An Atlas of Thyroid Hormone Receptors’ Target Genes in Mouse Tissues
Abstract
1. Introduction
2. Results
2.1. Definition of TR Target Genes
- (1)
- Genes that are upregulated after T3 treatment of hypothyroid mice. Initial T3 depletion is important because some genes are more sensitive to it than to an excess of T3 [14]. Hypothyroidism is obtained most of the time by pharmacological means with at least two weeks of treatment with propyl-thio-uracyl (PTU) or methimazole (MMI). In vitro, serum used for cell cultures should be depleted of T3. When a time-course analysis is performed, a rapid response, within hours, is a good indication that the transcriptional activation is a direct consequence of TR-mediated regulation;
- (2)
- Regulatory sequences are occupied by either TRα1 or TRβ1/2, as evidenced by immunoprecipitation of chromatin. Until now, the difficulty of raising high-quality antibodies against TR, which are not abundant proteins, has limited the production of ChIP-seq datasets for the liver [15,16]. This technical limitation is now commonly circumvented by using tagged receptors, for which several transgenic mice have been produced [17,18]. The availability of a ‘floxed’ construct [19] allows us to address chromatin occupancy by TRα1 in a single cell type within a heterogeneous tissue.
2.2. Presentation of the Atlas
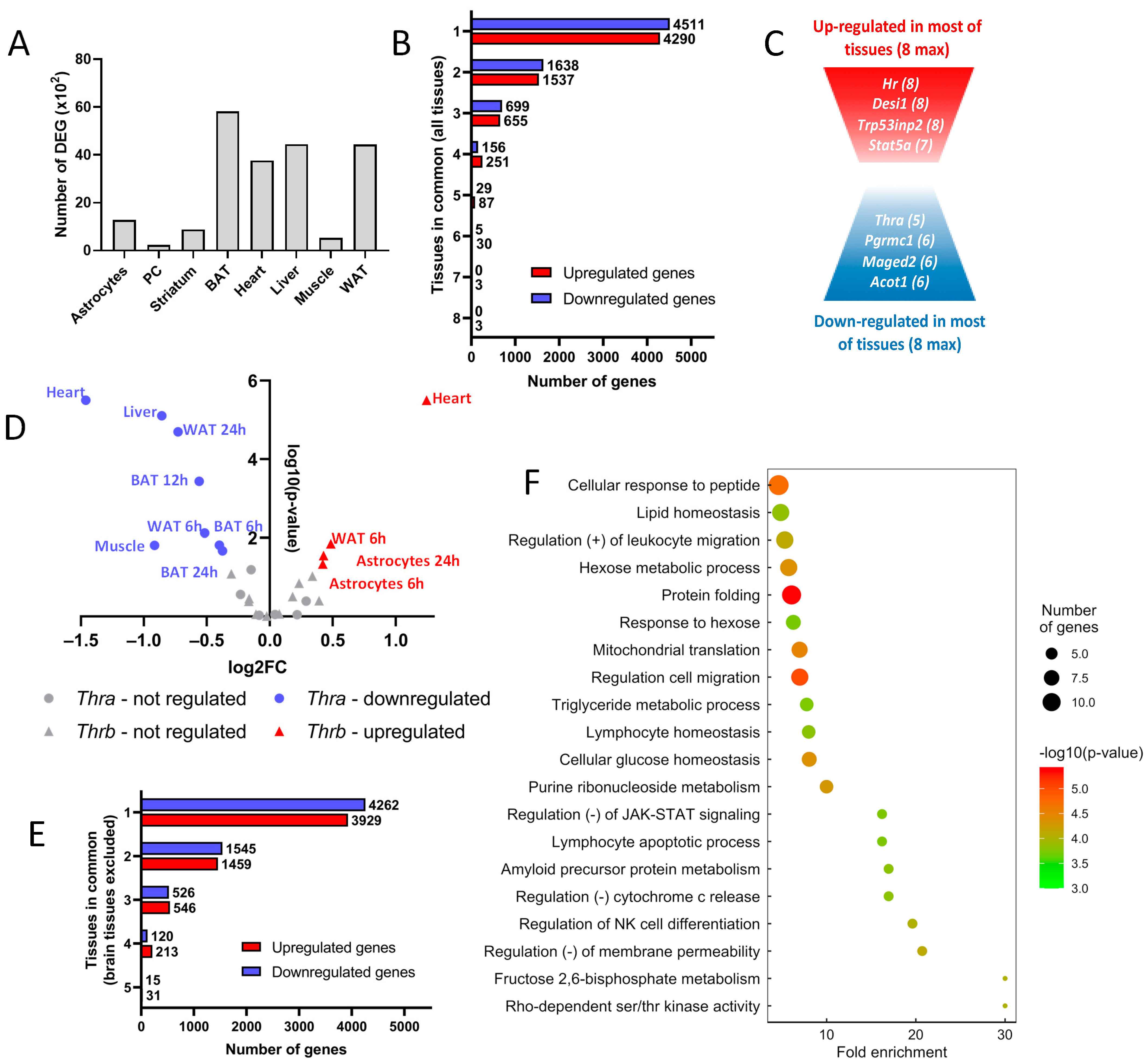
2.3. Differentially Expressed Genes
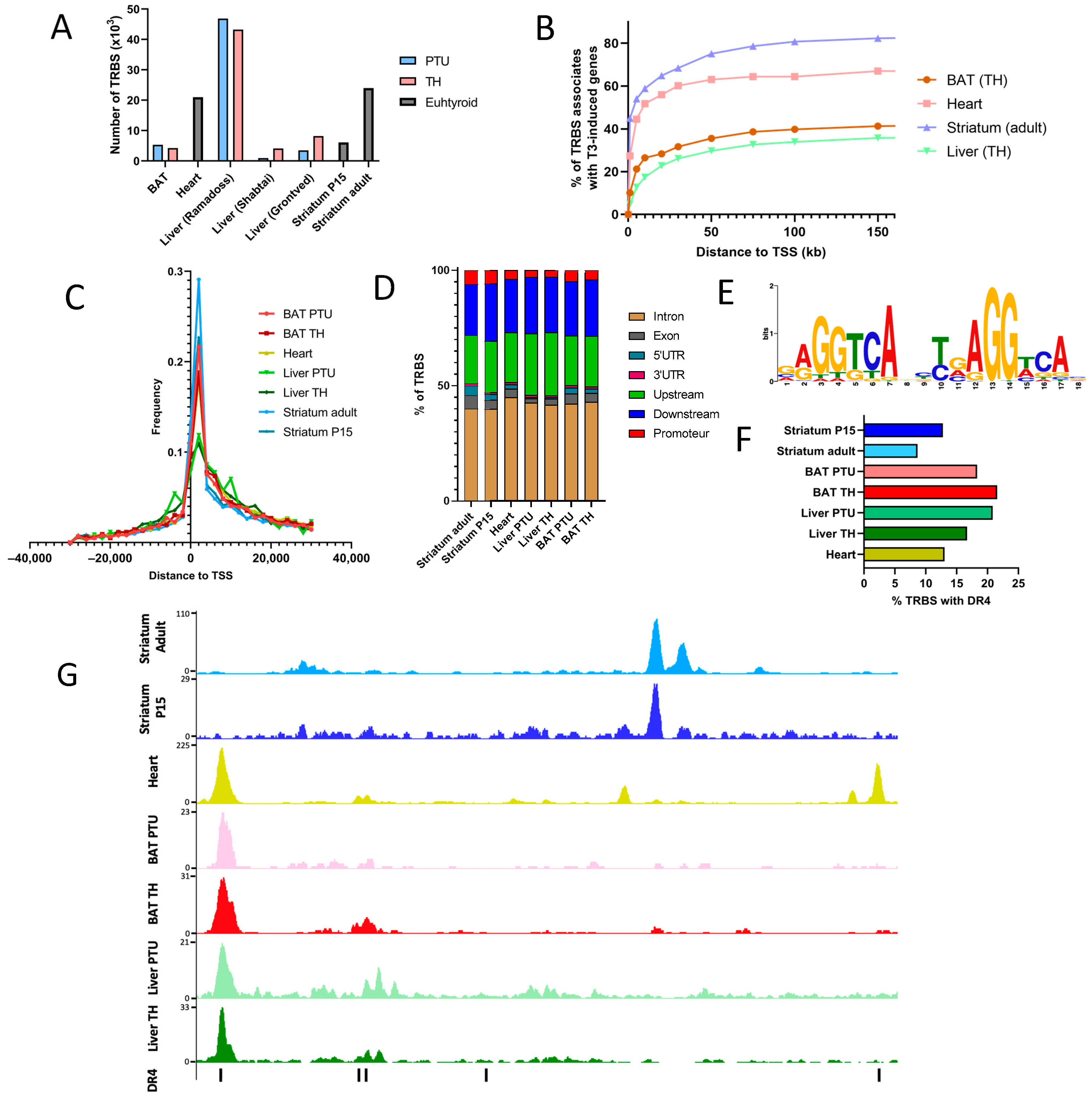
2.4. Chromatin Occupancy by TR
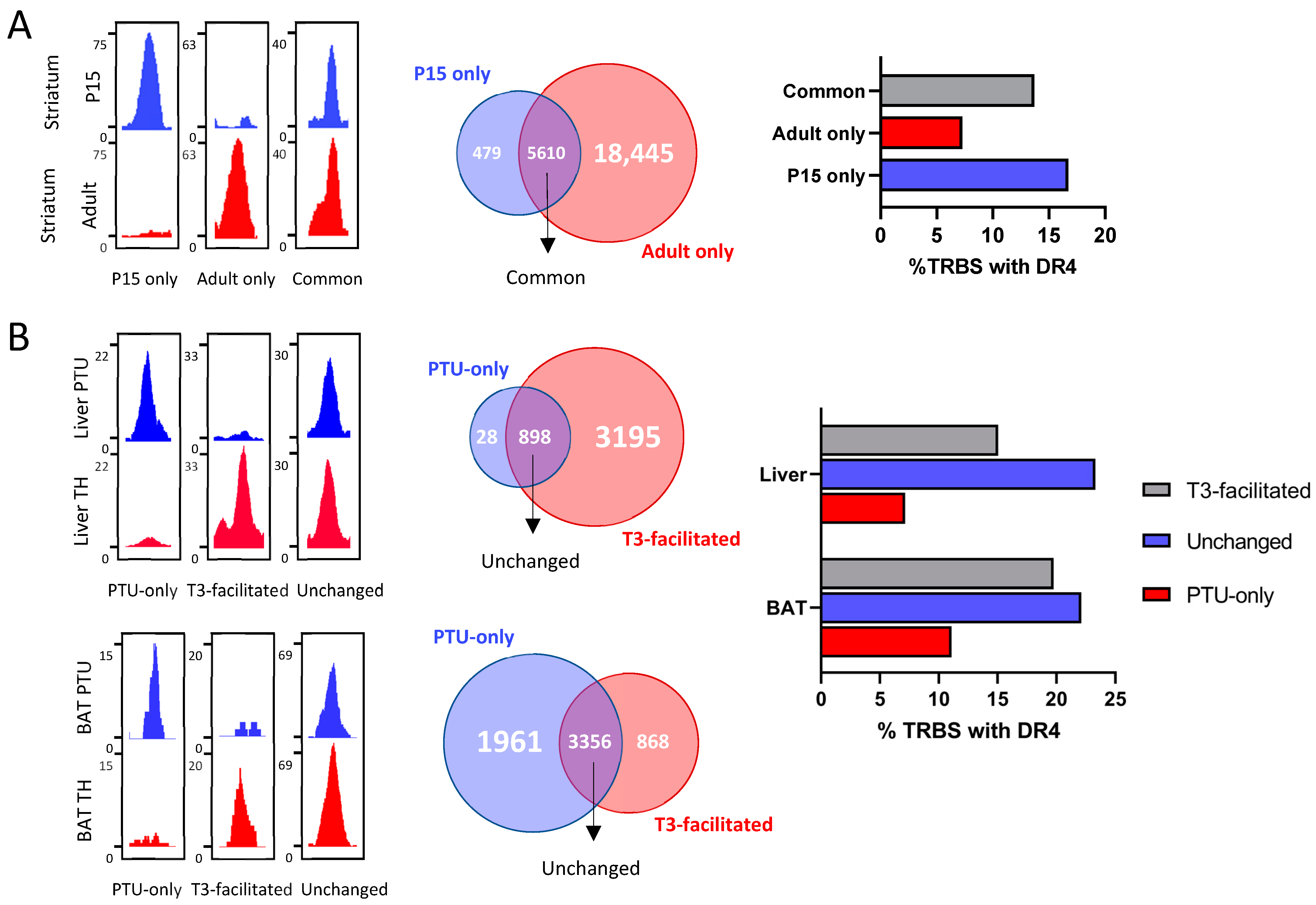
2.5. Evolution of Chromatin Occupancy during Cell Differentiation and after T3 Treatment
