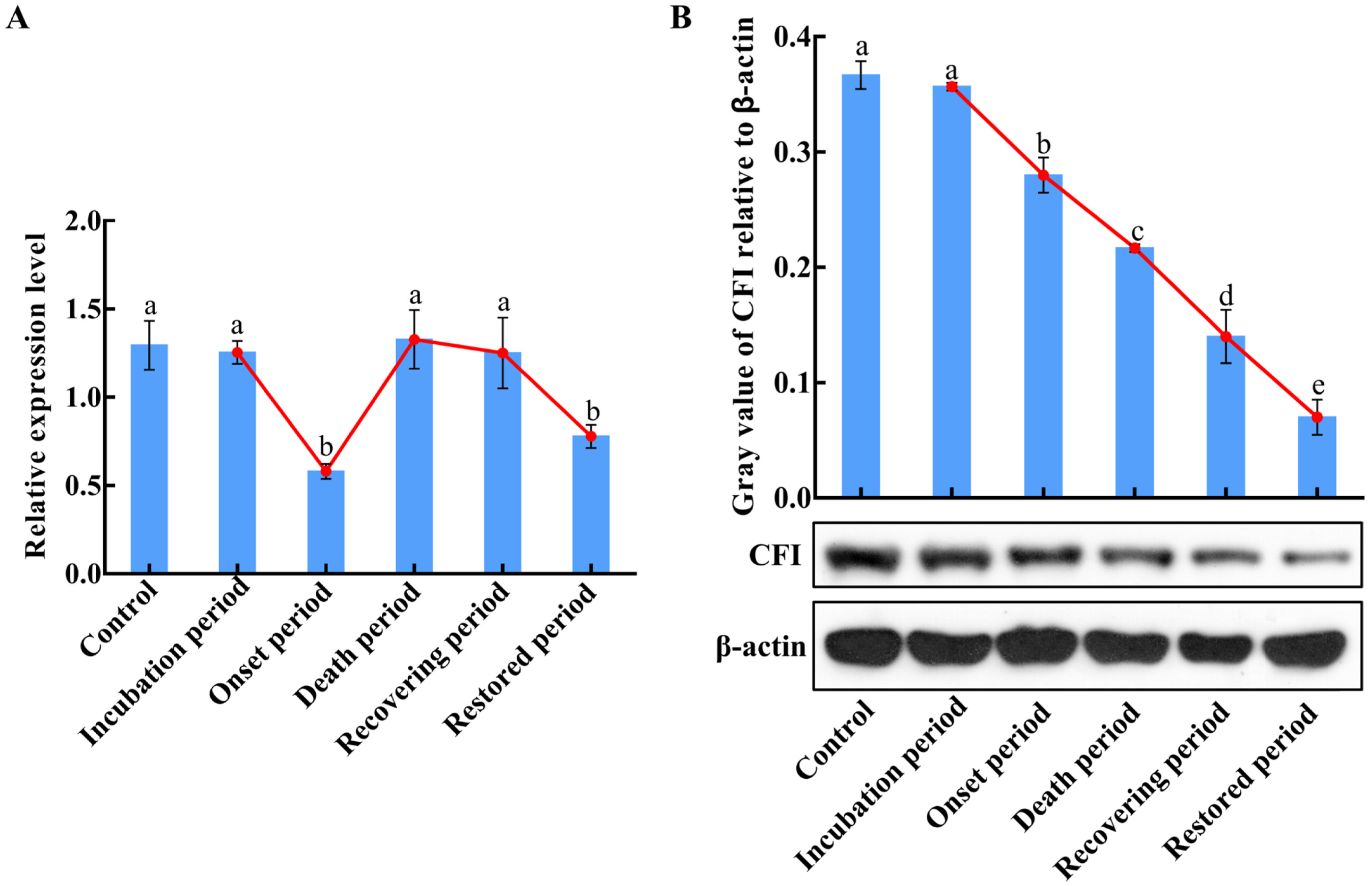
2.6. The mRNA and Protein Expression Changes of CiCFI in the Liver during GCRV Infection
2.7. The Fold Changes of CiCFI and CiC3b Proteins in the Serum during GCRV Infection
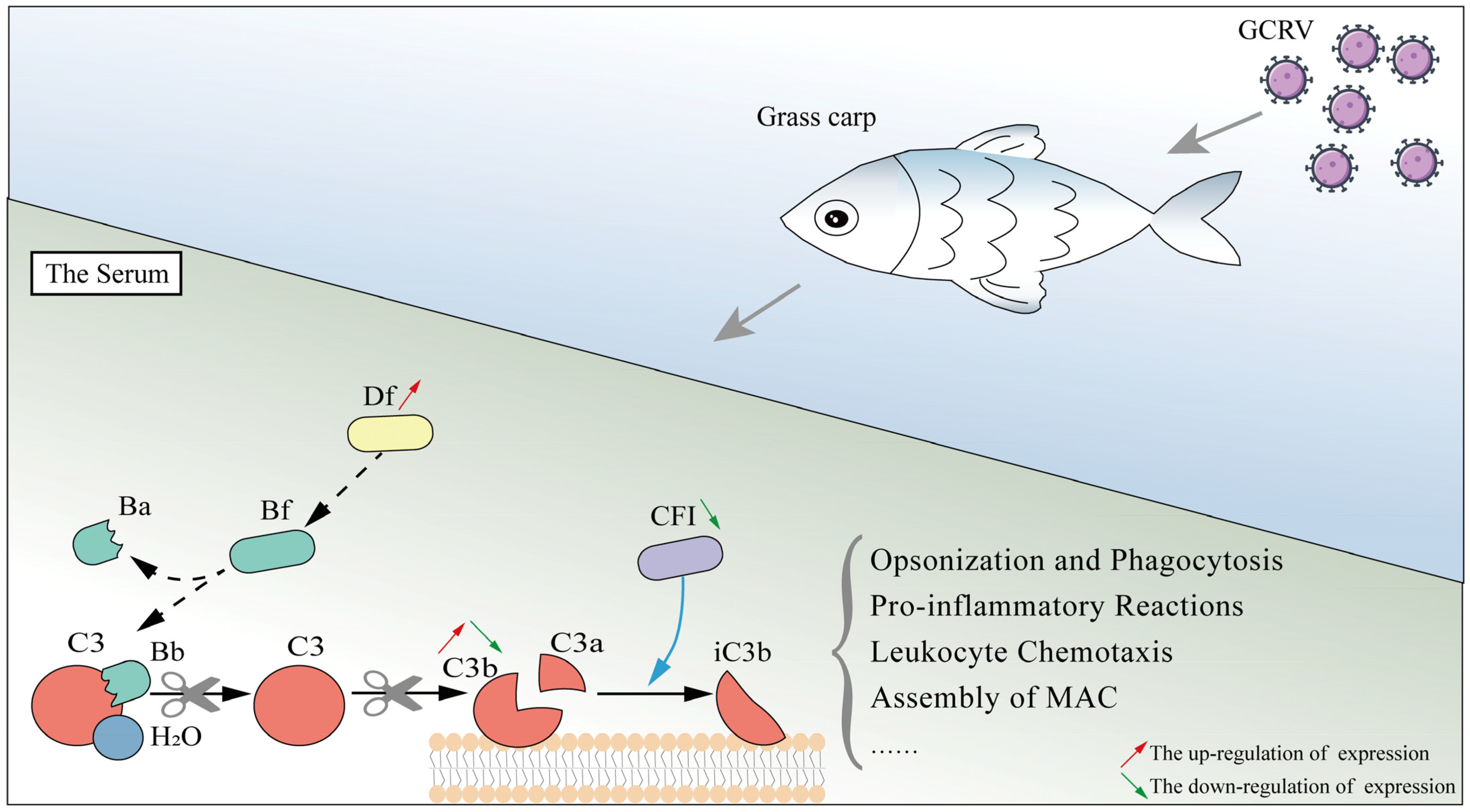
3. Discussion
4. Materials and Methods
4.1. Sequence Analysis of CiCFI
4.2. Structural and Phylogenetic Analysis of CiCFI
4.3. Prokaryotic Expression and Purification of Recombinant CiCFI Protein
4.4. Preparation and Validation of Polyclonal Antibodies
4.5. The Incubation of rCiCFI with Grass Carp Serum
4.6. The Expression Analysis of CiCFI mRNA and Protein Expression in Different Tissues
4.7. The Detection of CiCFI mRNA and Protein Level in the Liver after GCRV Infection
4.8. The Detection of CiCFI and CiC3b Protein Levels in the Serum after GCRV Infection
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarma, J.V.; Ward, P.A. The complement system. Cell Tissue Res. 2011, 343, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, R.A. Complement in health and disease. Hematol. Oncol. Clin. N. Am. 2015, 29, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, M. Evolution of the complement system. Subcell. Biochem. 2014, 80, 31–43. [Google Scholar] [PubMed]
- Conigliaro, P.; Triggianese, P.; Ballanti, E.; Perricone, C.; Perricone, R.; Chimenti, M.S. Complement, infection, and autoimmunity. Curr. Opin. Rheumatol. 2019, 31, 532–541. [Google Scholar] [CrossRef]
- Nakao, M.; Tsujikura, M.; Ichiki, S.; Vo, T.K.; Somamoto, T. The complement system in teleost fish: Progress of post-homolog-hunting researches. Dev. Comp. Immunol. 2011, 35, 1296–1308. [Google Scholar] [CrossRef]
- Merle, N.S.; Church, S.E.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement System Part I—Molecular Mechanisms of Activation and Regulation. Front. Immunol. 2015, 6, 262. [Google Scholar] [CrossRef]
- Pushpa, K.; Gireesh-Babu, P.; Rajendran, K.V.; Purushothaman, C.S.; Dasgupta, S.; Makesh, M. Molecular cloning, sequencing and tissue-level expression of complement C3 of Labeo rohita (Hamilton, 1822). Fish Shellfish Immunol. 2014, 40, 319–330. [Google Scholar] [CrossRef]
- Endo, Y.; Takahashi, M.; Fujita, T. Lectin complement system and pattern recognition. Immunobiology 2006, 211, 283–293. [Google Scholar] [CrossRef]
- Noris, M.; Remuzzi, G. Overview of complement activation and regulation. Semin. Nephrol. 2013, 33, 479–492. [Google Scholar] [CrossRef]
- Menger, M.; Aston, W.P. Isolation and characterization of factor I of the bovine complement system. Am. J. Vet. Res. 2003, 64, 989–993. [Google Scholar] [CrossRef]
