Cultivation of Cryopreserved Human Dental Pulp Stem Cells—A New Approach to Maintaining Dental Pulp Tissue
Abstract
1. Introduction
2. Results
2.1. Analysis of Results
2.1.1. The Impact of the Novel Cryopreservation Method on Cell Multiplication and Expansion
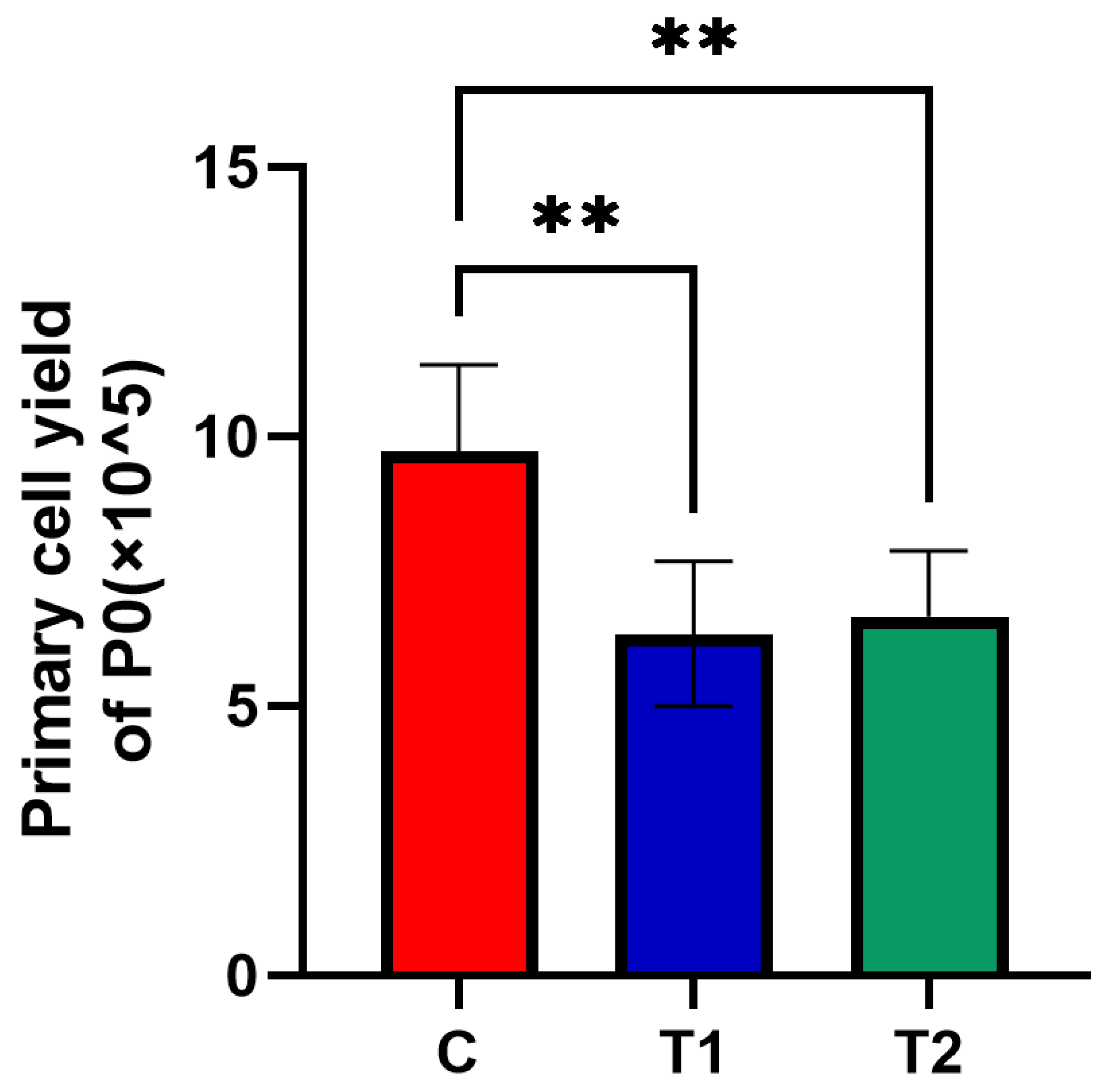
2.1.2. Effects of New Cryopreservation Strategy on the Primary Cell Yield
2.1.3. Identification of Specific Stem-Cell Markers
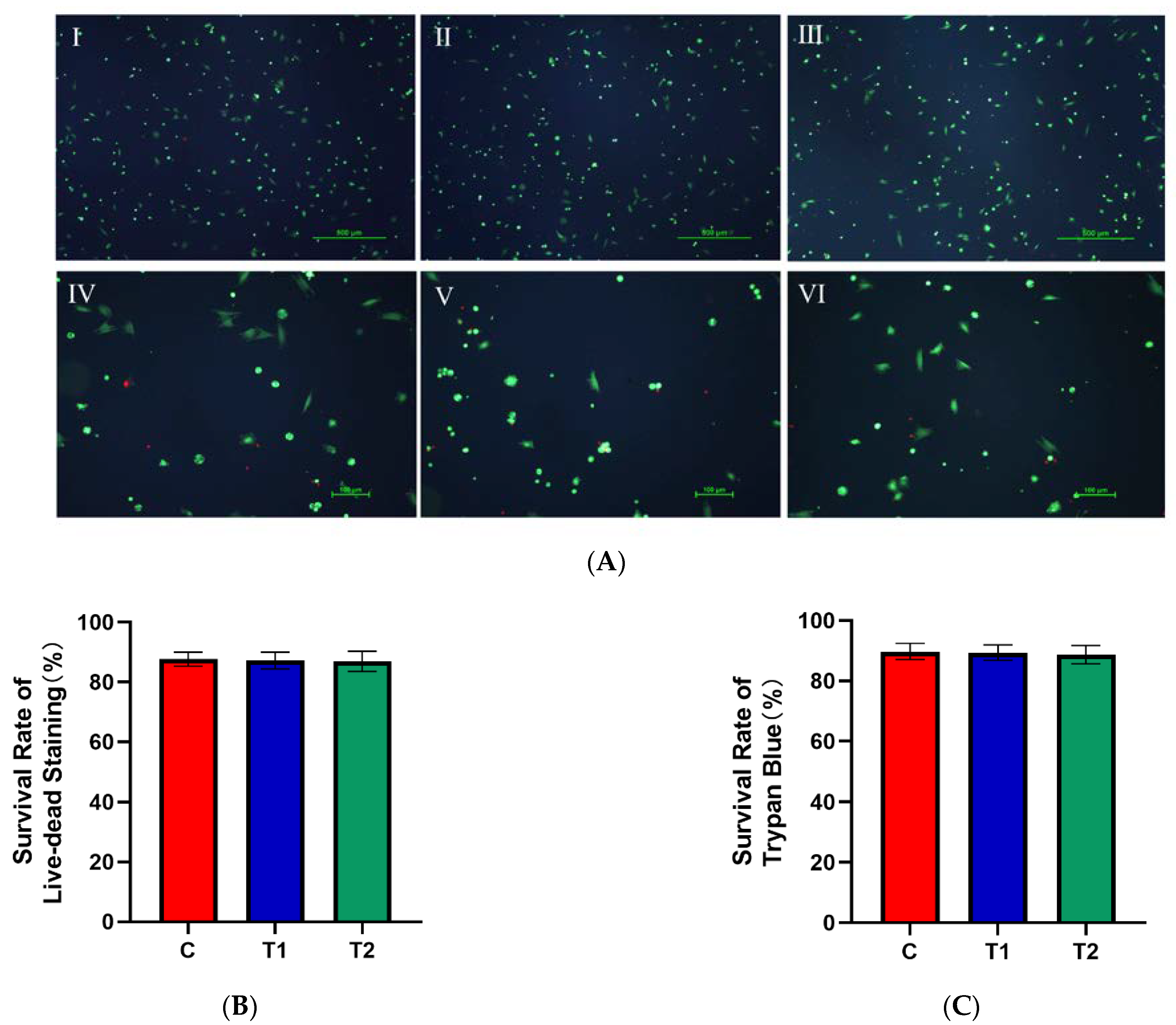
2.1.4. Effects of New Cryopreservation Strategy on the Cell Survival Rate after Trypan Blue and Live–Dead Staining
2.1.5. Effects of New Cryopreservation Strategy on the Primary Cell Proliferation
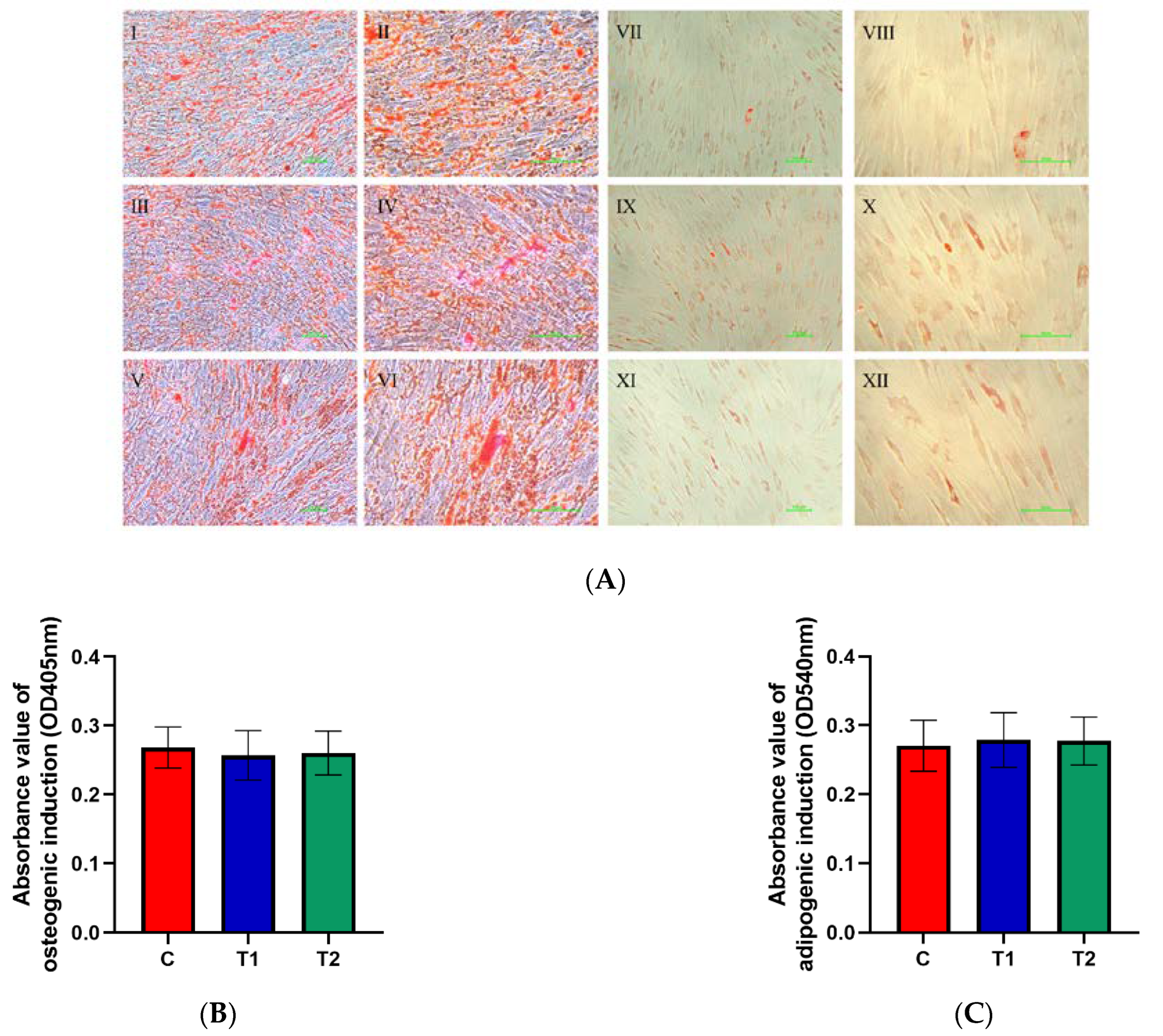
2.1.6. Effects of New Cryopreservation Strategy on the Differentiation Potential of Dental Pulp Cells
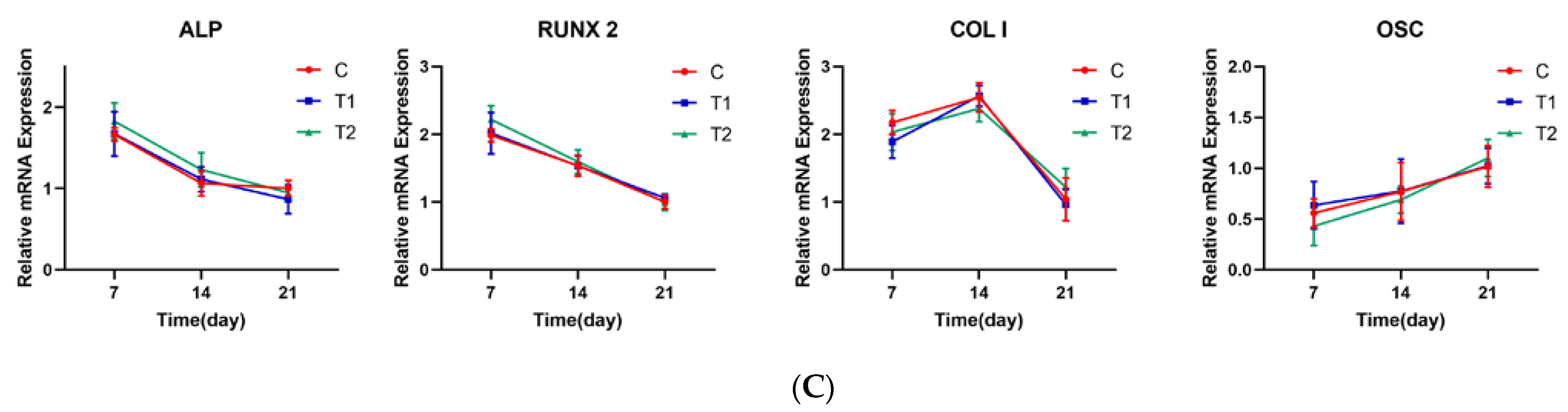
2.1.7. Representative Gene Expression Profile of LPL, PPARG, ALP, RUNX2, COL I and OSC in Each Group
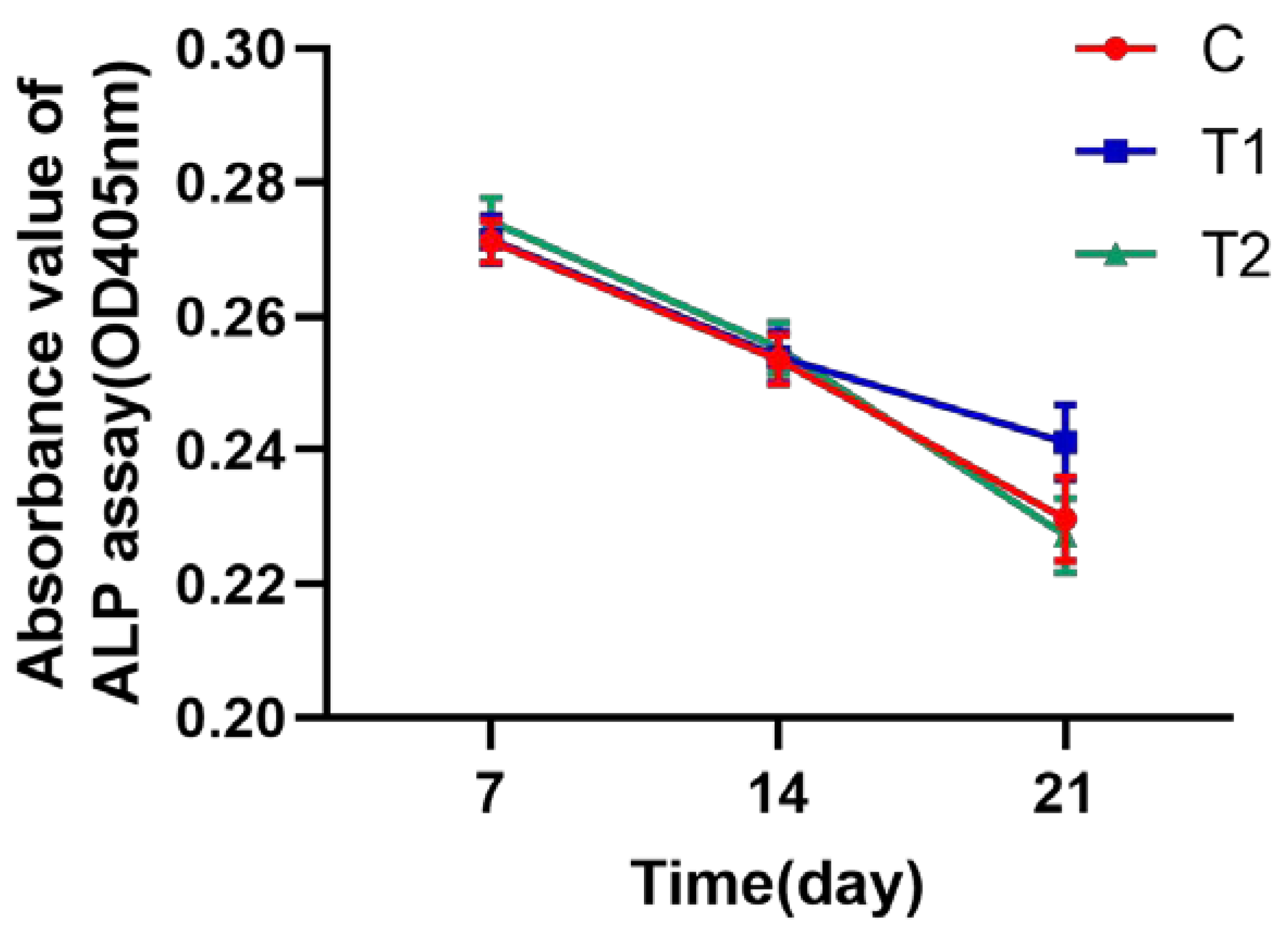
2.1.8. ALP Assay Test to Identify the Osteogenic Activity of hDPSCs
3. Discussion
4. Materials and Methods
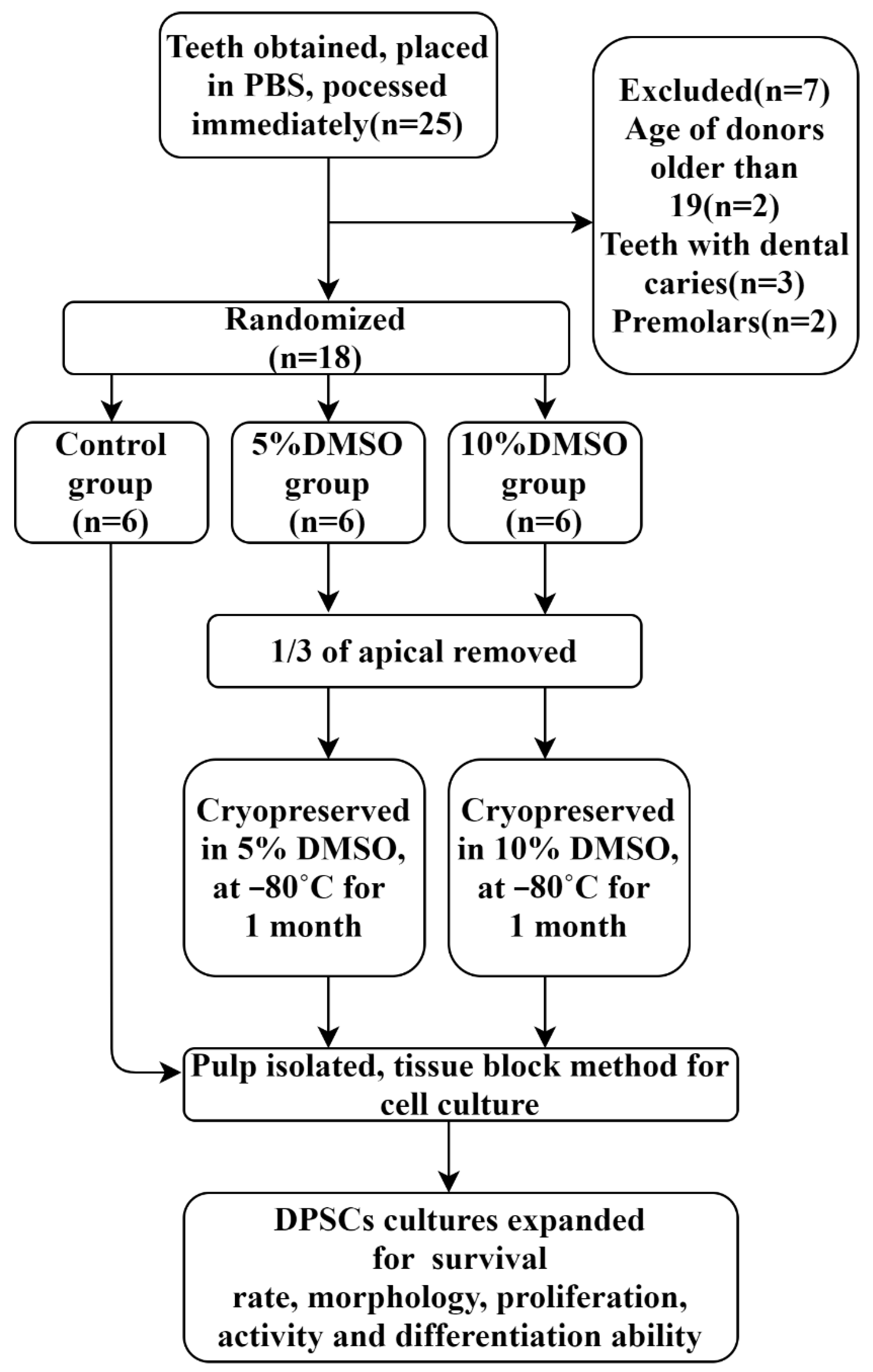
4.1. Collection of Samples
4.2. Cryopreservation
4.3. Culture of hDPSCs
4.4. Assessment of Primary Cellular Morphology and Recording of Primary Cell Growth Time
4.5. Cell Yield Computation
4.6. Flow Cytometry
4.7. Colony-Forming Efficiency
4.8. Cell Survival Rate
4.9. Proliferation Testing with MTS Assay
4.10. Differentiation Potential Assessment
4.11. Osteogenic Activity with ALP Assay
4.12. Gene Expression Detection
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoang, D.M.; Pham, P.T.; Bach, T.Q.; Ngo, A.T.L.; Nguyen, Q.T.; Phan, T.T.K.; Nguyen, G.H.; Le, P.T.T.; Hoang, V.T.; Forsyth, N.R.; et al. Stem cell-based therapy for human diseases. Signal Transduct. Target. Ther. 2022, 7, 272. [Google Scholar] [CrossRef]
