Development of a Transformation System and Locus Identification Pipeline for T-DNA in Chrysanthemum seticuspe, A Model Species for Hexaploid Cultivated Chrysanthemum
Abstract
:1. Introduction
2. Results and Discussion
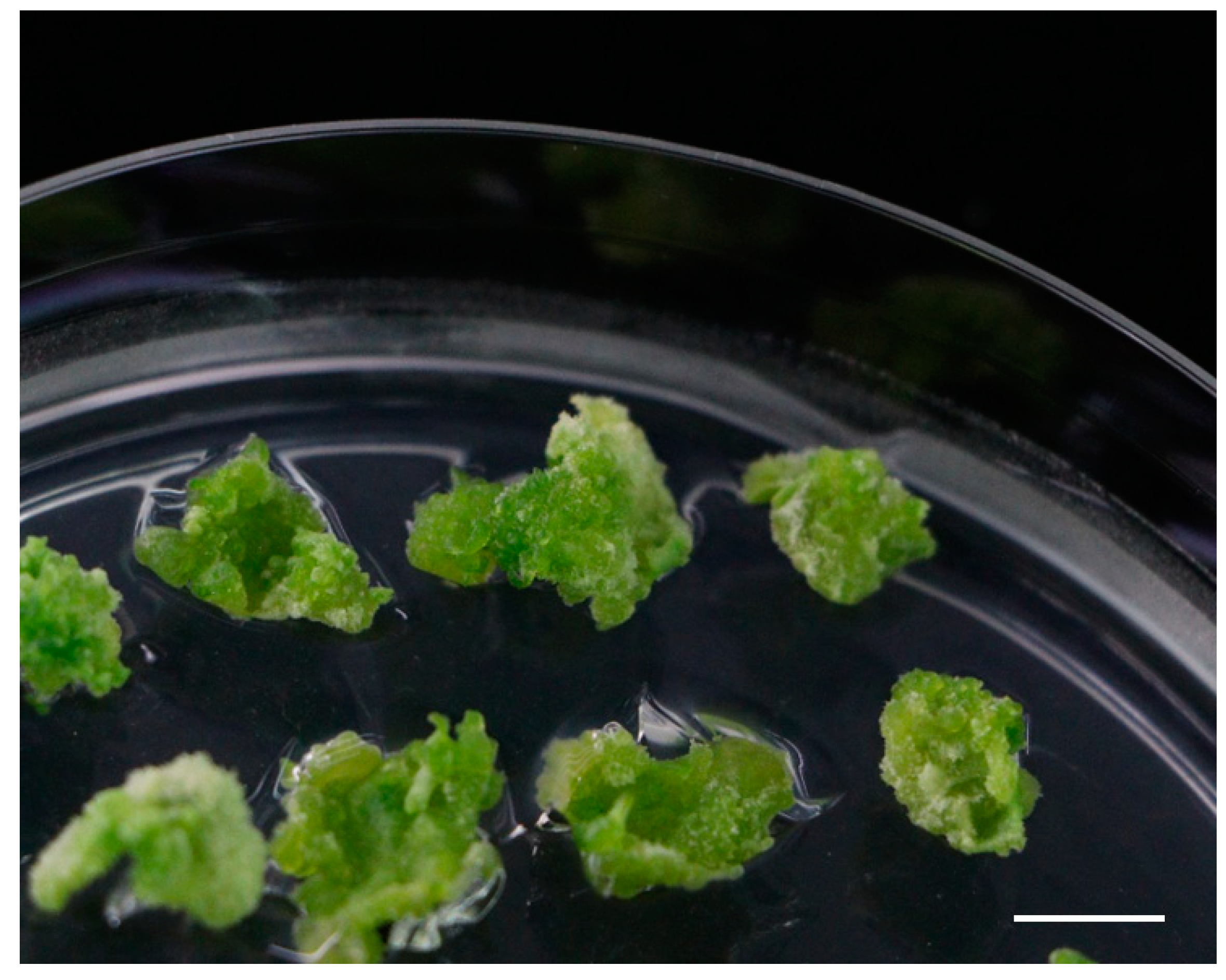
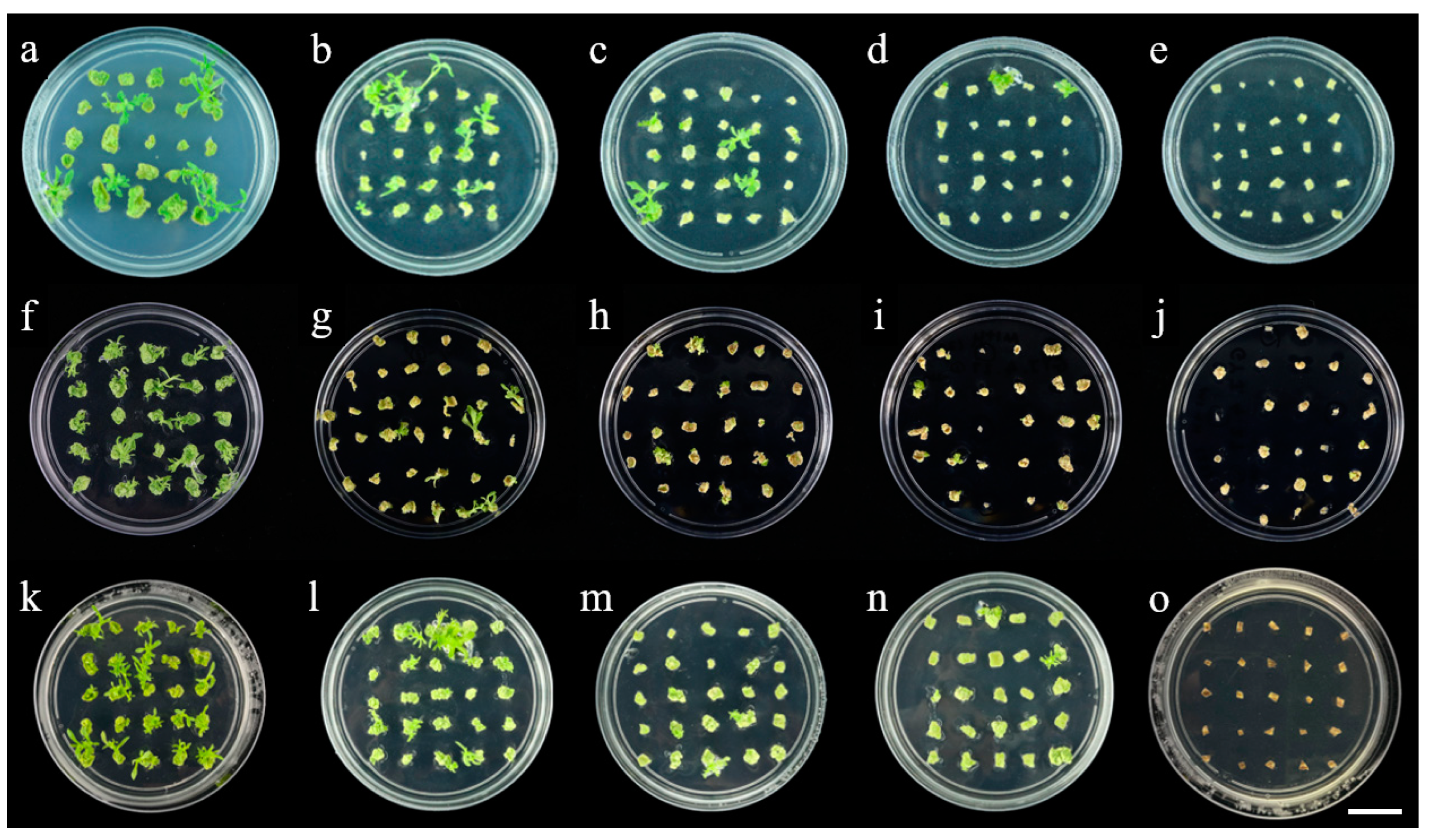
2.1. Role of Various Growth Regulators in Shoot Regeneration
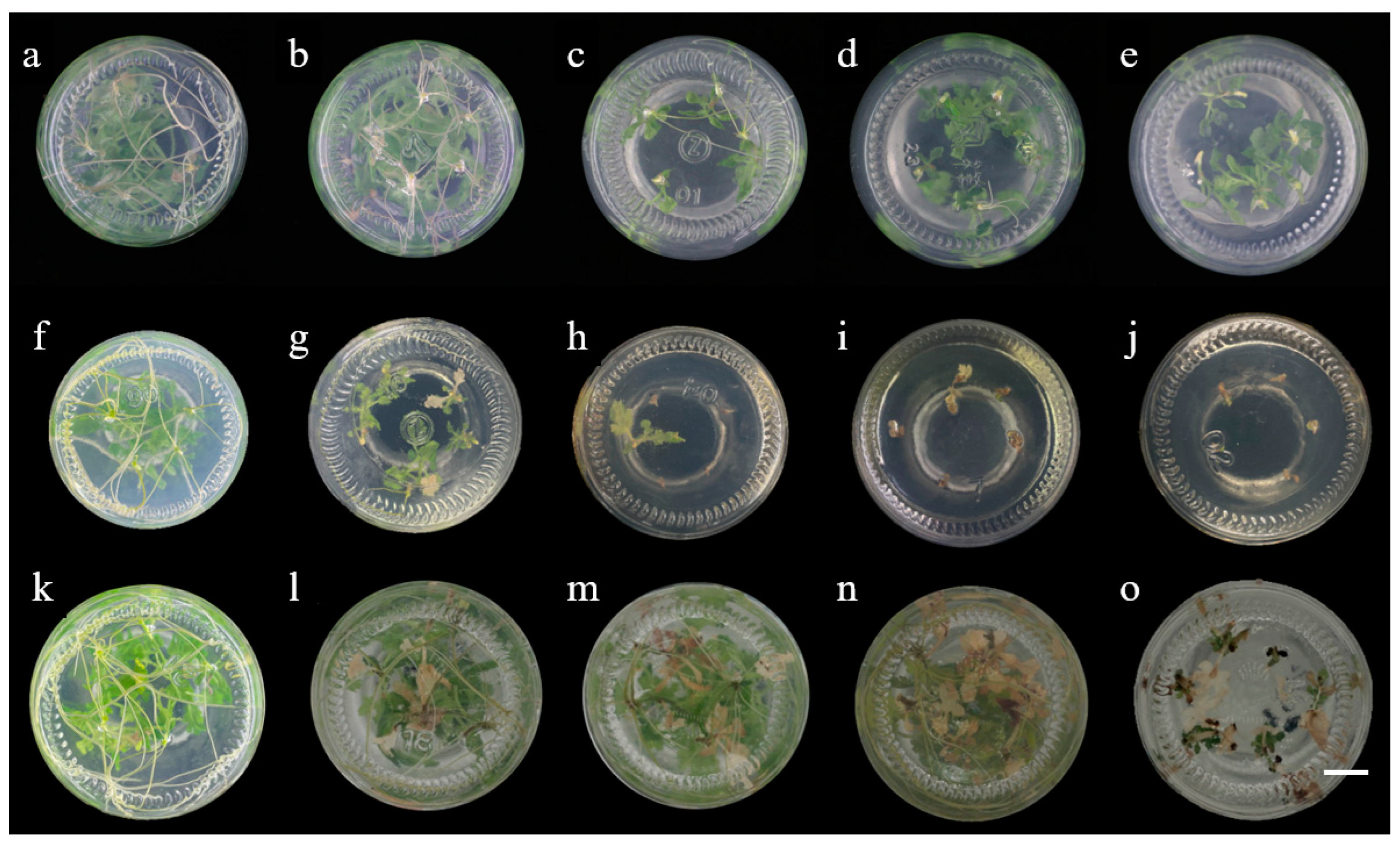
2.2. Effect of Various Antibiotics on Shoot Regeneration and Rooting
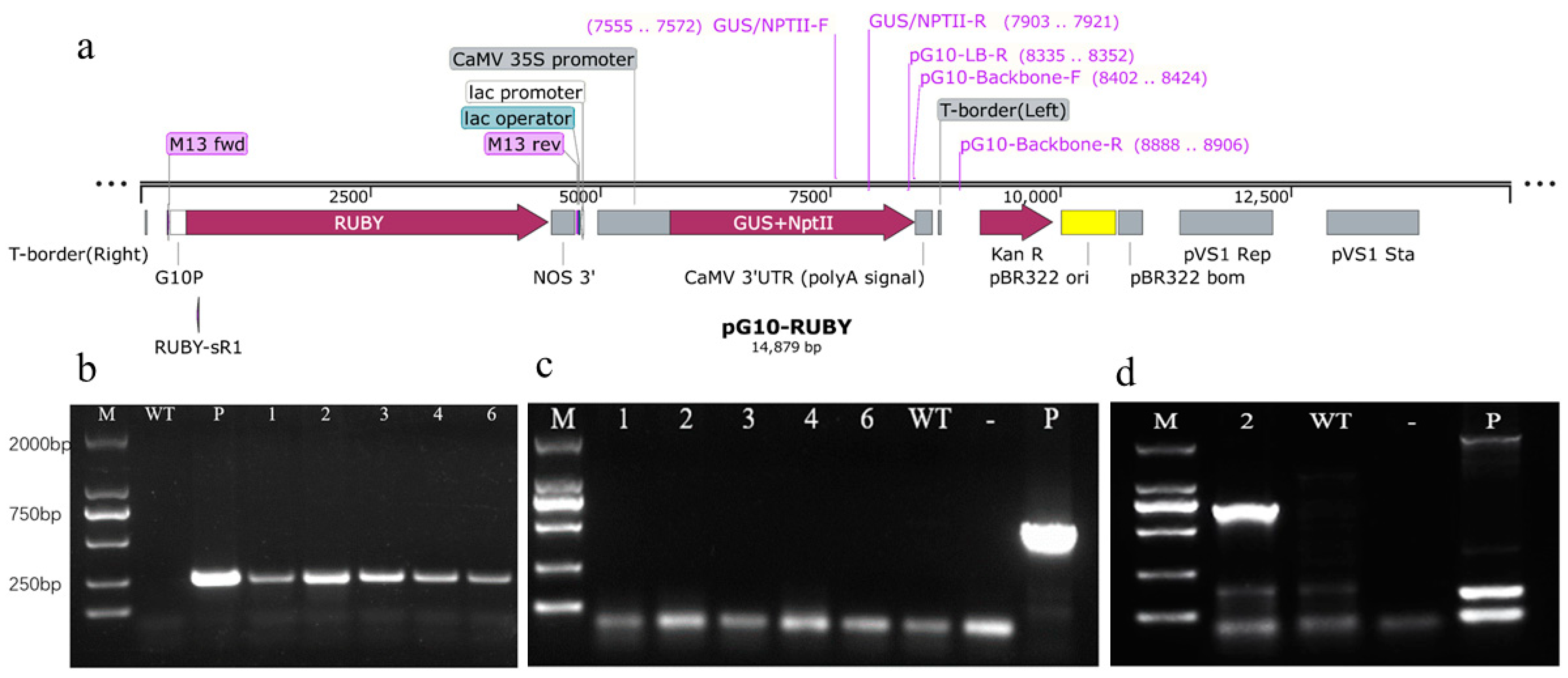
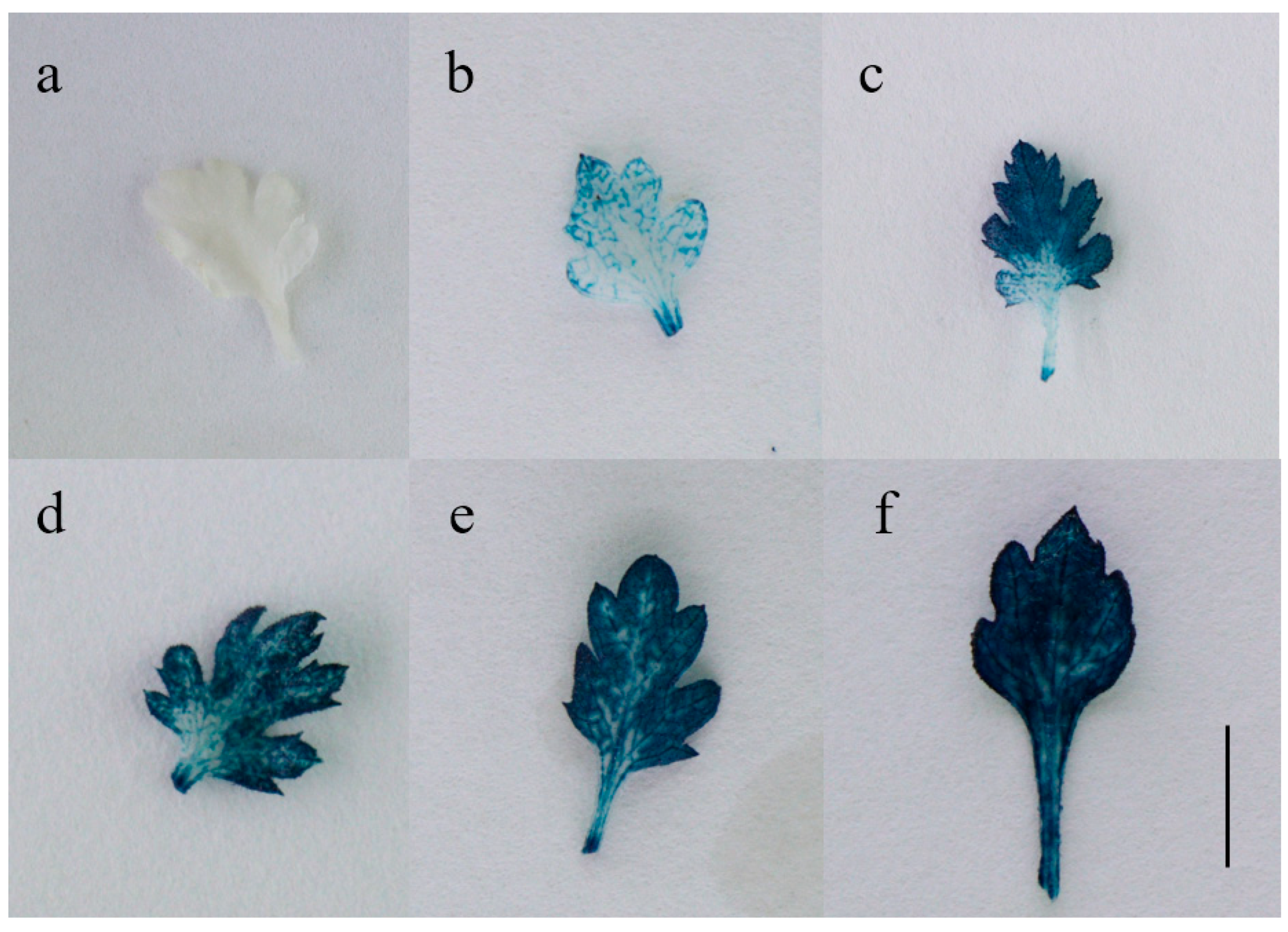
2.3. Molecular and Histochemical Analyses of Transgenic Plants
2.4. T-DNA Insertion Locus of pG10-2
3. Materials and Methods
3.1. Materials
3.2. Assessment of Growth Regulators of the Untransformed Plants
3.3. Assessment of Antibiotic Sensitivity of the Untransformed Plants
3.4. Agrobacterium-Mediated Transformation
3.5. PCR Detection
3.6. GUS Staining
3.7. Identification of the T-DNA Insertion Locus Using High-Throughput Sequencing
3.8. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Higuchi, Y.; Narumi, T.; Oda, A.; Nakano, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA 2013, 110, 17137–17142. [Google Scholar] [CrossRef]
- Oda, A.; Narumi, T.; Li, T.; Kando, T.; Higuchi, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J. Exp. Bot. 2012, 63, 1461–1477. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Taniguchi, K.; Masuda, Y.; Kozuka, T.; Aruga, Y.; Han, J.; Motohara, K.; Nakata, M.; Sumitomo, K.; Hisamatsu, T. A pure line derived from a self-compatible Chrysanthemum seticuspe mutant as a model strain in the genus Chrysanthemum. Plant Sci. 2019, 287, 110174. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Hirakawa, H.; Fukai, E.; Toyoda, A.; Kajitani, R.; Minakuchi, Y.; Itoh, T.; Higuchi, Y.; Kozuka, T.; Bono, H. A chromosome-level genome sequence of Chrysanthemum seticuspe, a model species for hexaploid cultivated chrysanthemum. Commun. Biol. 2021, 4, 1167. [Google Scholar] [CrossRef]