- Jusko, M.; Potempa, J.; Kantyka, T.; Bielecka, E.; Miller, H.K.; Kalinska, M.; Dubin, G.; Garred, P.; Shaw, L.N.; Blom, A.M. Staphylococcal proteases aid in evasion of the human complement system. J. Innate Immun. 2014, 6, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.C.; Sim, R.B.; Lea, S.M.; Fremeaux-Bacchi, V.; Blom, A.M. Complement factor I in health and disease. Mol. Immunol. 2011, 48, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, P.J. The story of complement factor I. Immunobiology 2019, 224, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Fraczek, L.A.; Martin, B.K. Transcriptional control of genes for soluble complement cascade regulatory proteins. Mol. Immunol. 2010, 48, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Terado, T.; Nonaka, M.I.; Nonaka, M.; Kimura, H. Conservation of the modular structure of complement factor I through vertebrate evolution. Dev. Comp. Immunol. 2002, 26, 403–413. [Google Scholar] [CrossRef]
- Roversi, P.; Johnson, S.; Caesar, J.J.; McLean, F.; Leath, K.J.; Tsiftsoglou, S.A.; Morgan, B.P.; Harris, C.L.; Sim, R.B.; Lea, S.M. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proc. Natl. Acad. Sci. USA 2011, 108, 12839–12844. [Google Scholar] [CrossRef]
- Ricklin, D.; Hajishengallis, G.; Yang, K.; Lambris, J.D. Complement: A key system for immune surveillance and homeostasis. Nat. Immunol. 2010, 11, 785–797. [Google Scholar] [CrossRef]
- Wu, B.; Ouyang, Z.; Lyon, C.J.; Zhang, W.; Clift, T.; Bone, C.R.; Li, B.; Zhao, Z.; Kimata, J.T.; Yu, X.G.; et al. Plasma Levels of Complement Factor I and C4b Peptides Are Associated with HIV Suppression. ACS Infect. Dis. 2017, 3, 880–885. [Google Scholar] [CrossRef]
- Densen, P. Complement deficiencies and meningococcal disease. Clin. Exp. Immunol. 1991, 86, 57–62. [Google Scholar] [CrossRef]
- Cunnion, K.M.; Buescher, E.S.; Hair, P.S. Serum complement factor I decreases Staphylococcus aureus phagocytosis. J. Lab. Clin. Med. 2005, 146, 279–286. [Google Scholar] [CrossRef]
- Alosaimi, B.; Mubarak, A.; Hamed, M.E.; Almutairi, A.Z.; Alrashed, A.A.; AlJuryyan, A.; Enani, M.; Alenzi, F.Q.; Alturaiki, W. Complement Anaphylatoxins and Inflammatory Cytokines as Prognostic Markers for COVID-19 Severity and In-Hospital Mortality. Front. Immunol. 2021, 12, 668725. [Google Scholar] [CrossRef] [PubMed]
- Abernathy, J.W.; Lu, J.; Liu, H.; Kucuktas, H.; Liu, Z. Molecular characterization of complement factor I reveals constitutive expression in channel catfish. Fish Shellfish Immunol. 2009, 27, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Gong, Q.; Wen, Z.; Yuan, D. Molecular characterization and expression of complement factor I in Pelteobagrus vachellii during Aeromonas hydrophila infection. Dev. Comp. Immunol. 2018, 82, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Li, X.; Chen, Y.; Lu, Y.; Yu, M.; Chen, X.; Zhang, W.; Zeng, Y.; Sun, L.; Chen, S.; et al. Complement factor I from flatfish half-smooth tongue (Cynoglossus semilaevis) exhibited anti-microbial activities. Dev. Comp. Immunol. 2015, 53, 199–209. [Google Scholar] [CrossRef]
