Karyotype Analysis, Genomic and Fluorescence In Situ Hybridization (GISH and FISH) Reveal the Ploidy and Parental Origin of Chromosomes in Paeonia Itoh Hybrids
Abstract
:1. Introduction
2. Results
2.1. Genome Size and Ploidy Analysis of Different Materials in Paeonia L.
2.2. Karyotype Analysis of Peonies among Different Taxa
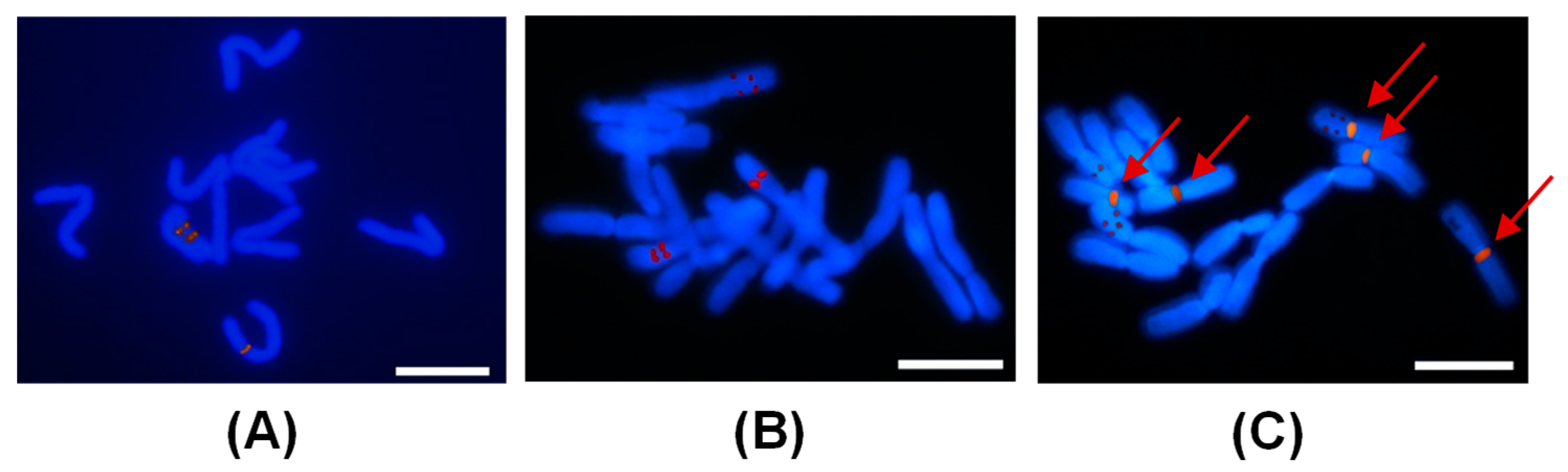
2.3. GISH Analysis of Peonies among Different Taxa
2.4. FISH Analysis of Peonies among Different Taxa
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Flow Cytometric Measurement
4.3. Karyotype Analysis
4.4. GISH and FISH
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Zhang, X.; Zhao, X. Tree Peony of China; Encyclopedia of China Publishing House: Beijing, China, 2011. [Google Scholar]
- Kamenetsky-Goldstein, R.; Yu, X. Cut peony industry: The first 30 years of research and new horizons. Hortic. Res. 2022, 9, uhac079. [Google Scholar] [CrossRef] [PubMed]
- Marković, T.; Prijić, Ž.; Xue, J.; Zhang, X.; Radanović, D.; Ren, X.; Filipović, V.; Lukić, M.; Gordanić, S. The seed traits associated with dormancy and germination of herbaceous peonies, focusing on species Native in Serbia and China. Horticulturae 2022, 8, 585. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, M.; Li, S.; Chen, Q.; da Silva, J.A.T.; Wang, A.; Yu, X.; Wang, L. Germplasm resources and genetic breeding of Paeonia: A systematic review. Hortic. Res. 2020, 7, 107. [Google Scholar] [CrossRef] [PubMed]
- Page, M. Paeonia Itoh hybrids. Plantsman 2005, 4, 36–39. [Google Scholar]
- te Beest, M.; Le Roux, J.J.; Richardson, D.M.; Brysting, A.K.; Suda, J.; Kubesova, M.; Pysek, P. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 2012, 109, 19–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Sreedasyam, A.; Ando, A.; Song, Q.; De Santiago, L.M.; Hulse-Kemp, A.M.; Ding, M.; Ye, W.; Kirkbride, R.C.; Jenkins, J.; et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020, 52, 525–533. [Google Scholar] [CrossRef]
- Hoopes, G.; Meng, X.; Hamilton, J.P.; Achakkagari, S.R.; Guesdes, F.d.A.F.; Bolger, M.E.; Coombs, J.J.; Esselink, D.; Kaiser, N.R.; Kodde, L.; et al. Phased, chromosome-scale genome assemblies of tetraploid potato reveal a complex genome, transcriptome, and predicted proteome landscape underpinning genetic diversity. Mol. Plant 2022, 15, 520–536. [Google Scholar] [CrossRef]
- Awan, M.J.A.; Rasheed, A.; Saeed, N.A.; Mansoor, S. Aegilops tauschi presents a genetic roadmap for hexaploid wheat improvement. Trends Genet. 2022, 38, 307–309. [Google Scholar] [CrossRef]