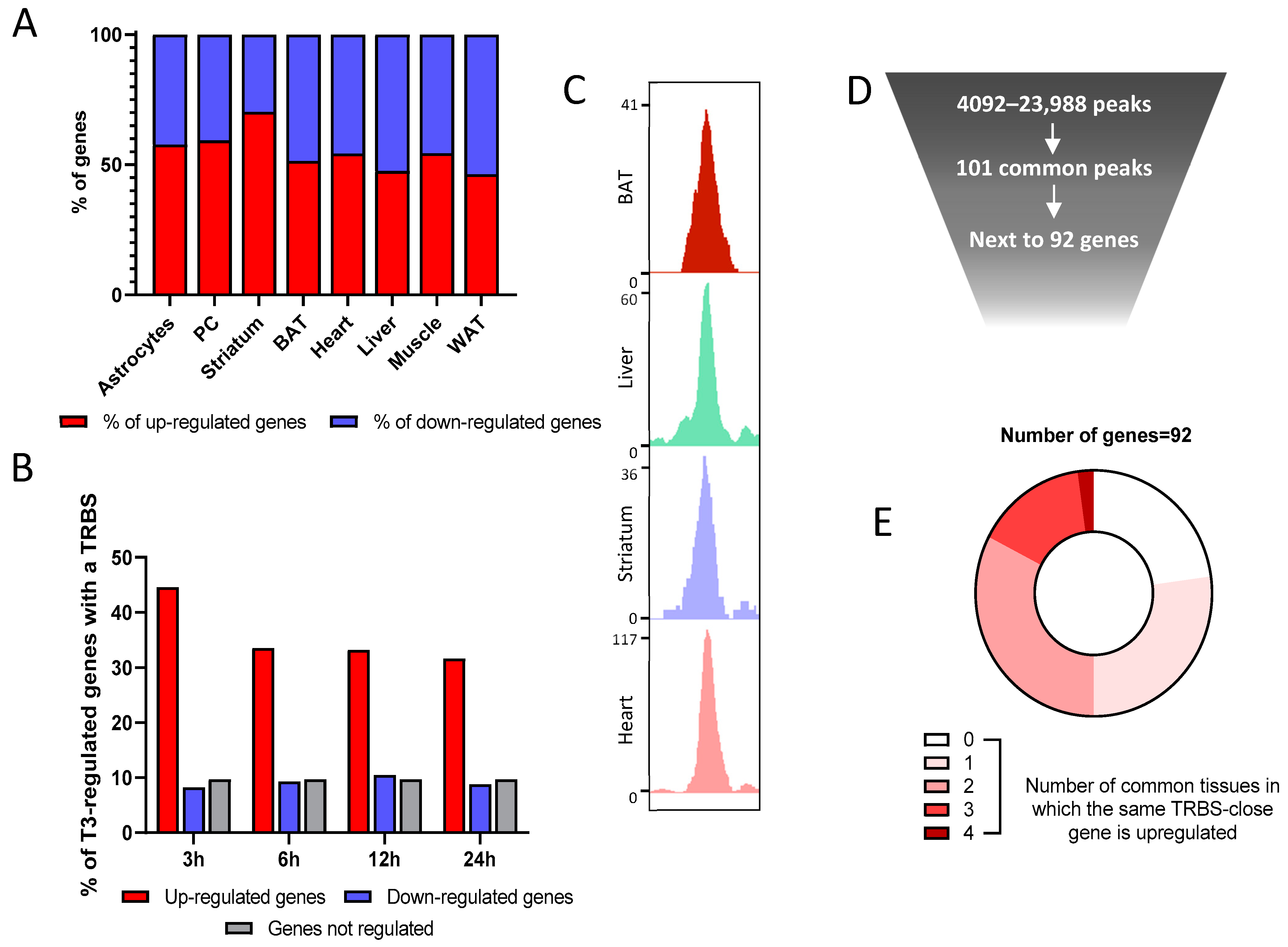
2.6. Combination of Transcriptome and Cistrome Data
3. Discussion
- (1)
- Many TR target genes encode extracellular proteins: growth factors, neurotrophins, etc. Therefore, in a tissue, the cellular response to these factors is difficult to separate from a direct, cell-autonomous response to T3. This has been notably exemplified in the brain, where interaction between neighboring cell types is a common theme [20,36]. In mice, the availability of Thra and Thrb “floxed” alleles enables us to address this point. TR target genes are expected to be downregulated when the T3 response is selectively blocked in the cell type considered. This provides an additional and useful criterion to define TR target genes [20,37];
- (2)
- Cross-species comparisons among mammals or vertebrates could outline gene regulations that are the most relevant for the conserved function of T3. Only a few attempts have been made in this direction, but the regulation of Klf9 by TR is clearly conserved between mammals and amphibians [38] and crucial for neuronal maturation [39];
- (3)
- Other genomic assays are required to address the influence of chromatin remodeling on cistrome and define the consequences of TR binding, especially on histone tail modifications [40]. This should help to address the possibility that there are many “silent TRBS”;
- (4)
- The development of high-throughput functional assays is needed to demonstrate the ability of TRBS to mediate T3 transactivation. Recently, a proof of principle has been obtained that the so-called synthetic STARR-seq approach can address this question in vitro [28].