- Anastasiou, V.; Mikrou, A.; Papanastasiou, A.D.; Zarkadis, I.K. The molecular identification of factor H and factor I molecules in rainbow trout provides insights into complement C3 regulation. Fish Shellfish Immunol. 2011, 31, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.B.; Jin, C.D.; Li, M.F. The trypsin-like serine protease domain of Paralichthys olivaceus complement factor I regulates complement activation and inhibits bacterial growth. Fish Shellfish Immunol. 2020, 97, 18–26. [Google Scholar] [CrossRef]
- Nakao, M.; Hisamatsu, S.; Nakahara, M.; Kato, Y.; Smith, S.L.; Yano, T. Molecular cloning of the complement regulatory factor I isotypes from the common carp (Cyprinus carpio). Immunogenetics 2003, 54, 801–806. [Google Scholar] [CrossRef]
- Fisheries Bureau of Ministry of Agriculture in China. China Fishery Statistical Yearbook of 2021; China Agriculture Press: Beijing, China, 2021.
- He, L.; Zhang, A.; Xiong, L.; Li, Y.; Huang, R.; Liao, L.; Zhu, Z.; Wang, A.Y. Deep Circular RNA Sequencing Provides Insights into the Mechanism Underlying Grass Carp Reovirus Infection. Int. J. Mol. Sci. 2017, 18, 1977. [Google Scholar] [CrossRef]
- Wang, H.; Ding, C.; Wang, J.; Zhao, X.; Jin, S.; Liang, J.; Luo, H.; Li, D.; Li, R.; Li, Y.; et al. Molecular cloning and expression analysis of coagulation factor VIII and plasminogen involved in immune response to GCRV, and immunity activity comparison of grass carp Ctenopharyngodon idella with different viral resistance. Fish Shellfish Immunol. 2019, 86, 794–804. [Google Scholar] [CrossRef]
- Roh, H.; Kim, A.; Kim, N.; Lee, Y.; Kim, D.H. Multi-Omics Analysis Provides Novel Insight into Immuno-Physiological Pathways and Development of Thermal Resistance in Rainbow Trout Exposed to Acute Thermal Stress. Int. J. Mol. Sci. 2020, 21, 9198. [Google Scholar] [CrossRef]
- Chu, P.; He, L.; Huang, R.; Liao, L.; Li, Y.; Zhu, Z.; Hu, W.; Wang, Y. Autophagy Inhibits Grass Carp Reovirus (GCRV) Replication and Protects Ctenopharyngodon idella Kidney (CIK) Cells from Excessive Inflammatory Responses after GCRV Infection. Biomolecules 2020, 10, 1296. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.H.; Zhong, L.; Liu, Q.L.; Xiao, T.Y.; Su, J.M.; Chen, K.J.; Wang, H.Q.; Dai, Y.J.; Chen, J. Characterization of grass carp spleen transcriptome during GCRV infection. Genet. Mol. Res. 2016, 15, gmr6650. [Google Scholar] [CrossRef] [PubMed]