- Hirakawa, H.; Sumitomo, K.; Hisamatsu, T.; Nagano, S.; Shirasawa, K.; Higuchi, Y.; Kusaba, M.; Koshioka, M.; Nakano, Y.; Yagi, M. De novo whole-genome assembly in Chrysanthemum seticuspe, a model species of Chrysanthemums, and its application to genetic and gene discovery analysis. DNA Res. 2019, 26, 195–203. [Google Scholar] [CrossRef]
- Shinoyama, H.; Anderson, N.; Furuta, H.; Mochizuki, A.; Nomura, Y.; Singh, R.; Datta, S.K.; Wang, B.; Teixeira da Silva, J. Chrysanthemum biotechnology. Floric. Ornam. Plant Biotechnol. Adv. Top. Issues 2006, 2, 140–163. [Google Scholar]
- Song, A.; An, J.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 2014, 80, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cui, Y.; Wang, B.; Li, Y.; Zhang, M.; Dai, S.; Huang, H. Genotype screening of recipient resources with high regeneration and transformation efficiency in chrysanthemum. Phyton 2022, 91, 869–888. [Google Scholar]
- Kumar, R.V.; Sharma, V.; Chattopadhyay, B.; Chakraborty, S. An improved plant regeneration and Agrobacterium-mediated transformation of red pepper (Capsicum annuum L.). Physiol. Mol. Biol. Plants 2012, 18, 357–364. [Google Scholar] [CrossRef]
- Naing, A.H.; Ai, T.N.; Jeon, S.M.; Lim, S.H.; Kim, C.K. An efficient protocol for Agrobacterium-mediated genetic transformation of recalcitrant chrysanthemum cultivar Shinma. Acta Physiol. Plant. 2016, 38, 38. [Google Scholar] [CrossRef]