4. Materials and Methods
4.1. Transcriptome Analysis
4.2. Cistrome Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tata, J.R. Amphibian Metamorphosis as a Model for the Developmental Actions of Thyroid Hormone. Mol. Cell. Endocrinol. 2006, 246, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Gandrillon, O.; Ferrand, N.; Michaille, J.J.; Roze, L.; Zile, M.H.; Samarut, J. C-ErbA Alpha/T3R and RARs Control Commitment of Hematopoietic Self-Renewing Progenitor Cells to Apoptosis or Differentiation and Are Antagonized by the v-ErbA Oncogene. Oncogene 1994, 9, 749–758. [Google Scholar] [PubMed]
- Robson, H.; Siebler, T.; Stevens, D.A.; Shalet, S.M.; Williams, G.R. Thyroid Hormone Acts Directly on Growth Plate Chondrocytes to Promote Hypertrophic Differentiation and Inhibit Clonal Expansion and Cell Proliferation. Endocrinology 2000, 141, 3887–3897. [Google Scholar] [CrossRef] [PubMed]
- Billon, N.; Tokumoto, Y.; Forrest, D.; Raff, M. Role of Thyroid Hormone Receptors in Timing Oligodendrocyte Differentiation. Dev. Biol. 2001, 235, 110–120. [Google Scholar] [CrossRef]
- Boukhtouche, F.; Brugg, B.; Wehrlé, R.; Bois-Joyeux, B.; Danan, J.-L.; Dusart, I.; Mariani, J. Induction of Early Purkinje Cell Dendritic Differentiation by Thyroid Hormone Requires RORα. Neural Dev. 2010, 5, 18. [Google Scholar] [CrossRef]
- Wrutniak-Cabello, C.; Casas, F.; Cabello, G. Thyroid Hormone Action in Mitochondria. J. Mol. Endocrinol. 2001, 26, 67–77. [Google Scholar] [CrossRef]
- Flamant, F. Futures Challenges in Thyroid Hormone Signaling Research. Front. Endocrinol. (Lausanne) 2016, 7, 58. [Google Scholar] [CrossRef]
- Velasco, L.F.R.; Togashi, M.; Walfish, P.G.; Pessanha, R.P.; Moura, F.N.; Barra, G.B.; Nguyen, P.; Rebong, R.; Yuan, C.; Simeoni, L.A.; et al. Thyroid Hormone Response Element Organization Dictates the Composition of Active Receptor. J. Biol. Chem. 2007, 282, 12458–12466. [Google Scholar] [CrossRef]
- Yen, P.M. Physiological and Molecular Basis of Thyroid Hormone Action. Physiol. Rev. 2001, 81, 1097–1142. [Google Scholar] [CrossRef]
- Chatonnet, F.; Flamant, F.; Morte, B. A Temporary Compendium of Thyroid Hormone Target Genes in Brain. Biochim Biophys Acta 2015, 1849, 122–129. [Google Scholar] [CrossRef]
- Gil-Ibáñez, P.; Bernal, J.; Morte, B. Thyroid Hormone Regulation of Gene Expression in Primary Cerebrocortical Cells: Role of Thyroid Hormone Receptor Subtypes and Interactions with Retinoic Acid and Glucocorticoids. PLoS ONE 2014, 9, e91692. [Google Scholar] [CrossRef] [PubMed]
- Gil-Ibañez, P.; García-García, F.; Dopazo, J.; Bernal, J.; Morte, B. Global Transcriptome Analysis of Primary Cerebrocortical Cells: Identification of Genes Regulated by Triiodothyronine in Specific Cell Types. Cereb. Cortex 2017, 27, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Morte, B.; Gil-Ibáñez, P.; Bernal, J. Regulation of Gene Expression by Thyroid Hormone in Primary Astrocytes: Factors Influencing the Genomic Response. Endocrinology 2018, 159, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Yen, P.M.; Feng, X.; Flamant, F.; Chen, Y.; Walker, R.L.; Weiss, R.E.; Chassande, O.; Samarut, J.; Refetoff, S.; Meltzer, P.S. Effects of Ligand and Thyroid Hormone Receptor Isoforms on Hepatic Gene Expression Profiles of Thyroid Hormone Receptor Knockout Mice. EMBO Rep. 2003, 4, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Grøntved, L.; Waterfall, J.J.; Kim, D.W.; Baek, S.; Sung, M.-H.; Zhao, L.; Park, J.W.; Nielsen, R.; Walker, R.L.; Zhu, Y.J.; et al. Transcriptional Activation by the Thyroid Hormone Receptor through Ligand-Dependent Receptor Recruitment and Chromatin Remodelling. Nat. Commun. 2015, 6, 7048. [Google Scholar] [CrossRef] [PubMed]