- Gronthos, S.; Brahim, J.; Li, W.; Fisher, L.W.; Cherman, N.; Boyde, A.; DenBesten, P.; Robey, P.G.; Shi, S. Stem Cell Properties of Human Dental Pulp Stem Cells. J. Dent. Res. 2002, 81, 531–535. [Google Scholar] [CrossRef]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef]
- Huang, G.T.-J.; Gronthos, S.; Shi, S. Mesenchymal Stem Cells Derived from Dental Tissues vs. Those from Other Sources: Their Biology and Role in Regenerative Medicine. J. Dent. Res. 2009, 88, 792–806. [Google Scholar] [CrossRef]
- La Noce, M.; Stellavato, A.; Vassallo, V.; Cammarota, M.; Laino, L.; Desiderio, V.; Del Vecchio, V.; Nicoletti, G.F.; Tirino, V.; Papaccio, G.; et al. Hyaluronan-Based Gel Promotes Human Dental Pulp Stem Cells Bone Differentiation by Activating YAP/TAZ Pathway. Cells 2021, 10, 2899. [Google Scholar] [CrossRef]
- Kawashima, N.; Noda, S.; Yamamoto, M.; Okiji, T. Properties of Dental Pulp–derived Mesenchymal Stem Cells and the Effects of Culture Conditions. J. Endod. 2017, 43, S31–S34. [Google Scholar] [CrossRef]
- Jo, Y.-Y.; Lee, H.-J.; Kook, S.-Y.; Choung, H.-W.; Park, J.-Y.; Chung, J.-H.; Choung, Y.-H.; Kim, E.-S.; Yang, H.-C.; Choung, P.-H. Isolation and Characterization of Postnatal Stem Cells from Human Dental Tissues. Tissue Eng. 2007, 13, 767–773. [Google Scholar] [CrossRef]
- Wei, X.; Ling, J.; Wu, L.; Liu, L.; Xiao, Y. Expression of Mineralization Markers in Dental Pulp Cells. J. Endod. 2007, 33, 703–708. [Google Scholar] [CrossRef]
- Chang, C.-C.; Chang, K.-C.; Tsai, S.-J.; Chang, H.-H.; Lin, C.-P. Neurogenic differentiation of dental pulp stem cells to neuron-like cells in dopaminergic and motor neuronal inductive media. J. Formos. Med. Assoc. 2014, 113, 956–965. [Google Scholar] [CrossRef]
- Huang, G.T.-J.; Shagramanova, K.; Chan, S.W. Formation of Odontoblast-Like Cells from Cultured Human Dental Pulp Cells on Dentin In Vitro. J. Endod. 2006, 32, 1066–1073. [Google Scholar] [CrossRef]
- Tatullo, M.; Marrelli, M.; Shakesheff, K.M.; White, L.J. Dental pulp stem cells: Function, isolation and applications in regenerative medicine. J. Tissue Eng. Regen. Med. 2015, 9, 1205–1216. [Google Scholar] [CrossRef]