- Fan, M.; Gao, Y.; Wu, Z.; Zhang, Q. Linkage map development by EST-SSR Markers and QTL analysis for inflorescence and leaf traits in Chrysanthemum (Chrysanthemum morifolium Ramat.). Plants 2020, 9, 1342. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 765. [Google Scholar] [CrossRef]
- Evtushenko, E.V.; Lipikhina, Y.A.; Stepochkin, P.I.; Vershinin, A.V. Cytogenetic and molecular characteristics of rye genome in octoploid triticale (× Triticosecale Wittmack). Comp. Cytogenet. 2019, 13, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, S.A.E.C.; Evans, L.J.; Kirkland, L.; Rader, R. A global review of watermelon pollination biology and ecology: The increasing importance of seedless cultivars. Sci. Hortic. 2020, 271, 109493. [Google Scholar] [CrossRef]
- Farshadfar, M. Chromosome location of ISSR markers and genes controlling seed germination under drought stress in wheat-barley disomic addition lines. Cell. Mol. Biol. 2020, 66, 101–108. [Google Scholar] [CrossRef]
- Guerra, M. Chromosome numbers in plant cytotaxonomy: Concepts and implications. Cytogenet. Genome Res. 2008, 120, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Sun, H.; Li, Z. Chromosome data mining and its application in plant diversity research. Plant Sci. J. 2019, 37, 260–269. [Google Scholar]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold Ltd.: London, UK, 1971; pp. 87–93. [Google Scholar]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Baltisberger, M.; Horandl, E. Karyotype evolution supports the molecular phylogeny in the genus Ranunculus (Ranunculaceae). Perspect. Plant Ecol. Evol. Syst. 2016, 18, 1–14. [Google Scholar] [CrossRef]
- Ebrahimzadeh-Vesal, R.; Shokrgozar, M.A.; Nayernia, K.; Teimoori-Toolabi, L.; Estiar, M.A.; Miryounesi, M.; Nourashrafeddin, S.; Modarressi, M.H. MicroRNA profiling during germline differentiation of mouse embryonic stem cells. Cell. Mol. Biol. 2015, 61, 84–91. [Google Scholar]
- Huang, X.Q.; Ju, H.R.; Song, G.H. Evaluation of basal hormone levels and androgen receptor gene mutations in individuals with recurrent abortion. Cell. Mol. Biol. 2021, 67, 274–281. [Google Scholar] [CrossRef]
- Nkongolo, K.K.; Mehes-Smith, M. Karyotype evolution in the Pinaceae: Implication with molecular phylogeny. Genome 2012, 55, 735–753. [Google Scholar] [CrossRef]
- Saensouk, S.; Saensouk, P. Karyotype analysis of three species of Allium (Amaryllidaceae) from thailand. Biodiversitas J. Biol. Divers. 2021, 22, 3458–3466. [Google Scholar] [CrossRef]