- Hönes, G.S.; Rakov, H.; Logan, J.; Liao, X.-H.; Werbenko, E.; Pollard, A.S.; Præstholm, S.M.; Siersbæk, M.S.; Rijntjes, E.; Gassen, J.; et al. Noncanonical Thyroid Hormone Signaling Mediates Cardiometabolic Effects in Vivo. Proc. Natl. Acad. Sci. USA 2017, 114, E11323–E11332. [Google Scholar] [CrossRef]
- Minakhina, S.; Bansal, S.; Zhang, A.; Brotherton, M.; Janodia, R.; De Oliveira, V.; Tadepalli, S.; Wondisford, F.E. A Direct Comparison of Thyroid Hormone Receptor Protein Levels in Mice Provides Unexpected Insights into Thyroid Hormone Action. Thyroid 2020, 30, 1193–1204. [Google Scholar] [CrossRef]
- Shabtai, Y.; Nagaraj, N.K.; Batmanov, K.; Cho, Y.-W.; Guan, Y.; Jiang, C.; Remsberg, J.; Forrest, D.; Lazar, M.A. A Coregulator Shift, Rather than the Canonical Switch, Underlies Thyroid Hormone Action in the Liver. Genes Dev. 2021, 35, 367–378. [Google Scholar] [CrossRef]
- Hirose, K.; Payumo, A.Y.; Cutie, S.; Hoang, A.; Zhang, H.; Guyot, R.; Lunn, D.; Bigley, R.B.; Yu, H.; Wang, J.; et al. Evidence for Hormonal Control of Heart Regenerative Capacity during Endothermy Acquisition. Science 2019, 364, 184–188. [Google Scholar] [CrossRef]
- Richard, S.; Guyot, R.; Rey-Millet, M.; Prieux, M.; Markossian, S.; Aubert, D.; Flamant, F. A Pivotal Genetic Program Controlled by Thyroid Hormone during the Maturation of GABAergic Neurons. iScience 2020, 23, 100899. [Google Scholar] [CrossRef]
- Minerath, R.A.; Dewey, C.M.; Hall, D.D.; Grueter, C.E. Regulation of Cardiac Transcription by Thyroid Hormone and Med13. J. Mol. Cell. Cardiol. 2019, 129, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Nicolaisen, T.S.; Klein, A.B.; Dmytriyeva, O.; Lund, J.; Ingerslev, L.R.; Fritzen, A.M.; Carl, C.S.; Lundsgaard, A.-M.; Frost, M.; Ma, T.; et al. Thyroid Hormone Receptor α in Skeletal Muscle Is Essential for T3-Mediated Increase in Energy Expenditure. FASEB J. 2020, 34, 15480–15491. [Google Scholar] [CrossRef] [PubMed]
- Ramadoss, P.; Abraham, B.J.; Tsai, L.; Zhou, Y.; Costa-e-Sousa, R.H.; Ye, F.; Bilban, M.; Zhao, K.; Hollenberg, A.N. Novel Mechanism of Positive versus Negative Regulation by Thyroid Hormone Receptor Β1 (TRβ1) Identified by Genome-Wide Profiling of Binding Sites in Mouse Liver. J. Biol. Chem. 2014, 289, 1313–1328. [Google Scholar] [CrossRef]
- Thompson, C.C. Thyroid Hormone-Responsive Genes in Developing Cerebellum Include a Novel Synaptotagmin and a Hairless Homolog. J. Neurosci. 1996, 16, 7832–7840. [Google Scholar] [CrossRef]
- Tata, J.R. Autoinduction of Nuclear Hormone Receptors during Metamorphosis and Its Significance. Insect Biochem. Mol. Biol. 2000, 30, 645–651. [Google Scholar] [CrossRef]
- Rastinejad, F.; Perlmann, T.; Evans, R.M.; Sigler, P.B. Structural Determinants of Nuclear Receptor Assembly on DNA Direct Repeats. Nature 1995, 375, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Le Dily, F.; Beato, M. Signaling by Steroid Hormones in the 3D Nuclear Space. Int J. Mol. Sci 2018, 19, 306. [Google Scholar] [CrossRef]