- Karaöz, E.; Demircan, P.C.; Sağlam, Ö.; Aksoy, A.; Kaymaz, F.; Duruksu, G. Human dental pulp stem cells demonstrate better neural and epithelial stem cell properties than bone marrow-derived mesenchymal stem cells. Histochem. Cell Biol. 2011, 136, 455–473. [Google Scholar] [CrossRef]
- Govindasamy, V.; Ronald, V.S.; Abdullah, A.N.; Nathan, K.G.; Aziz, Z.A.C.A.; Abdullah, M.; Musa, S.; Kasim, N.H.A.; Bhonde, R.R. Differentiation of Dental Pulp Stem Cells into Islet-like Aggregates. J. Dent. Res. 2011, 90, 646–652. [Google Scholar] [CrossRef]
- Dasari, V.R.; Veeravalli, K.K.; Dinh, D.H. Mesenchymal stem cells in the treatment of spinal cord injuries: A review. World J. Stem Cells 2014, 6, 120–133. [Google Scholar] [CrossRef]
- Király, M.; Porcsalmy, B.; Pataki, A.; Kádár, K.; Jelitai, M.; Molnár, B.; Hermann, P.; Gera, I.; Grimm, W.-D.; Ganss, B.; et al. Simultaneous PKC and cAMP activation induces differentiation of human dental pulp stem cells into functionally active neurons. Neurochem. Int. 2009, 55, 323–332. [Google Scholar] [CrossRef]
- Ikeda, E.; Yagi, K.; Kojima, M.; Yagyuu, T.; Ohshima, A.; Sobajima, S.; Tadokoro, M.; Katsube, Y.; Isoda, K.; Kondoh, M.; et al. Multipotent cells from the human third molar: Feasibility of cell-based therapy for liver disease. Differentiation 2008, 76, 495–505. [Google Scholar] [CrossRef]
- Taghipour, Z.; Karbalaie, K.; Kiani, A.; Niapour, A.; Bahramian, H.; Nasr-Esfahani, M.H.; Baharvand, H. Transplantation of Undifferentiated and Induced Human Exfoliated Deciduous Teeth-Derived Stem Cells Promote Functional Recovery of Rat Spinal Cord Contusion Injury Model. Stem Cells Dev. 2012, 21, 1794–1802. [Google Scholar] [CrossRef]
- Sakai, K.; Yamamoto, A.; Matsubara, K.; Nakamura, S.; Naruse, M.; Yamagata, M.; Sakamoto, K.; Tauchi, R.; Wakao, N.; Imagama, S.; et al. Human dental pulp-derived stem cells promote locomotor recovery after complete transection of the rat spinal cord by multiple neuro-regenerative mechanisms. J. Clin. Investig. 2012, 122, 80–90. [Google Scholar] [CrossRef]