- Younis, A.; Ramzan, F.; Hwang, Y.-J.; Lim, K.-B. FISH and GISH: Molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Rep. 2015, 34, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Schwarzacher, T.; Leitch, A.R.; Bennett, M.D.; Heslop-Harrison, J.S. In situ localization of parental genomes in a wide hybrid. Ann. Bot. 1989, 64, 315–324. [Google Scholar] [CrossRef]
- Kato, A.; Vega, J.M.; Han, F.P.; Lamb, J.C.; Birchler, J.A. Advances in plant chromosome identification and cytogenetic techniques. Curr. Opin. Plant Biol. 2005, 8, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Arantes Chaves, A.L.; Mendes Carvalho, P.H.; Mendes Ferreira, M.T.; Gandolfi Benites, F.R.; Techio, V.H. Genomic constitution, allopolyploidy, and evolutionary proposal for Cynodon Rich. based on GISH. Protoplasma 2022, 259, 999–1011. [Google Scholar] [CrossRef]
- Khrustaleva, L.; Mardini, M.; Kudryavtseva, N.; Alizhanova, R.; Romanov, D.; Sokolov, P.; Monakhos, G. The power of genomic in situ hybridization (GISH) in interspecific breeding of bulb onion (Allium cepa L.) resistant to downy mildew (Peronospora destructor Berk. Casp.). Plants 2019, 8, 36. [Google Scholar] [CrossRef]
- Harrison, G.E.; Heslopharrison, J.S. Centromeric repetitive DNA-sequences in the genus Brassica. Theor. Appl. Genet. 1995, 90, 157–165. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.; Schwarzacher, T.; Anamthawat-Jonsson, K.; Leitch, A.R.; Shi, M.; Leitch, I.J. In situ hybridization with automated chromosome denaturation. Tech. J. Methods Cell Mol. Biol. 1991, 3, 109–115. [Google Scholar]
- Lee, H.-I.; Younis, A.; Hwang, Y.-J.; Kang, Y.-I.; Lim, K.-B. Molecular cytogenetic analysis and phylogenetic relationship of 5S and 45S ribosomal DNA in Sinomartagon lilium Species by fluorescence in Situ Hybridization (FISH). Hortic. Environ. Biotechnol. 2014, 55, 514–523. [Google Scholar] [CrossRef]
- Said, M.; Hribova, E.; Danilova, T.V.; Karafiatova, M.; Cizkova, J.; Friebe, B.; Dolezel, J.; Gill, B.S.; Vrana, J. The Agropyron cristatum karyotype, chromosome structure and cross-genome homoeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes. Theor. Appl. Genet. 2018, 131, 2213–2227. [Google Scholar] [CrossRef]
- Garcia, S.; Kovarik, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the Plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Roa, F.; Guerra, M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, J.; da Silva, J.A.T.; Yu, X. 2017 Variation in ploidy and karyological diversity in different herbaceous peony cultivar groups. J. Am. Soc. Hortic. Sci. 2017, 142, 272–278. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Lu, J.; Zhang, Z.; Hu, J.; Huang, S.; Jin, W. Comparison of the distribution of the repetitive DNA sequences in three variants of Cucumis sativus reveals their phylogenetic relationships. J. Genet. Genom. 2011, 38, 39–45. [Google Scholar] [CrossRef]