- Petty, L.M.; Harberd, N.P.; Carré, I.A.; Thomas, B.; Jackson, S.D. Expression of the Arabidopsis gai gene under its own promoter causes a reduction in plant height in chrysanthemum by attenuation of the gibberellin response. Plant Sci. 2003, 164, 175–182. [Google Scholar] [CrossRef]
- Shinoyama, H.; Kazuma, T.; Komano, M.; NOMURA, Y.; TSUCHIYA, T. An Efficient Transformation System in Chrysanthemum [Dendranthema × grandiflorum (Ramat.) Kitamura] for Stable and Non-chimeric Expression of Foreign Genes. Plant Biotechnol. 2002, 19, 335–343. [Google Scholar] [CrossRef] [Green Version]
- Toguri, T.; Ogawa, T.; Kakitani, M.; Tukahara, M.; Yoshioka, M. Agrobacterium-mediated transformation of chrysanthemum (Dendranthema grandiflora) plants with a disease resistance gene (pac1). Plant Biotechnol. 2003, 20, 121–127. [Google Scholar] [CrossRef]
- Deroles, S.; Smith, M.A.L.; Lee, C. Factors affecting transformation of cell cultures from three dicotyledonous pigment-producing species using microprojectile bombardment. Plant Cell Tissue Organ Cult. 2002, 70, 69–76. [Google Scholar] [CrossRef]
- Chen, J.; Wang, L.; Chen, J.; Huang, J.; Liu, F.; Guo, R.; Yang, L.; Grabon, A.; Zhao, K.; Kong, F.; et al. Agrobacterium tumefaciens-mediated transformation system for the important medicinal plant Dendrobium catenatum Lindl. Vitr. Cell. Dev. Biol.-Plant 2018, 54, 228–239. [Google Scholar] [CrossRef]
- da Silva, M.L.; Paim Pinto, D.L.; Passos, A.B.; Marcelino-Guimarães, F.C.; Rossi, A.A.B.; Krause, W.; de Carvalho, I.F.; Batista, D.S.; Rocha, D.I.; Otoni, W.C. Novel and efficient transformation of wild passion fruit (Passiflora cincinnata Mast.) using sonication-assisted Agrobacterium-mediated transformation. Vitr. Cell. Dev. Biol.-Plant 2021, 57, 380–386. [Google Scholar] [CrossRef]
