Towards a Mechanistic Model of Tau-Mediated Pathology in Tauopathies: What Can We Learn from Cell-Based In Vitro Assays?
Abstract
1. Introduction
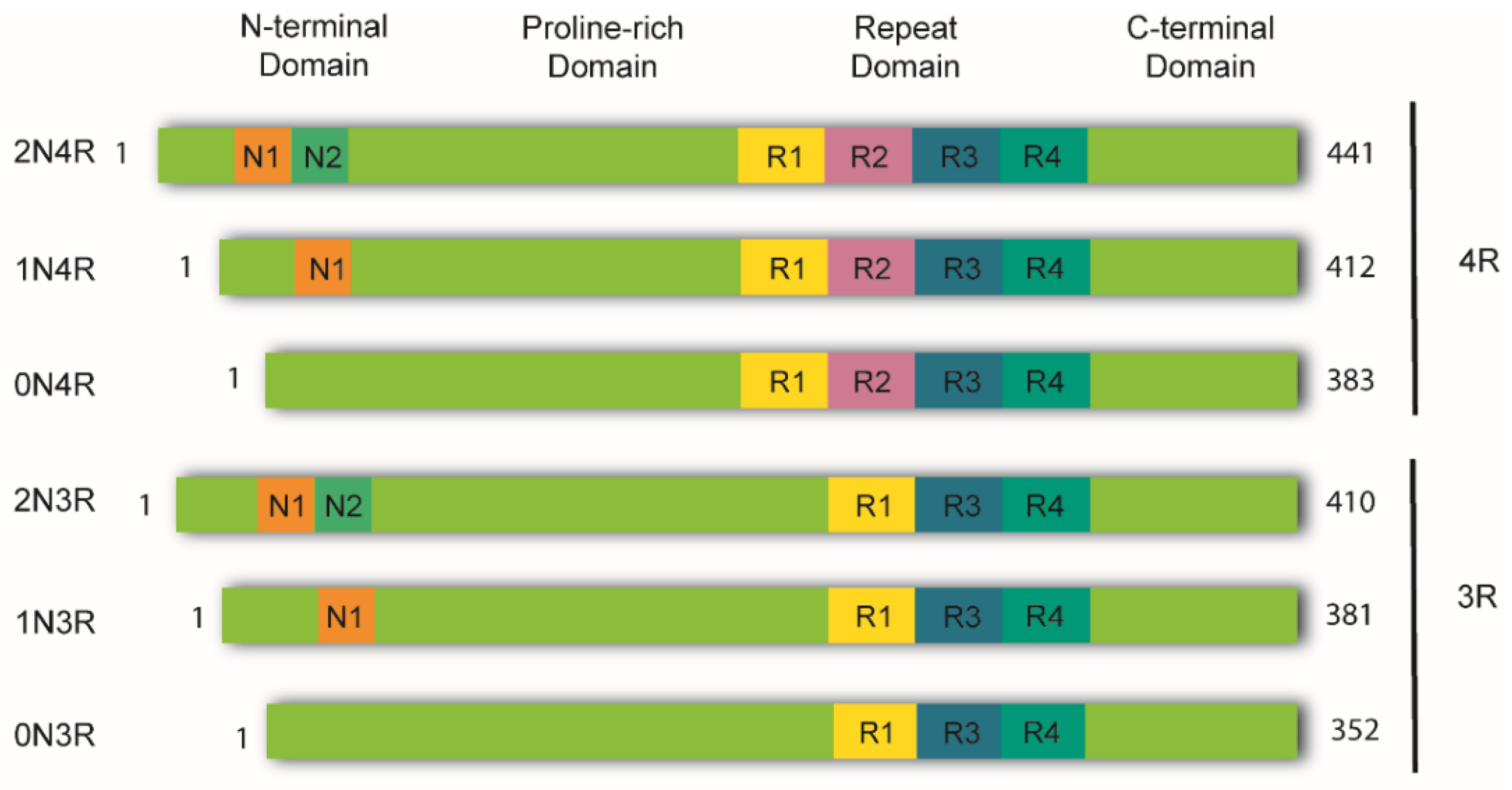
2. Tau Structure and Processing
3. The “Prion-like” Nature of Tau and Its Strains
4. Principal Approaches in the Rodent Model’s Scenery for Tau Pathology
5. Cellular Models of Tau Pathology: Aggregation, Seeding, and Spreading
5.1. In Vitro Modelling of Tau Aggregation: Seminal Models
5.2. Cellular Models of Tau Seeding: Cellular Internalization of Proteopathic Tau Seeds
Cell-Based Assays: Proteopathic Seeding
5.3. Cellular Models of Tau Spreading and Serial Propagation
5.4. Tauopathies in Primary Neural Cells: The Use of Microfluidic Devices in Experimental Design
5.5. iPSCs for Modeling and Studying Tauopathies In Vitro
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ballatore, C.; Lee, V.M.-Y.; Trojanowski, J.Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 2007, 8, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, D.W.; Hwo, S.-Y.; Kirschner, M.W. Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. J. Mol. Biol. 1977, 116, 207–225. [Google Scholar] [CrossRef]
- Sanders, D.W.; Kaufman, S.K.; DeVos, S.L.; Sharma, A.M.; Mirbaha, H.; Li, A.; Barker, S.J.; Foley, A.C.; Thorpe, J.R.; Serpell, L.C.; et al. Distinct Tau Prion Strains Propagate in Cells and Mice and Define Different Tauopathies. Neuron 2014, 82, 1271–1288. [Google Scholar] [CrossRef] [PubMed]
- Binder, L.I.; Frankfurter, A.; Rebhun, L.I. The distribution of tau in the mammalian central nervous system. J. Cell Biol. 1985, 101, 1371–1378. [Google Scholar] [CrossRef]
- LoPresti, P.; Szuchet, S.; Papasozomenos, S.C.; Zinkowski, R.P.; Binder, L.I. Functional implications for the microtubule-associated protein tau: Localization in oligodendrocytes. Proc. Natl. Acad. Sci. USA 1995, 92, 10369–10373. [Google Scholar] [CrossRef]