- Greilhuber, J.; Dolezel, J.; Lysak, M.A.; Bennett, M.D. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Ann. Bot. 2005, 95, 255–260. [Google Scholar] [CrossRef]
- Puttick, M.N.; Clark, J.; Donoghue, P.C.J. Size is not everything: Rates of genome size evolution, not C-value, correlate with speciation in angiosperms. Proc. R. Soc. B 2015, 282, 20152289. [Google Scholar] [CrossRef]
- Sassone, A.B.; Lopez, A.; Hojsgaard, D.H.; Giussani, L.M. A novel indicator of karyotype evolution in the tribe Leucocoryneae (Allioideae, Amaryllidaceae). J. Plant Res. 2018, 131, 211–223. [Google Scholar] [CrossRef]
- Li, J.; Zhu, K.; Wang, Q.; Chen, X. Genome size variation and karyotype diversity in eight taxa of Sorbus sensu stricto (Rosaceae) from China. Comp. Cytogenet. 2021, 15, 137–148. [Google Scholar]
- Wendel, J.F. The wondrous cycles of polyploidy in plants. Am. J. Bot. 2015, 102, 1753–1756. [Google Scholar] [CrossRef]
- Sun, W.; Wang, H.; Wu, R.; Sun, H.; Li, Z. Karyomorphology of three endemic plants (Brassicaceae: Euclidieae and Arabideae) from the Qinghai-Tibet Plateau and its significance. Plant Divers. 2020, 42, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Al-Shehbaz, I.; Yue, J.; Sun, H. New insights into the taxonomy of tribe Euclidieae (Brassicaceae), evidence from nrITS sequence data. PhytoKeys 2018, 100, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Winterfeld, G.; Ley, A.; Hoffmann, M.H.; Paule, J.; Rosser, M. Dysploidy and polyploidy trigger strong variation of chromosome numbers in the prayer-plant family (Marantaceae). Plant Syst. Evol. 2020, 306, 36. [Google Scholar] [CrossRef]
- Oginuma, K.; Tobe, H. Karyomorphology and chromosome evolution in Nyssaceae (Cornales). Cytologia 2021, 86, 29–34. [Google Scholar] [CrossRef]
- Silva, G.S.; Souza, M.M. Genomic in situ hybridization in plants. Genet. Mol. Res. 2013, 12, 2953–2965. [Google Scholar] [CrossRef]
- Lim, K.Y.; Matyasek, R.; Kovarik, A.; Leitch, A.R. Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 2004, 82, 599–606. [Google Scholar] [CrossRef]
- Majka, J.; Majka, M.; Kwiatek, M.; Wisniewska, H. Similarities and differences in the nuclear genome organization within Pooideae species revealed by comparative genomic in situ hybridization (GISH). J. Appl. Genet. 2017, 58, 151–161. [Google Scholar] [CrossRef]