- Kim, C.; Dai, W. Agrobacterium-mediated transformation of purple raspberry (Rubus occidentalis × R. idaeus) with the PtFIT (FER-like iron deficiency–induced transcription factor 1) gene. Vitr. Cell. Dev. Biol.-Plant 2022, 58, 343–350. [Google Scholar] [CrossRef]
- Aida, R.; Ohira, K.; Tanaka, Y.; Yoshida, K.; Kishimoto, S.; Shibata, M.; Ohmiya, A. Efficient transgene expression in chrysanthemum, Dendranthema grandiflorum (Ramat.) Kitamura, by using the promoter of a gene for chrysanthemum chlorophyll-a/b-binding protein. Breed. Sci. 2004, 54, 51–58. [Google Scholar] [CrossRef]
- Ledger, S.E.; Deroles, S.C.; Given, N.K. Regeneration and Agrobacterium-mediated transformation of chrysanthemum. Plant Cell Rep. 1991, 10, 195–199. [Google Scholar] [CrossRef] [PubMed]
- de Jong, J.; Mertens, M.M.; Rademaker, W. Stable expression of the GUS reporter gene in chrysanthemum depends on binary plasmid T-DNA. Plant Cell Rep. 1994, 14, 59–64. [Google Scholar] [CrossRef]
- Urban, L.A.; Sherman, J.M.; Moyer, J.W.; Daub, M.E. High frequency shoot regeneration and Agrobacterium-mediated transformation of chrysanthemum (Dendranthema grandiflora). Plant Sci. 1994, 98, 69–79. [Google Scholar] [CrossRef]
- Gaba, V.P. Plant growth regulators in plant tissue culture and development. In Plant Development and Biotechnology; CRC Press: Boca Raton, FL, USA, 2005; pp. 87–99. [Google Scholar]
- Park, S.H.; Kim, G.H.; Jeong, B.R. Adventitious shoot regeneration in chrysanthemum as affected by plant growth regulators, sucrose, and dark period. Hortic. Environ. Biotechnol. 2005, 46, 335–340. [Google Scholar]