- Volanakis, J.E.; Narayana, S.V. Complement factor D, a novel serine protease. Protein Sci. 1996, 5, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.H.; Xiao, T.Y.; Qin, B.B.; Xu, B.H.; Lv, Z.; Wang, H.Q. Functional Identification of Complement Factor D and Analysis of Its Expression during GCRV Infection in Grass Carp (Ctenopharyngodon idella). Int. J. Mol. Sci. 2021, 22, 12011. [Google Scholar] [CrossRef]
- Cheema, N.; Herbst, A.; McKenzie, D.; Aiken, J.M. Apoptosis and necrosis mediate skeletal muscle fiber loss in age-induced mitochondrial enzymatic abnormalities. Aging Cell 2015, 14, 1085–1093. [Google Scholar] [CrossRef]
- Martin, M.; Leffler, J.; Smolag, K.I.; Mytych, J.; Bjork, A.; Chaves, L.D.; Alexander, J.J.; Quigg, R.J.; Blom, A.M. Factor H uptake regulates intracellular C3 activation during apoptosis and decreases the inflammatory potential of nucleosomes. Cell Death Differ. 2016, 23, 903–911. [Google Scholar] [CrossRef]
- Kraiczy, P.; Skerka, C.; Zipfel, P.F.; Brade, V. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: A new protein family involved in complement resistance. Wien. Klin. Wochenschr. 2002, 114, 568–573. [Google Scholar]
- Giang, J.; Seelen, M.A.J.; van Doorn, M.B.A.; Rissmann, R.; Prens, E.P.; Damman, J. Complement Activation in Inflammatory Skin Diseases. Front. Immunol. 2018, 9, 639. [Google Scholar] [CrossRef]
- Mödinger, Y.; Rapp, A.; Pazmandi, J.; Vikman, A.; Holzmann, K.; Haffner-Luntzer, M.; Huber-Lang, M.; Ignatius, A. C5aR1 interacts with TLR2 in osteoblasts and stimulates the osteoclast-inducing chemokine CXCL10. J. Cell Mol. Med. 2018, 22, 6002–6014. [Google Scholar] [CrossRef]
- Corvillo, F.; Okrój, M.; Nozal, P.; Melgosa, M.; Sánchez-Corral, P.; López-Trascasa, M. Nephritic Factors: An Overview of Classification, Diagnostic Tools and Clinical Associations. Front. Immunol. 2019, 10, 886. [Google Scholar] [CrossRef]
- Inafuku, S.; Klokman, G.; Connor, K.M. The Alternative Complement System Mediates Cell Death in Retinal Ischemia Reperfusion Injury. Front. Mol. Neurosci. 2018, 11, 278. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.C.; Shih, C.S.; Dachenhausen, S.G. Coelomocytes express SpBf, a homologue of factor B, the second component in the sea urchin complement system. J. Immunol. 1998, 161, 6784–6793. [Google Scholar]
- Ai, S.; Zheng, J.; Qiu, C.X.; Lu, X.L.; Li, X.W. Urinary proteomics analysis based on mass spectrometry and identification of therapeutic targets of Shenkangling interventions in rats with adriamycin nephropathy using iTRAQ. Am. J. Transl. Res. 2018, 10, 2115–2125. [Google Scholar]