- D’Aquino, R.; De Rosa, A.; Laino, G.; Caruso, F.; Guida, L.; Rullo, R.; Checchi, V.; Laino, L.; Tirino, V.; Papaccio, G. Human dental pulp stem cells: From biology to clinical applications. J. Exp. Zoöl. Part B Mol. Dev. Evol. 2008, 312B, 408–415. [Google Scholar] [CrossRef]
- Kerkis, I.; Ambrosio, C.E.; Kerkis, A.; Martins, D.S.; Zucconi, E.; Fonseca, S.A.S.; Cabral, R.M.; Maranduba, C.M.C.; Gaiad, T.P.; Morini, A.C.; et al. Early transplantation of human immature dental pulp stem cells from baby teeth to golden retriever muscular dystrophy (GRMD) dogs: Local or systemic? J. Transl. Med. 2008, 6, 35. [Google Scholar] [CrossRef]
- Yamamura, Y.; Yamada, H.; Sakurai, T.; Ide, F.; Inoue, H.; Muramatsu, T.; Mishima, K.; Hamada, Y.; Saito, I. Treatment of salivary gland hypofunction by transplantation with dental pulp cells. Arch. Oral Biol. 2013, 58, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Pilbauerova, N.; Schmidt, J.; Soukup, T.; Ivancakova, R.K.; Suchanek, J. The Effects of Cryogenic Storage on Human Dental Pulp Stem Cells. Int. J. Mol. Sci. 2021, 22, 4432. [Google Scholar] [CrossRef]
- Ferrúa, C.P.; Centeno, E.G.Z.; da Rosa, L.C.; Amaral, C.C.D.; Severo, R.F.; Sarkis-Onofre, R.; Nascimento, G.G.; Cordenonzi, G.; Bast, R.K.; Demarco, F.F.; et al. How has dental pulp stem cells isolation been conducted? A scoping review. Braz. Oral Res. 2017, 31, e87. [Google Scholar] [CrossRef]
- Collart-Dutilleul, P.-Y.; Chaubron, F.; De Vos, J.; Cuisinier, F.J. Allogenic banking of dental pulp stem cells for innovative therapeutics. World J. Stem Cells 2015, 7, 1010–1021. [Google Scholar]
- Hilkens, P.; Driesen, R.B.; Wolfs, E.; Gervois, P.; Vangansewinkel, T.; Ratajczak, J.; Dillen, Y.; Bronckaers, A.; Lambrichts, I. Cryopreservation and Banking of Dental Stem Cells. Biobanking Cryopreserv. Stem Cells 2016, 951, 199–235. [Google Scholar]