- Xu, X.; Legay, S.; Berni, R.; Hausman, J.F.; Guerriero, G. Transcriptomic Changes in Internode Explants of Stinging Nettle during Callogenesis. Int. J. Mol. Sci. 2021, 22, 12319. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Huang, M.E.; Pua, E.-C. High frequency shoot regeneration from leaf disc explants of garland chrysanthemum (Chrysanthemum coronarium L.) in vitro. Plant Sci. 1997, 126, 219–226. [Google Scholar] [CrossRef]
- Choi, D.; Kim, J.; Lim, H.; Kim, H.; Choi, J.; Choi, Y.; Seo, S. Plant Regeneration from Leaf Explants and Efficient Agrobacterium-mediated Transformation System of Chrysanthemum (Dendranthema grandiflorum). In XXVI International Horticultural Congress: Asian Plants with Unique Horticultural Potential: Genetic Resources, Cultural 620; ISHS: Leuven, Belgium, 2002; pp. 333–338. [Google Scholar]
- Nahid, J.S.; Shyamali, S.; Kazumi, H. High frequency shoot regeneration from petal explants of Chrysanthemum morifolium Ramat. in vitro. Pak. J. Biol. Sci. PJBS 2007, 10, 3356–3361. [Google Scholar]
- Haider, S.; Gao, Y.; Gao, Y. Standardized Genetic Transformation Protocol for Chrysanthemum cv.‘Jinba’with TERMINAL FLOWER 1 Homolog CmTFL1a. Genes 2020, 11, 860. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Gao, Y. Establishment of a regeneration system for the genetic transformation of ground-cover Chrysanthemum ‘Jindinghongxin’. Int. Conf. Germplasm Ornam. 2012, 2012, 213–218. [Google Scholar] [CrossRef]
- Sjahril, R.; Jamaluddin, I.; Nadir, M.; Dungga, N. Effect of Selection Agents to Chrysanthemum (Chrysanthemum morifolium) Callus Growth after Agrobacterium-Mediated Genetic Transformation; IOP Publishing: Bristol, UK, 2018; Volume 157, p. 012044. [Google Scholar]
- Rojo, F.P.; Seth, S.; Erskine, W.; Kaur, P. An improved protocol for Agrobacterium-mediated transformation in subterranean Clover (Trifolium subterraneum L.). Int. J. Mol. Sci. 2021, 22, 4181. [Google Scholar] [CrossRef]