- Benabdelmouna, A.; Shi, Y.; Abirached-Darmency, M.; Darmency, H. Genomic in situ hybridization (GISH) discriminates between the A and the B genomes in diploid and tetraploid Setaria species. Genome 2001, 44, 685–690. [Google Scholar] [CrossRef]
- Li, Z.; Li, M.; Huang, R.; Wu, X.; Song, Y. Genomic in situ hybridization (GISH) discriminates the A, B and C genomes in Brassica allotetraploid species. Chin. J. Oil Crop Sci. 2002, 24, 10–14. [Google Scholar]
- Snowdon, R.J.; Kohler, W.; Friedt, W.; Kohler, A. Genomic in situ hybridization in Brassica amphidiploids and interspecific hybrids. Theor. Appl. Genet. 1997, 95, 1320–1324. [Google Scholar] [CrossRef]
- She, C.-W.; Jiang, X.-H. Karyotype analysis of Lablab purpureus (L.) sweet using fluorochrome banding and fluorescence in situ hybridisation with rDNA probes. Czech J. Genet. Plant Breed. 2015, 51, 110–116. [Google Scholar] [CrossRef]
- He, J.; Lin, S.; Yu, Z.; Song, A.; Guan, Z.; Fang, W.; Chen, S.; Zhang, F.; Jiang, J.; Chen, F.; et al. Identification of 5S and 45S rDNA sites in Chrysanthemum species by using oligonucleotide fluorescence in situ hybridization (Oligo-FISH). Mol. Biol. Rep. 2021, 48, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.M.; Sang, T. Chromosomal structural rearrangement of Paeonia brownii and P. californica revealed by fluorescence in situ hybridization. Genome 1998, 41, 848–853. [Google Scholar] [CrossRef]
- Zhang, D.M.; Sang, T. Physical mapping of ribosomal RNA genes in peonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization: Implications for phylogeny and concerted evolution. Am. J. Bot. 1999, 86, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Sliwinska, E.; Loureiro, J.; Leitch, I.J.; Smarda, P.; Bainard, J.; Bures, P.; Chumova, Z.; Horova, L.; Koutecky, P.; Lucanova, M.; et al. Application-based guidelines for best practices in plant flow cytometry. Cytom. Part A 2022, 101, 749–781. [Google Scholar] [CrossRef]
- Roux, N.; Toloza, A.; Radecki, Z.; Zapata-Arias, F.J.; Dolezel, J. Rapid detection of aneuploidy in Musa using flow cytometry. Plant Cell Rep. 2003, 21, 483–490. [Google Scholar] [CrossRef]
- Dolezel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef]
- Paszko, B. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst. Evol. 2006, 258, 39–48. [Google Scholar] [CrossRef]
- Peruzzi, L.; Leitch, I.J.; Caparelli, K.F. Chromosome diversity and evolution in Liliaceae. Ann. Bot. 2009, 103, 459–475. [Google Scholar] [CrossRef]