- Flamant, F.; Zekri, Y.; Guyot, R. Functional Definition of Thyroid Hormone Response Elements Based on a Synthetic STARR-Seq Screen. Endocrinology 2022, 163, bqac084. [Google Scholar] [CrossRef]
- Emont, M.P.; Jacobs, C.; Essene, A.L.; Pant, D.; Tenen, D.; Colleluori, G.; Di Vincenzo, A.; Jørgensen, A.M.; Dashti, H.; Stefek, A.; et al. A Single-Cell Atlas of Human and Mouse White Adipose Tissue. Nature 2022, 603, 926–933. [Google Scholar] [CrossRef]
- Hoffman, J.A.; Papas, B.N.; Trotter, K.W.; Archer, T.K. Single-Cell RNA Sequencing Reveals a Heterogeneous Response to Glucocorticoids in Breast Cancer Cells. Commun. Biol. 2020, 3, 126. [Google Scholar] [CrossRef]
- Belton, J.-M.; McCord, R.P.; Gibcus, J.H.; Naumova, N.; Zhan, Y.; Dekker, J. Hi-C: A Comprehensive Technique to Capture the Conformation of Genomes. Methods 2012, 58, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Van de Werken, H.J.G.; Landan, G.; Holwerda, S.J.B.; Hoichman, M.; Klous, P.; Chachik, R.; Splinter, E.; Valdes-Quezada, C.; Oz, Y.; Bouwman, B.A.M.; et al. Robust 4C-Seq Data Analysis to Screen for Regulatory DNA Interactions. Nat. Methods 2012, 9, 969–972. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhang, Z.; Wang, K. 3C and 3C-Based Techniques: The Powerful Tools for Spatial Genome Organization Deciphering. Mol. Cytogenet. 2018, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Paquette, M.A.; Atlas, E.; Wade, M.G.; Yauk, C.L. Thyroid Hormone Response Element Half-Site Organization and Its Effect on Thyroid Hormone Mediated Transcription. PLoS ONE 2014, 9, e101155. [Google Scholar] [CrossRef] [PubMed]
- Chatonnet, F.; Guyot, R.; Benoît, G.; Flamant, F. Genome-Wide Analysis of Thyroid Hormone Receptors Shared and Specific Functions in Neural Cells. Proc. Natl. Acad. Sci. USA 2013, 110, E766–E775. [Google Scholar] [CrossRef]
- Fauquier, T.; Chatonnet, F.; Picou, F.; Richard, S.; Fossat, N.; Aguilera, N.; Lamonerie, T.; Flamant, F. Purkinje Cells and Bergmann Glia Are Primary Targets of the TRα1 Thyroid Hormone Receptor during Mouse Cerebellum Postnatal Development. Development 2014, 141, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Flamant, F.; Cheng, S.-Y.; Hollenberg, A.N.; Moeller, L.C.; Samarut, J.; Wondisford, F.E.; Yen, P.M.; Refetoff, S. Thyroid Hormone Signaling Pathways: Time for a More Precise Nomenclature. Endocrinology 2017, 158, 2052–2057. [Google Scholar] [CrossRef]
- Bagamasbad, P.D.; Bonett, R.M.; Sachs, L.; Buisine, N.; Raj, S.; Knoedler, J.R.; Kyono, Y.; Ruan, Y.; Ruan, X.; Denver, R.J. Deciphering the Regulatory Logic of an Ancient, Ultraconserved Nuclear Receptor Enhancer Module. Mol. Endocrinol. 2015, 29, 856–872. [Google Scholar] [CrossRef]
- Avci, H.X.; Lebrun, C.; Wehrlé, R.; Doulazmi, M.; Chatonnet, F.; Morel, M.-P.; Ema, M.; Vodjdani, G.; Sotelo, C.; Flamant, F.; et al. Thyroid Hormone Triggers the Developmental Loss of Axonal Regenerative Capacity via Thyroid Hormone Receptor A1 and Krüppel-like Factor 9 in Purkinje Cells. Proc. Natl. Acad. Sci. USA 2012, 109, 14206–14211. [Google Scholar] [CrossRef]