- Ahmed, R.I.; Ding, A.; Xie, M.; Kong, Y. Progress in optimization of Agrobacterium-mediated transformation in sorghum (Sorghum bicolor). Int. J. Mol. Sci. 2018, 19, 2983. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chen, F.; Chen, S.; Fang, W.; Miao, H.; Luo, H. Transformation of Lycoris longituba agglutinin gene to cut chrysanthemum and identification of aphid resistance in the transgenic plants. Acta Bot. Boreali-Occident. Sin. 2009, 29, 2318–2325. [Google Scholar]
- Kumar, S.; Raj, S.K.; Sharma, A.K.; Varma, H.N. Genetic transformation and development of Cucumber mosaic virus resistant transgenic plants of Chrysanthemum morifolium cv. Kundan. Sci. Hortic. 2012, 134, 40–45. [Google Scholar] [CrossRef]
- Murashige, T. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- He, Y.; Zhang, T.; Sun, H.; Zhan, H.; Zhao, Y. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 2020, 7, 1–6. [Google Scholar] [CrossRef]
- Sjahril, R.; Mii, M. High-efficiency Agrobacterium-mediated transformation of Phalaenopsis using meropenem, a novel antibiotic to eliminate Agrobacterium. J. Hortic. Sci. Biotechnol. 2006, 81, 458–464. [Google Scholar] [CrossRef]
- Takatsu, Y.; Nishizawa, Y.; Hibi, T.; Akutsu, K. Transgenic chrysanthemum (Dendranthema grandiflorum (Ramat.) Kitamura) expressing a rice chitinase gene shows enhanced resistance to gray mold (Botrytis cinerea). Sci. Hortic. 1999, 82, 113–123. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]

| Types of Antibiotics | Concentration of Antibiotic (mg·L−1) | Callus Induction Rate (%) | Budding Leaf Disc Rate (%) | Budding Rate (%) |
|---|---|---|---|---|
| Kanamycin | 0 | 100 ± 0 a | 77.33 ± 5.81 b | 114.67 ± 8.11 a |
| 4.0 | 96.00 ± 2.31 ab | 22.67 ± 7.06 c | 36.00 ± 6.11 b | |