- Czernicka, M.; Mscichowska, A.; Klein, M.; Muras, P.; Grzebelus, E. Paternity determination of interspecific rhododendron hybrids by genomic in situ hybridization (GISH). Genome 2010, 53, 277–284. [Google Scholar] [CrossRef]
- Doyle, J.J. A rapid total DNA preparation procedure for fresh plant tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Brammer, S.P.; Vasconcelos, S.; Poersch, L.B.; Oliveira, A.R.; Brasileiro-Vidal, A.C. Genomic in situ Hybridization in Triticeae: A Methodological Approach. In Plant Breeding from Laboratories to Fields; Andersen, S.B., Ed.; IntechOpen: London, UK, 2013. [Google Scholar]
- Altinordu, F.; Peruzzi, L.; Yu, Y.; He, X. A tool for the analysis of chromosomes: KaryoType. Taxon 2016, 65, 586–592. [Google Scholar] [CrossRef]
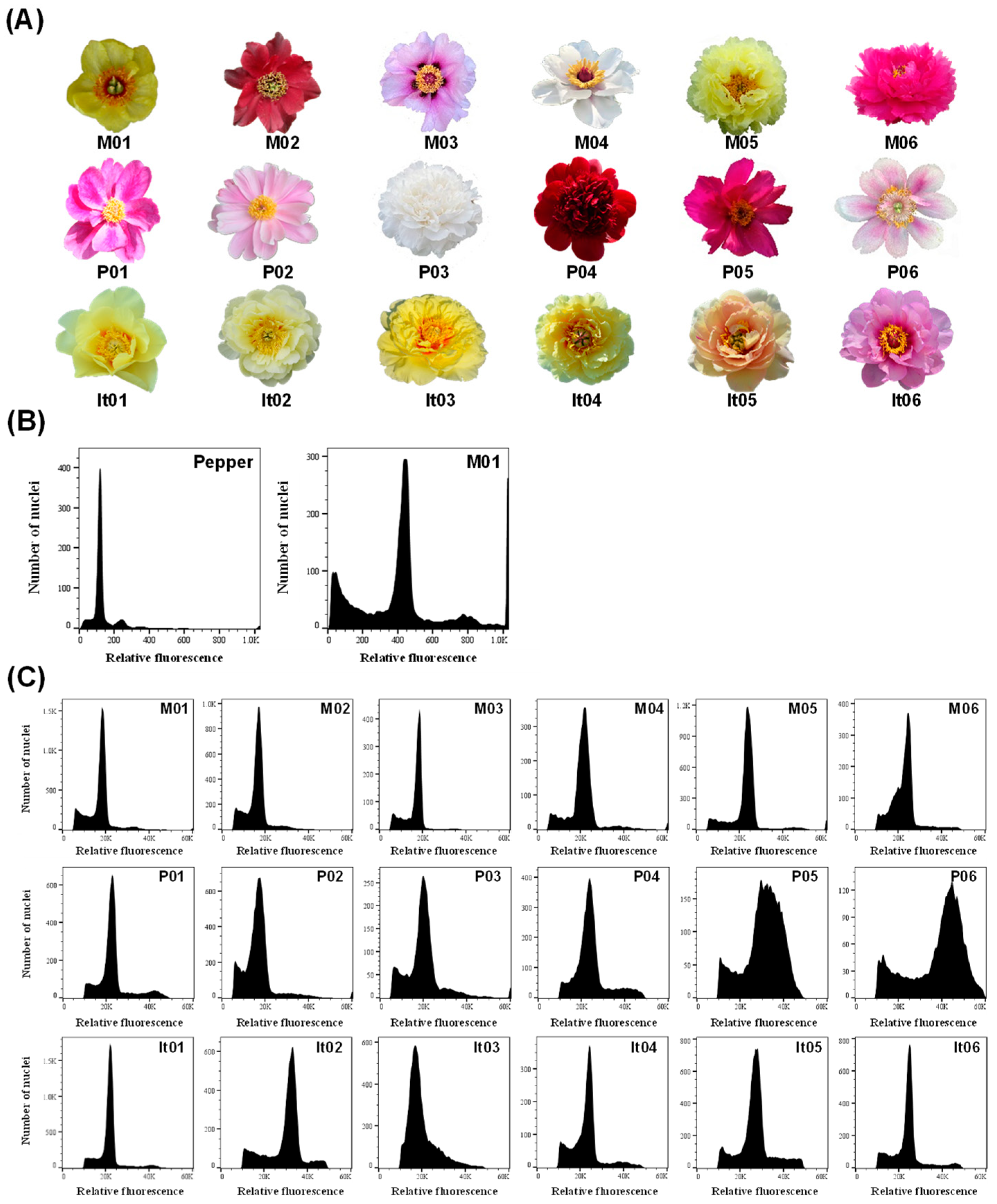

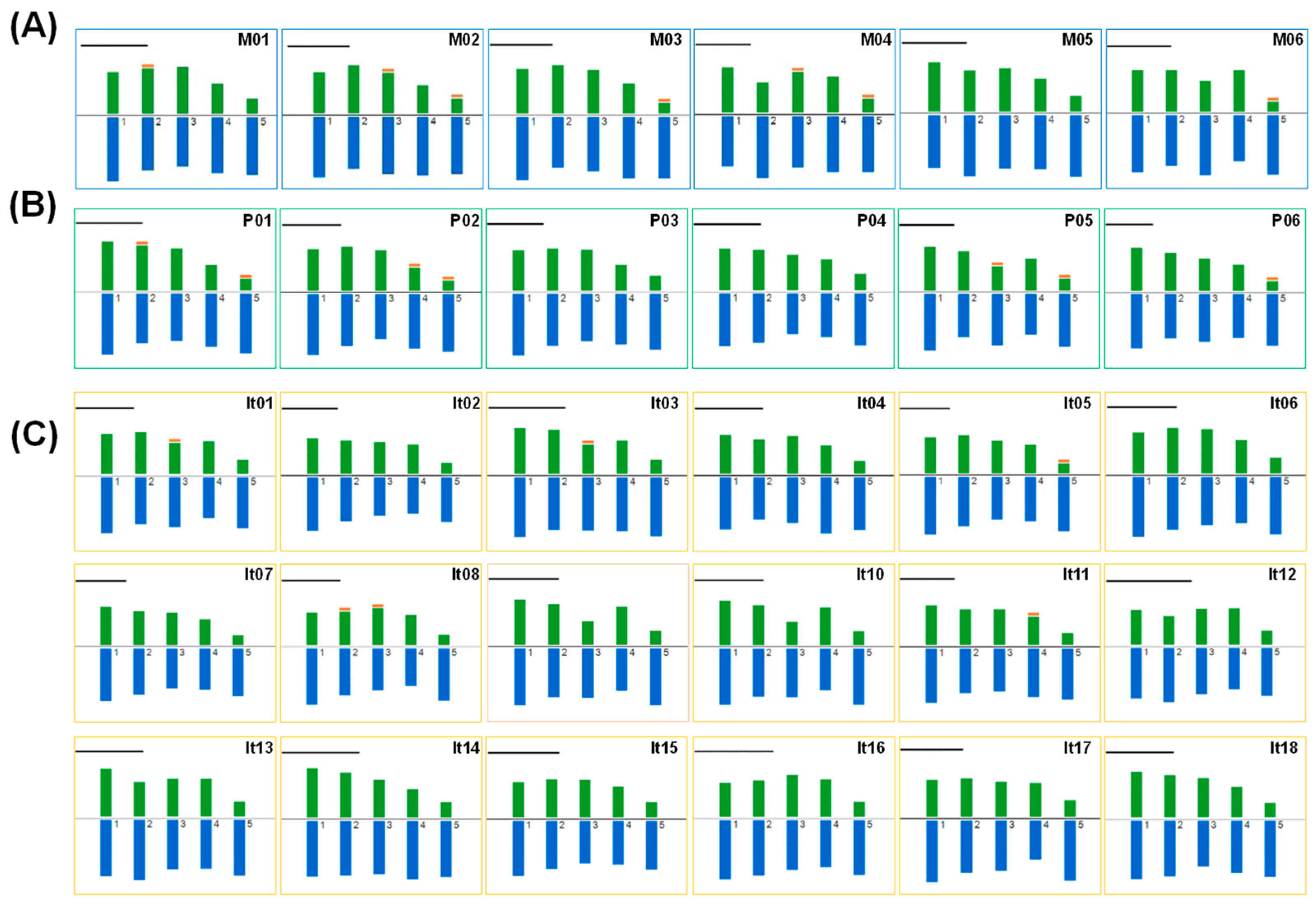




| Taxa | Code | Name | Indication a | Relative Ploidy b | Genome Size (Gb) c |
|---|---|---|---|---|---|
| Sect. Moutan | M01 | P. delavayi var. lutea | 18,823 | 2.00 | 12.45 |
| M02 | P. delavayi | 16,778 | 1.78 | 11.08 | |
| M03 | P. rockii | 17,966 | 1.91 | 11.89 | |
| M04 | P. ostii | 21,475 | 2.28 | 14.19 | |
| M05 | P. × lemoinei ‘High Noon’ | 23,902 | 2.54 | 15.81 | |
| M06 | P. suffruticosa ‘Luo Yang Hong’ | 23,928 | 2.54 | 15.81 | |
| Sect. Paeonia | P01 | P. veitchii | 22,341 | 2.37 | 14.75 |
| P02 | P. lactiflora ‘Bai Shao’ | 16,945 | 1.80 | 11.21 | |
| P03 | P. lactiflora “Yang Fei Chu Yu’ | 16,271 | 1.73 | 10.77 | |
| P04 | P. sp. ‘Red Charm’ | 19,997 | 2.12 | 13.20 | |
| P05 | P. mairei | 31,416 | 3.34 | 20.79 | |
| P06 | P. officinalis | 32,695 | 3.47 | 21.60 | |
| Itoh hybrid | It01 | ‘Going Bananas’ | 34,352 | 3.65 | 22.72 |
| It02 | ‘Bartzella’ | 32,167 | 3.42 | 21.29 | |
| It03 | ‘Viking Full Moon’ | 24,582 | 2.61 | 16.25 | |
| It04 | ‘Garden Treasure’ | 23,634 | 2.51 | 15.62 | |
| It05 | ‘Prairie Charm’ | 26,888 | 2.86 | 17.80 | |
| It06 | ‘Morning Lilac’ | 24,645 | 2.62 | 16.31 |
| Taxon | Code | Name | Ss’C | Ploidy | HKF | THL (μm) | RL (%) | MAR | XCI (%) | CVCL | CVCI | AI | L/S |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sect. Moutan | M01 | P. delavayi var. lutea | 2A | 2n = 2x = 10 | 6m * + 2sm + 2st | 68.38 | 15.87~23.11 | 1.87 | 38.25 | 13.86 | 28.95 | 4.01 | 1.50 |
| M02 | P. delavayi | 2A | 2n = 2x = 10 | 6m ** + 2sm + 2st ** | 73.90 | 15.66~22.26 | 2.02 | 36.57 | 13.85 | 29.09 | 4.03 | 1.46 | |
| M03 | P. rockii | 2A | 2n = 2x = 10 | 6m + 2sm + 2st * | 75.13 | 15.45~22.90 | 2.28 | 36.56 | 14.02 | 36.26 | 5.08 | 1.54 | |
| M04 | P. ostii | 2A | 2n = 2x = 10 | 6m * + 2sm + 2st * | 80.94 | 15.79~21.77 | 2.02 | 36.70 | 11.95 | 30.10 | 3.60 | 1.44 | |