- Perry, B.C.; Zhou, D.; Wu, X.; Yang, F.-C.; Byers, M.A.; Chu, T.-M.G.; Hockema, J.J.; Woods, E.J.; Goebel, W.S. Collection, cryopreservation, and characterization of human dental pulp-derived mesenchymal stem cells for banking and clinical use. Tissue Eng. Part C Methods 2008, 14, 149–156. [Google Scholar] [CrossRef]
- Zambelli, A.; Poggi, G.; Da Prada, G.; Pedrazzoli, P.; Cuomo, A.; Miotti, D.; Perotti, C.; Preti, P.; Della Cuna, G.R. Clinical toxicity of cryopreserved circulating progenitor cells infusion. Anticancer Res. 1999, 18, 4705–4708. [Google Scholar]
- Sato, M.; Kawase-Koga, Y.; Yamakawa, D.; Fujii, Y.; Chikazu, D. Bone Regeneration Potential of Human Dental Pulp Stem Cells Derived from Elderly Patients and Osteo-Induced by a Helioxanthin Derivative. Int. J. Mol. Sci. 2020, 21, 7731. [Google Scholar] [CrossRef]
- Graziano, A.; Papaccio, G.; Laino, G.; Graziano, A. Dental pulp stem cells: A promising tool for bone regeneration. Stem Cell Rev. 2008, 4, 21–26. [Google Scholar] [CrossRef]
- Liu, L.; Rando, T.A. Manifestations and mechanisms of stem cell aging. J. Cell Biol. 2011, 193, 257–266. [Google Scholar] [CrossRef]
- Alt, E.U.; Senst, C.; Murthy, S.N.; Slakey, D.P.; Dupin, C.L.; Chaffin, A.E.; Kadowitz, P.J.; Izadpanah, R. Aging alters tissue resident mesenchymal stem cell properties. Stem Cell Res. 2012, 8, 215–225. [Google Scholar] [PubMed]
- Wu, W.; Niklason, L.; Steinbacher, D.M. The Effect of Age on Human Adipose-Derived Stem Cells. Plast. Reconstr. Surg. 2013, 131, 27–37. [Google Scholar]
- Smith, J.A.; Daniel, R. Stem cells and aging: A chicken-or-the-egg issue? Aging Dis. 2012, 3, 260–268. [Google Scholar]