- Præstholm, S.M.; Siersbæk, M.S.; Nielsen, R.; Zhu, X.; Hollenberg, A.N.; Cheng, S.-Y.; Grøntved, L. Multiple Mechanisms Regulate H3 Acetylation of Enhancers in Response to Thyroid Hormone. PLoS Genet. 2020, 16, e1008770. [Google Scholar] [CrossRef]
- Zekri, Y.; Agnol, L.D.; Flamant, F. In Vitro Assessment of Pesticides Capacity to Act as Agonists/Antagonists of the Thyroid Hormone Nuclear Receptors. iScience 2021, 24, 102957. [Google Scholar] [CrossRef] [PubMed]
- Gil-Ibañez, P.; Morte, B.; Bernal, J. Role of Thyroid Hormone Receptor Subtypes α and β on Gene Expression in the Cerebral Cortex and Striatum of Postnatal Mice. Endocrinology 2013, 154, 1940–1947. [Google Scholar] [CrossRef] [PubMed]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif Analysis of Large DNA Datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for Occurrences of a given Motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
| Organ/Cell Type | RNA-Seq Datasets | ChIP-Seq Datasets | ||||
|---|---|---|---|---|---|---|
| Publication | Age | Hypothyroidism | Treatment | TR and Publication | Conditions | |
| Striatum | Unpublished GSE210976 | 8 w | - | T3/T4 unique IP injection 20 (T3) and 200 (T4) μg/mL 12 and 24 h | GS-TRα–adult GABAergic neurons Unpublished GSE210975 | 8 w Euthyroid |
| GS-TRα–P15 GABAergic neurons [20] GSE143933 | 8 w Euthyroid | |||||
| Astrocytes (in vitro) | [13] GSE110372 | P3 | TH-deprived serum for 24 h | T3 culture medium 10 nM 6 and 24 h | - | |
| PC cells (in vitro) | [12] GSE68949 | E17.5 | TH-deprived serum for 24 h | T3 culture medium 10 nM 24 h | - | |
| Heart | [21] GSE124117 | 8 w | PTU in food (4 weeks) | T3 daily IP injection 1 ug/g of BW Achieved 72 h | GS-TRα Cardiomyocytes [19] GSE125414 | P14 Euthyroid |
| Skeletal muscle | [22] GSE146336 | Adult | - | T3 daily SC injection 100 nmol/kg BW 7 days | - | |
| BAT | Unpublished GSE201136 | 8 w | PTU in food (2 weeks) | T3/T4 unique IP injection 20 (T3) and 200 (T4) μg/mL 3, 6, 12 and 24 h | GS-TRα Brown adipocytes Unpublished GSE201136 | 8 w PTU and PTU + T3/T4 |
| WAT | Unpublished GSE210976 | 8 w | PTU in food (2 weeks) | T3/T4 unique IP injection 20 (T3) and 200 (T4) μg/mL 6 and 24 h | - | |
| Liver | [18] GSE159648 | 12–14 w | MMI in drinking water (4 weeks) | T3 in drinking water 5 μg/mL 7 days | TRβ1 [15] GSE65947 | Adult PTU and PTU + T3 |
| Biotin-TRβ1 [23] GSE52613 | Adult PTU and PTU + T3 | |||||
| HA-TRβ1 [18] GSE159648 | Adult PTU and PTU + T3 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zekri, Y.; Guyot, R.; Flamant, F. An Atlas of Thyroid Hormone Receptors’ Target Genes in Mouse Tissues. Int. J. Mol. Sci. 2022, 23, 11444. https://doi.org/10.3390/ijms231911444
Zekri Y, Guyot R, Flamant F. An Atlas of Thyroid Hormone Receptors’ Target Genes in Mouse Tissues. International Journal of Molecular Sciences. 2022; 23(19):11444. https://doi.org/10.3390/ijms231911444
Chicago/Turabian StyleZekri, Yanis, Romain Guyot, and Frédéric Flamant. 2022. "An Atlas of Thyroid Hormone Receptors’ Target Genes in Mouse Tissues" International Journal of Molecular Sciences 23, no. 19: 11444. https://doi.org/10.3390/ijms231911444
APA StyleZekri, Y., Guyot, R., & Flamant, F. (2022). An Atlas of Thyroid Hormone Receptors’ Target Genes in Mouse Tissues. International Journal of Molecular Sciences, 23(19), 11444. https://doi.org/10.3390/ijms231911444