| M05 | P. × lemoinei ‘High Noon’ | 2A | 2n = 2x = 10 | 8m + 2st | 72.31 | 16.66~22.02 | 1.86 | 38.34 | 11.62 | 28.02 | 3.26 | 1.43 | |
| M06 | P. suffruticosa ‘Luo Yang Hong’ | 2A | 2n = 2x = 10 | 6m + 2sm + 2st * | 58.51 | 16.00~22.56 | 2.27 | 36.76 | 12.21 | 35.89 | 4.38 | 1.48 | |
| Sect. Paeonia | P01 | P. veitchii | 2A | 2n = 2x = 10 | 6m * + 2sm + 2st * | 66.28 | 16.06~24.81 | 2.20 | 37.53 | 16.97 | 36.55 | 6.20 | 1.60 |
| P02 | P. lactiflora ‘Bai Shao’ | 2A | 2n = 2x = 10 | 6m + 2sm ** + 2st * | 72.74 | 15.64~23.95 | 2.36 | 35.71 | 16.48 | 37.91 | 6.25 | 1.63 | |
| P03 | P. lactiflora “Yang Fei Chu Yu’ | 2A | 2n = 2x = 10 | 6m + 2sm + 2st | 76.75 | 16.30~23.76 | 1.97 | 36.97 | 15.14 | 28.41 | 4.30 | 1.59 | |
| P04 | P. sp. ‘Red Charm’ | 2A | 2n = 3x = 15 | 10m + 3sm + 2st | 60.11 | 16.94~23.42 | 1.69 | 40.25 | 13.65 | 23.54 | 3.21 | 1.67 | |
| P05 | P. mairei | 2A | 2n = 4x = 20 | 12m + 4sm * + 4st * | 71.81 | 16.31~25.37 | 2.10 | 36.89 | 17.02 | 32.48 | 5.53 | 1.76 | |
| P06 | P. officinalis | 2A | 2n = 4x = 20 | 12m + 4sm + 4st *** | 82.25 | 15.61~24.96 | 2.24 | 36.66 | 17.60 | 33.58 | 5.91 | 1.95 | |
| Itoh hybrid | It01 | ‘Going Bananas’ | 2A | 2n = 3x = 15 | 9m + 3sm ** + 3st | 69.70 | 16.17~23.73 | 1.85 | 38.48 | 14.98 | 25.57 | 3.83 | 1.77 |
| It02 | ‘Bartzella’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 64.56 | 15.50~24.94 | 1.91 | 38.33 | 17.65 | 27.74 | 4.90 | 1.89 | |
| It03 | ‘Viking Full Moon’ | 2A | 2n = 3x = 15 | 9m * + 3sm + 3st | 58.31 | 16.41~22.36 | 2.04 | 36.45 | 11.18 | 26.94 | 3.01 | 1.57 | |
| It04 | ‘Garden Treasure’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 58.99 | 16.34~22.79 | 1.99 | 37.15 | 12.13 | 29.54 | 3.58 | 1.57 | |
| It05 | ‘Prairie Charm’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st * | 79.05 | 16.32~23.68 | 2.25 | 35.95 | 14.84 | 32.19 | 4.78 | 1.62 | |
| It06 | ‘Morning Lilac’ | 2A | 2n = 3x = 15 | 12m + 3st | 64.07 | 16.44~22.60 | 1.79 | 39.60 | 13.50 | 26.34 | 3.56 | 1.65 | |
| It07 | ‘Cora Louise’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 73.35 | 15.72~24.74 | 2.09 | 36.83 | 17.14 | 29.97 | 5.14 | 1.74 | |
| It08 | ‘Julia Rose’ | 2A | 2n = 3x = 15 | 9m * + 3sm * + 3st | 64.68 | 16.56~23.38 | 2.16 | 37.16 | 13.68 | 33.19 | 4.54 | 1.62 | |
| It09 | ‘Court Jester’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 59.63 | 17.10~24.45 | 1.98 | 37.82 | 15.13 | 31.00 | 4.69 | 1.73 | |
| It10 | ‘Lemon Dream’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 66.70 | 15.50~23.27 | 2.06 | 36.17 | 14.26 | 29.08 | 4.15 | 1.87 | |