| 6.0 | 60.00 ± 2.31 d | 13.33 ± 7.06 cd | 16.00 ± 9.24 de | |
| 8.0 | 25.33 ± 3.53 f | 4.00 ± 4.00 de | 5.33 ± 5.33 de | |
| 10.0 | 8.00 ± 2.31 gh | 0 ± 0 | 0 ± 0 | |
| Hygromycin | 0 | 100 ± 0 a | 78.67 ± 3.53 ab | 128.00 ± 8.33 a |
| 1.0 | 87.67 ± 2.91 b | 9.43 ± 4.28 de | 12.62 ± 5.91 c | |
| 1.5 | 72.00 ± 5.29 c | 7.35 ± 2.15 de | 9.87 ± 3.34 de | |
| 2.0 | 42.67 ± 8.11 e | 3.51 ± 2.15 de | 3.51 ± 2.15 de | |
| 2.5 | 13.33 ± 2.40 g | 0 ± 0 | 0 ± 0 | |
| Glufosinate-ammonium | 0 | 100 ± 0 a | 89.33 ± 3.53 a | 118.67 ± 7.06 a |
| 0.3 | 85.33 ± 5.33 b | 24.00 ± 2.31 c | 33.33 ± 4.81 bc | |
| 0.4 | 54.67 ± 5.81 d | 13.33 ± 3.53 cd | 18.67 ± 4.81 cd | |
| 0.5 | 28.00 ± 4.62 f | 5.33 ± 3.53 de | 5.33 ± 3.53 de | |
| 0.6 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Types of Antibiotics | Concentration of Antibiotic (mg·L−1) | Frequency of Rooting (%) |
|---|---|---|
| Kanamycin | 0.00 | 100 ± 0 a |
| 6.00 | 33.3 ± 6.67 c | |
| 8.00 | 26.7 ± 6.67 c | |
| 10.00 | 14.4 ± 6.67 c | |
| 12.00 | 0 ± 0 | |
| Hygromycin | 0.00 | 100 ± 0 a |
| 1.50 | 13.3 ± 6.67 c | |
| 2.00 | 0 ± 0 | |
| 2.50 | 0 ± 0 | |
| 3.00 | 0 ± 0 | |
| Glufosinate-ammonium | 0.00 | 100 ± 0 a |
| 0.20 | 80 ± 11.55 b | |
| 0.30 | 60 ± 0 b | |
| 0.40 | 33.3 ± 6.67 c | |
| 0.50 | 0 ± 0 |
| Code | 6-BA (mg·L−1) | NAA (mg·L−1) | Budding Leaf Disc Rate (%) | Budding Rate (%) |
|---|---|---|---|---|
| M1 | 1 | 0.2 | 96.00 ± 2.31 a | 162.67 ± 16.38 a |
| M2 | 1 | 0.5 | 70.67 ± 9.33 bc | 92.00 ± 10.58 bc |
| M3 | 1 | 0.8 | 13.33 ± 3.53 f | 16.00 ± 4.00 e |
| M4 | 2 | 0.2 | 61.33 ± 8.74 bcd | 97.33 ± 20.95 bc |
| M5 | 2 | 0.5 | 78.67 ± 8.74 ab | 124.00 ± 6.93 b |
| M6 | 2 | 0.8 | 52.00 ± 2.31 cd | 90.67 ± 4.81 bc |
| M7 | 3 | 0.2 | 14.67 ± 1.33 f | 16.00 ± 0 e |