- Murray, P.E.; Stanley, H.R.; Matthews, J.B.; Sloan, A.J.; Smith, A.J. Age-related odontometric changes of human teeth. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontology 2002, 93, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Nada, O.A.; Kluwe, L.; Gosau, M.; Smeets, R.; Friedrich, R.E. Expansion of Human Dental Pulp Cells In Vitro Under Different Cryopreservation Conditions. Vivo 2020, 34, 2363–2370. [Google Scholar] [CrossRef] [PubMed]
- Raik, S.; Kumar, A.; Rattan, V.; Seth, S.; Kaur, A.; Charyya, S.B. Assessment of Post-thaw Quality of Dental Mesenchymal Stromal Cells After Long-Term Cryopreservation by Uncontrolled Freezing. Appl. Biochem. Biotechnol. 2019, 191, 728–743. [Google Scholar]
- Kumar, A.; Bhattacharyya, S.; Rattan, V. Effect of uncontrolled freezing on biological characteristics of human dental pulp stem cells. Cell Tissue Bank. 2015, 16, 513–522. [Google Scholar] [CrossRef]
- Woods, E.J.; Perry, B.C.; Hockema, J.J.; Larson, L.; Zhou, D.; Goebel, W.S. Optimized cryopreservation method for human dental pulp-derived stem cells and their tissues of origin for banking and clinical use. Cryobiology 2009, 59, 150–157. [Google Scholar] [CrossRef]
- Ginani, F.; Soares, D.M.; Rabêlo, L.M.; Rocha, H.A.O.; De Souza, L.B.; Barboza, C.A.G. Effect of a cryopreservation protocol on the proliferation of stem cells from human exfoliated deciduous teeth. Acta Odontol. Scand. 2016, 74, 598–604. [Google Scholar]
- Yanasse, R.H.; Marques, L.; Fukasawa, J.T.; Segato, R.; Kinoshita, A.; Matsumoto, M.A.; Felisbino, S.L.; Solano, B.; dos Santos, R.R. Xenotransplantation of human dental pulp stem cells in platelet-rich plasma for the treatment of full-thickness articular cartilage defects in a rabbit model. Exp. Ther. Med. 2019, 17, 4344–4356. [Google Scholar] [CrossRef]
- Wang, J.; Zuzzio, K.; Walker, C.L. Systemic Dental Pulp Stem Cell Secretome Therapy in a Mouse Model of Amyotrophic Lateral Sclerosis. Brain Sci. 2019, 9, 165. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, Y.; Nepal, N.; Li, G.; Yang, N.; Chen, H.; Lin, Q.; Ji, X.; Zhang, S.; Jin, S. Dental pulp stem cells overexpressing hepatocyte growth factor facilitate the repair of DSS-induced ulcerative colitis. Stem Cell Res. Ther. 2021, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Hata, M.; Omi, M.; Kobayashi, Y.; Nakamura, N.; Miyabe, M.; Ito, M.; Ohno, T.; Imanishi, Y.; Himeno, T.; Kamiya, H.; et al. Sustainable Effects of Human Dental Pulp Stem Cell Transplantation on Diabetic Polyneuropathy in Streptozotocine-Induced Type 1 Diabetes Model Mice. Cells 2021, 10, 2473. [Google Scholar] [CrossRef] [PubMed]
- Carter, K.; Worthington, S. Predictors of Third Molar Impaction: A Systematic Review and Meta-analysis. J. Dent. Res. 2016, 95, 267–276. [Google Scholar] [CrossRef]
- Ghaeminia, H.; Perry, J.; Nienhuijs, M.E.L.; Toedtling, V.; Tummers, M.; Hoppenreijs, T.J.M.; van der Sanden, W.J.M.; Mettes, T.G. Surgical removal versus retention for the management of asymptomatic disease-free impacted wisdom teeth. Cochrane Database Syst. Rev. 2016, 2016, CD003879. [Google Scholar] [CrossRef]
- McArdle, L.W.; Renton, T. The effects of NICE guidelines on the management of third molar teeth. Br. Dent. J. 2012, 213, E8. [Google Scholar] [CrossRef]
- Brokaw, W.C. The third molar question: When and why should we recommend removal? Va. Dent. J. 1991, 68, 18–21. [Google Scholar]
- Tate, T.E. Impactions: Observe or treat? W. V. Dent. J. 1994, 68, 19–23. [Google Scholar]
- Omar, Z.; Short, L.; Banting, D.W.; Saltaji, H. Profile changes following extraction orthodontic treatment: A comparison of first versus second premolar extraction. Int. Orthod. 2018, 16, 91–104. [Google Scholar] [CrossRef]
- Eubanks, E.J.; Tarle, S.A.; Kaigler, D. Tooth Storage, Dental Pulp Stem Cell Isolation, and Clinical Scale Expansion without Animal Serum. J. Endod. 2014, 40, 652–657. [Google Scholar] [CrossRef]
- Huynh, N.C.-N.; Le, S.H.; Doan, V.N.; Ngo, L.T.Q.; Tran, H.L.B. Simplified conditions for storing and cryopreservation of dental pulp stem cells. Arch. Oral Biol. 2017, 84, 74–81. [Google Scholar] [CrossRef] [PubMed]