- Tsuru, H.; Osaka, M.; Hiraoka, Y.; Yoshida, M. HFD-induced hepatic lipid accumulation and inflammation are decreased in Factor D deficient mouse. Sci. Rep. 2020, 10, 17593. [Google Scholar] [CrossRef] [PubMed]
- Holland, M.C.; Lambris, J.D. The complement system in teleosts. Fish Shellfish Immunol. 2002, 12, 399–420. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Ma, A.; Chi, X.; Li, Q.; Pang, Y.; Su, P. A novel complement factor I involving in the complement system immune response from Lampetra morii. Fish Shellfish Immunol. 2020, 98, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.C.; Nita, I.; Mansson, L.; Groeneveld, T.W.; Trouw, L.A.; Villoutreix, B.O.; Blom, A.M. Analysis of binding sites on complement factor I that are required for its activity. J. Biol. Chem. 2010, 285, 6235–6245. [Google Scholar] [CrossRef]
- Tsiftsoglou, S.A.; Willis, A.C.; Li, P.; Chen, X.; Mitchell, D.A.; Rao, Z.; Sim, R.B. The catalytically active serine protease domain of human complement factor I. Biochemistry 2005, 44, 6239–6249. [Google Scholar] [CrossRef]
- Geerlings, M.J.; Volokhina, E.B.; de Jong, E.K.; van de Kar, N.; Pauper, M.; Hoyng, C.B.; van den Heuvel, L.P.; den Hollander, A.I. Genotype-phenotype correlations of low-frequency variants in the complement system in renal disease and age-related macular degeneration. Clin. Genet. 2018, 94, 330–338. [Google Scholar] [CrossRef]
- Ullman, C.G.; Haris, P.I.; Smith, K.F.; Sim, R.B.; Emery, V.C.; Perkins, S.J. Beta-sheet secondary structure of an LDL receptor domain from complement factor I by consensus structure predictions and spectroscopy. FEBS Lett. 1995, 371, 199–203. [Google Scholar] [CrossRef]
- Ullman, C.G.; Perkins, S.J. The Factor I and follistatin domain families: The return of a prodigal son. Biochem. J. 1997, 326, 939–941. [Google Scholar] [CrossRef] [PubMed]
- Haefliger, J.A.; Tschopp, J.; Vial, N.; Jenne, D.E. Complete primary structure and functional characterization of the sixth component of the human complement system. Identification of the C5b-binding domain in complement C6. J. Biol. Chem. 1989, 264, 18041–18051. [Google Scholar] [CrossRef]
- Lambris, J.D.; Lao, Z.; Oglesby, T.J.; Atkinson, J.P.; Hack, C.E.; Becherer, J.D. Dissection of CR1, factor H, membrane cofactor protein, and factor B binding and functional sites in the third complement component. J. Immunol. 1996, 156, 4821–4832. [Google Scholar] [PubMed]
- Ricklin, D.; Reis, E.S.; Mastellos, D.C.; Gros, P.; Lambris, J.D. Complement component C3—The “Swiss Army Knife” of innate immunity and host defense. Immunol. Rev. 2016, 274, 33–58. [Google Scholar] [CrossRef]