| M8 | 3 | 0.5 | 29.33 ± 9.61 ef | 41.33 ± 8.11 de |
| M9 | 3 | 0.8 | 42.67 ± 9.33 de | 61.33 ± 15.03 cd |
| Primer Name | Sequence (5′–3′) | Amplicon Length | Application |
|---|---|---|---|
| GUS/NPTII-F | GGGCAACAAGCCGAAAGA | 252 bp | Evaluate transgenic plants of Gojo-0 |
| GUS/NPTII-R | TGAGCGTCGCAGAACATTACAT | ||
| pG10-Backbone-F | GCCTTCTTGACGAGTTCTTCTGA | 505 bp | Evaluate pG10-RUBY vector backbone |
| pG10-Backbone-R | CATGCTACCCTCCGCGAGA | ||
| Cse2.0_LG3-F | AGGGTGGTGGTCATTTAGCCT | ~700 bp | Evaluate insertion site of line 2 |
| pG10-LB-R | TGGGCTGACCGCTTCCTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhang, J.; Li, P.; Gao, Y.; Yu, Q.; Sun, D.; Zhang, L.; Wang, S.; Tian, J.; Wang, Z.; et al. Development of a Transformation System and Locus Identification Pipeline for T-DNA in Chrysanthemum seticuspe, A Model Species for Hexaploid Cultivated Chrysanthemum. Int. J. Mol. Sci. 2022, 23, 11426. https://doi.org/10.3390/ijms231911426
Zhang J, Zhang J, Li P, Gao Y, Yu Q, Sun D, Zhang L, Wang S, Tian J, Wang Z, et al. Development of a Transformation System and Locus Identification Pipeline for T-DNA in Chrysanthemum seticuspe, A Model Species for Hexaploid Cultivated Chrysanthemum. International Journal of Molecular Sciences. 2022; 23(19):11426. https://doi.org/10.3390/ijms231911426
Chicago/Turabian StyleZhang, Jiali, Jing Zhang, Peiling Li, Yuan Gao, Qi Yu, Daojin Sun, Lingling Zhang, Siqi Wang, Jing Tian, Zhenxing Wang, and et al. 2022. "Development of a Transformation System and Locus Identification Pipeline for T-DNA in Chrysanthemum seticuspe, A Model Species for Hexaploid Cultivated Chrysanthemum" International Journal of Molecular Sciences 23, no. 19: 11426. https://doi.org/10.3390/ijms231911426