| It11 | ‘Sequestered Sunshine’ | 2A | 2n = 3x = 15 | 9m + 3sm * + 3st | 72.00 | 15.97~24.01 | 1.98 | 37.48 | 14.29 | 28.81 | 4.12 | 1.63 | |
| It12 | ‘Yellow Crown’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 45.84 | 15.80~21.96 | 1.91 | 37.39 | 12.30 | 25.44 | 3.13 | 1.66 | |
| It13 | ‘Ballarena de Saval’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 65.58 | 15.85~23.47 | 1.89 | 37.90 | 14.04 | 26.75 | 3.76 | 1.63 | |
| It14 | ‘Sugar Plum Fairy’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 58.35 | 15.75~23.21 | 1.90 | 37.61 | 14.28 | 27.90 | 3.98 | 1.69 | |
| It15 | ‘Lollipop’ | 2A | 2n = 3x = 15 | 10m + 3sm + 2st | 55.73 | 16.19~22.76 | 1.76 | 39.04 | 12.95 | 22.56 | 2.92 | 1.59 | |
| It16 | ‘Magical Mystery Tour’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 54.54 | 16.15~21.69 | 1.92 | 37.27 | 11.49 | 26.12 | 3.00 | 1.67 | |
| It17 | ‘Scarlet Heaven’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 67.17 | 17.19~23.26 | 1.89 | 37.66 | 12.29 | 25.05 | 3.08 | 1.65 | |
| It18 | ‘Dark Eyes’ | 2A | 2n = 3x = 15 | 9m + 3sm + 3st | 64.69 | 16.03~23.60 | 1.87 | 38.12 | 14.86 | 26.90 | 4.00 | 1.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, L.; Chen, T.; Zhao, X.; Wang, S.; Ren, X.; Xue, J.; Zhang, X. Karyotype Analysis, Genomic and Fluorescence In Situ Hybridization (GISH and FISH) Reveal the Ploidy and Parental Origin of Chromosomes in Paeonia Itoh Hybrids. Int. J. Mol. Sci. 2022, 23, 11406. https://doi.org/10.3390/ijms231911406
Cui L, Chen T, Zhao X, Wang S, Ren X, Xue J, Zhang X. Karyotype Analysis, Genomic and Fluorescence In Situ Hybridization (GISH and FISH) Reveal the Ploidy and Parental Origin of Chromosomes in Paeonia Itoh Hybrids. International Journal of Molecular Sciences. 2022; 23(19):11406. https://doi.org/10.3390/ijms231911406
Chicago/Turabian StyleCui, Litao, Tai Chen, Xin Zhao, Shunli Wang, Xiuxia Ren, Jingqi Xue, and Xiuxin Zhang. 2022. "Karyotype Analysis, Genomic and Fluorescence In Situ Hybridization (GISH and FISH) Reveal the Ploidy and Parental Origin of Chromosomes in Paeonia Itoh Hybrids" International Journal of Molecular Sciences 23, no. 19: 11406. https://doi.org/10.3390/ijms231911406
APA StyleCui, L., Chen, T., Zhao, X., Wang, S., Ren, X., Xue, J., & Zhang, X. (2022). Karyotype Analysis, Genomic and Fluorescence In Situ Hybridization (GISH and FISH) Reveal the Ploidy and Parental Origin of Chromosomes in Paeonia Itoh Hybrids. International Journal of Molecular Sciences, 23(19), 11406. https://doi.org/10.3390/ijms231911406