- Janssen, B.J.; Huizinga, E.G.; Raaijmakers, H.C.; Roos, A.; Daha, M.R.; Nilsson-Ekdahl, K.; Nilsson, B.; Gros, P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 2005, 437, 505–511. [Google Scholar] [CrossRef]
- Qiu, W.Q.; Luo, S.; Ma, S.A.; Saminathan, P.; Li, H.; Gunnersen, J.M.; Gelbard, H.A.; Hammond, J.W. The Sez6 Family Inhibits Complement by Facilitating Factor I Cleavage of C3b and Accelerating the Decay of C3 Convertases. Front. Immunol. 2021, 12, 607641. [Google Scholar] [CrossRef]
- Xue, X.; Wu, J.; Ricklin, D.; Forneris, F.; Di Crescenzio, P.; Schmidt, C.Q.; Granneman, J.; Sharp, T.H.; Lambris, J.D.; Gros, P. Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses. Nat. Struct. Mol. Biol. 2017, 24, 643–651. [Google Scholar] [CrossRef]
- Nakao, M.; Miura, C.; Itoh, S.; Nakahara, M.; Okumura, K.; Mutsuro, J.; Yano, T. A complement C3 fragment equivalent to mammalian C3d from the common carp (Cyprinus carpio): Generation in serum after activation of the alternative pathway and detection of its receptor on the lymphocyte surface. Fish Shellfish Immunol. 2004, 16, 139–149. [Google Scholar] [CrossRef]
- Morris, K.M.; Aden, D.P.; Knowles, B.B.; Colten, H.R. Complement biosynthesis by the human hepatoma-derived cell line HepG2. J. Clin. Investig. 1982, 70, 906–913. [Google Scholar] [CrossRef]
- Kunnath-Muglia, L.M.; Chang, G.H.; Sim, R.B.; Day, A.J.; Ezekowitz, R.A. Characterization of Xenopus laevis complement factor I structure—Conservation of modular structure except for an unusual insert not present in human factor I. Mol. Immunol. 1993, 30, 1249–1256. [Google Scholar] [CrossRef]
- Shin, D.H.; Webb, B.M.; Nakao, M.; Smith, S.L. Characterization of shark complement factor I gene(s): Genomic analysis of a novel shark-specific sequence. Mol. Immunol. 2009, 46, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Schlaf, G.; Rothermel, E.; Oppermann, M.; Schieferdecker, H.L.; Jungermann, K.; Götze, O. Rat complement factor I: Molecular cloning, sequencing and expression in tissues and isolated cells. Immunology 1999, 98, 464–474. [Google Scholar] [CrossRef]
- Chi, H.; Zhou, K.; Shen, L.; Xu, J.; Li, J.; Chen, S.; Wu, X.; Tung, T.H.; Shen, B.; Zhu, H. The evaluation of the immune status of COVID-19 recovered subjects with persistent abnormal lung CT after one year: A longitudinal cohort study. Int. Immunopharmacol. 2022, 110, 109019. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, R.L. The complement system: Its importance in the host response to viral infection. Microbiol. Rev. 1982, 46, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Peng, Q.; Song, S.; Shi, D.; Zhang, X.; Guo, W.; Li, Y.; Zhou, J.; Zhu, X.; Zhao, Y.; et al. Nonstructural Protein 1 of Variant PEDV Plays a Key Role in Escaping Replication Restriction by Complement C3. J. Virol. 