| Primer | Direction | Sequence | Length of Products (bp) |
|---|---|---|---|
| LPL | Forward | ACAAGAGAGAACCAGACTCCAA | 76 |
| Reverse | GCGGACACTGGGTAATGCT | ||
| PPAR-γ | Forward | GGGATCAGCTCCGTGGATCT | 186 |
| Reverse | TGCACTTTGGTACTCTTGAAGTT | ||
| ALP | Forward | ACTGGTACTCAGACAACGAGAT | 97 |
| Reverse | ACGTCAATGTCCCTGATGTTATG | ||
| RUNX 2 | Forward | TGGTTACTGTCATGGCGGGTA | 97 |
| Reverse | TCTCAGATCGTTGAACCTTGCTA | ||
| Type I collagen | Forward | GGACACAATGGATTGCAAGG | 441 |
| Reverse | AACCACTGCTCCACTCTGG | ||
| Osteocalcin | Forward | GGCGCTACCTGTATCAATGG | 110 |
| Reverse | GTGGTCAGCCAACTCGTCA | ||
| GAPDH | Forward | GAGTCAACGGATTTGGTCGT | 185 |
| Reverse | GACAAGCTTCCCGTTCTCAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Yan, M.; Aarabi, G.; Peters, U.; Freytag, M.; Gosau, M.; Smeets, R.; Beikler, T. Cultivation of Cryopreserved Human Dental Pulp Stem Cells—A New Approach to Maintaining Dental Pulp Tissue. Int. J. Mol. Sci. 2022, 23, 11485. https://doi.org/10.3390/ijms231911485
Wang W, Yan M, Aarabi G, Peters U, Freytag M, Gosau M, Smeets R, Beikler T. Cultivation of Cryopreserved Human Dental Pulp Stem Cells—A New Approach to Maintaining Dental Pulp Tissue. International Journal of Molecular Sciences. 2022; 23(19):11485. https://doi.org/10.3390/ijms231911485
Chicago/Turabian StyleWang, Wang, Ming Yan, Ghazal Aarabi, Ulrike Peters, Marcus Freytag, Martin Gosau, Ralf Smeets, and Thomas Beikler. 2022. "Cultivation of Cryopreserved Human Dental Pulp Stem Cells—A New Approach to Maintaining Dental Pulp Tissue" International Journal of Molecular Sciences 23, no. 19: 11485. https://doi.org/10.3390/ijms231911485
APA StyleWang, W., Yan, M., Aarabi, G., Peters, U., Freytag, M., Gosau, M., Smeets, R., & Beikler, T. (2022). Cultivation of Cryopreserved Human Dental Pulp Stem Cells—A New Approach to Maintaining Dental Pulp Tissue. International Journal of Molecular Sciences, 23(19), 11485. https://doi.org/10.3390/ijms231911485