2022, e01024-22. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.H.; Lv, L.G.; Xiao, T.Y.; Liu, Q.L.; Su, H.; Liu, Y.; Ni, J.J. Cloning of the full-length cDNA of the gene encoding complement C5 from grass carp (Ctenopharyngodon idella) and its expression in different tissues by following grass carp reovirus infection. Aquacult. Int. 2021, 29, 2035–2048. [Google Scholar] [CrossRef]
- Zimmer, J.; Hobkirk, J.; Mohamed, F.; Browning, M.J.; Stover, C.M. On the Functional Overlap between Complement and Anti-Microbial Peptides. Front. Immunol. 2015, 5, 689. [Google Scholar] [CrossRef]
- Meng, X.; Shen, Y.; Wang, S.; Xu, X.; Dang, Y.; Zhang, M.; Li, L.; Zhang, J.; Wang, R.; Li, J. Complement component 3 (C3): An important role in grass carp (Ctenopharyngodon idella) experimentally exposed to Aeromonas hydrophila. Fish Shellfish Immunol. 2019, 88, 189–197. [Google Scholar] [CrossRef]
- de Vries, S.P.; Bootsma, H.J. Differential gene expression of Moraxella catarrhalis upon exposure to human serum. Genom. Data 2014, 2, 312–313. [Google Scholar] [CrossRef]
- Del Tordello, E.; Vacca, I.; Ram, S.; Rappuoli, R.; Serruto, D. Neisseria meningitidis NaIP cleaves human complement C3, facilitating degradation of C3b and survival in human serum. Proc. Natl. Acad. Sci. USA 2014, 111, 427–432. [Google Scholar] [CrossRef]
- Chen, D.D.; Li, J.H.; Yao, Y.Y.; Zhang, Y.A. Aeromonas hydrophila suppresses complement pathways via degradation of complement C3 in bony fish by metalloprotease. Fish Shellfish Immunol. 2019, 94, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Coulthard, L.G.; Woodruff, T.M. Is the Complement Activation Product C3a a Proinflammatory Molecule? Re-evaluating the Evidence and the Myth. J. Immunol. 2015, 194, 3542–3548. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.M.; Cabezas-Falcon, S.; Dubowsky, J.G.; Hulme-Jones, J.; Gordon, D.L. Dengue virus and the complement alternative pathway. FEBS Lett. 2020, 594, 2543–2555. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Xiao, T.; Jin, S.; Wang, J.; Wang, J.; Luo, H.; Li, R.; Sun, T.; Zou, J.; Li, Y. Characterization and immune function of the interferon-β promoter stimulator-1 in the barbel chub, Squaliobarbus curriculus. Dev. Comp. Immunol. 2020, 104, 103571. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Zhang, D.; Jiang, Q.; Sun, R.; Wang, H.; Zhang, H.; Song, L. A novel multi-domain C1qDC protein from Zhikong scallop Chlamys farreri provides new insights into the function of invertebrate C1qDC proteins. Dev. Comp. Immunol. 2015, 52, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






| Species | Accession Number |
|---|---|
| Lethenteron camtschaticum | BAG66070.1 |
| Homo sapiens | NP_000195.2 |
| Pongo abelii | XP_024101436.1 |
| Bos taurus | NP_001033185.1 |
| Mus musculus | NP_031712.2 |
| Rattus norvegicus | NP_077071.1 |
| Gallus gallus | NP_001258947.1 |
| Xenopus laevis | NP_001079421.1 |
| Chiloscyllium plagiosum | AHA61785.1 |
| Ginglymostoma cirratum | ABV21980.1 |
| Oncorhynchus mykiss | XP_021449058.1 |
| Esox lucius | XP_028973634.1 |
| Oryzias latipes | XP_004079594.1 |
| Cynoglossus semilaevis | AKN79751.1 |
| Echeneis naucrate | XP_029381915.1 |
| Monopterus albus | XP_020477827.1 |
| Oreochromis niloticus | XP_005459189.1 |
| Anabas testudineus | XP_026225021.1 |
| Perca flavescens | XP_028420661.1 |
| Ictalurus punctatus | ACV87004.1 |
| Tachysurus vachellii | ATC38299.1 |
| Tachysurus fulvidraco | XP_027018314.1 |
| Electrophorus electricus | XP_026852291.1 |
| Cyprinus carpio | BAB88920.1/BAB88921.1 |
| Carassius gibelio | AGU16535.1 |
| Labeo rohita | RXN08433.1 |
| Danio rerio | XP_017209949.1 |
| Anabarilius grahami | ROL01506.1 |
| Primer Name | Primer Sequence 5′-3′ | Usage | Accession |
|---|---|---|---|
| CFI-IF-F | GATCTGGTTCCGCGTGGATCCCTGAAGGAACTATCAGAGCT | CDS amplification | HM776035.1 |
| CFI-IF-R | CTCGAGTCGACCCGGGAATTCCTGGTTATATTTGGTTACAG | CDS amplification | |
| CFI-F | CACATAACGTACTATTGGCAAC | qPCR | |
| CFI-R | CCGACATTGAGTGATGACCA | qPCR | |
| β-actin-F | GCTATGTGGCTCTTGACTTCG | qPCR | M25013.1 |
| β-actin-R | GGGCACCTGAACCTCTCATT | qPCR | |
| 18S rRNA-F | ATTTCCGACACGGAGAGG | qPCR | EU047719.1 |
| 18S rRNA-R | CATGGGTTTAGGATACGCTC | qPCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Lv, Z.; Xiao, T.; Zhang, X.; Ding, C.; Qin, B.; Xu, B.; Liu, Q. Functional and Expressional Analyses Reveal the Distinct Role of Complement Factor I in Regulating Complement System Activation during GCRV Infection in Ctenopharyngodon idella. Int. J. Mol. Sci. 2022, 23, 11369. https://doi.org/10.3390/ijms231911369
Liu Y, Lv Z, Xiao T, Zhang X, Ding C, Qin B, Xu B, Liu Q. Functional and Expressional Analyses Reveal the Distinct Role of Complement Factor I in Regulating Complement System Activation during GCRV Infection in Ctenopharyngodon idella. International Journal of Molecular Sciences. 2022; 23(19):11369. https://doi.org/10.3390/ijms231911369
Chicago/Turabian StyleLiu, Yi, Zhao Lv, Tiaoyi Xiao, Xuewen Zhang, Chunhua Ding, Beibei Qin, Baohong Xu, and Qiaolin Liu. 2022. "Functional and Expressional Analyses Reveal the Distinct Role of Complement Factor I in Regulating Complement System Activation during GCRV Infection in Ctenopharyngodon idella" International Journal of Molecular Sciences 23, no. 19: 11369. https://doi.org/10.3390/ijms231911369