- Shin, R.W.; Iwaki, T.; Kitamoto, T.; Tateishi, J. Hydrated autoclave pretreatment enhances tau immunoreactivity in formalin-fixed normal and Alzheimer’s disease brain tissues. Lab. Investig. 1991, 64, 693–702. [Google Scholar]
- Alonso, A.C.; Zaidi, T.; Grundke-Iqbal, I.; Iqbal, K. Role of abnormally phosphorylated tau in the breakdown of microtubules in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1994, 91, 5562–5566. [Google Scholar] [CrossRef]
- de Garcini, E.M.; Serrano, L.; Avila, J. Self assembly of microtubule associated protein tau into filaments resembling those found in alzheimer disease. Biochem. Biophys. Res. Commun. 1986, 141, 790–796. [Google Scholar] [CrossRef]
- Götz, J.; Halliday, G.; Nisbet, R.M. Molecular Pathogenesis of the Tauopathies. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 239–261. [Google Scholar] [CrossRef]
- Hernández, F.; Ferrer, I.; Pérez, M.; Zabala, J.C.; del Rio, J.A.; Avila, J. Tau Aggregation. Neuroscience 2022. [Google Scholar] [CrossRef]
- Höglinger, G.; Respondek, G.; Kovacs, G. New classification of tauopathies. Rev. Neurol. 2018, 174, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G. Tauopathies. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 145, pp. 355–368. [Google Scholar] [CrossRef]
- Chung, D.-E.C.; Roemer, S.; Petrucelli, L.; Dickson, D.W. Cellular and pathological heterogeneity of primary tauopathies. Mol. Neurodegener. 2021, 16, 57. [Google Scholar] [CrossRef] [PubMed]
- Dugger, B.N.; Whiteside, C.M.; Maarouf, C.L.; Walker, D.G.; Beach, T.G.; Sue, L.I.; Garcia, A.; Dunckley, T.; Meechoovet, B.; Reiman, E.M.; et al. The Presence of Select Tau Species in Human Peripheral Tissues and Their Relation to Alzheimer’s Disease. J. Alzheimer’s Dis. 2016, 54, 1249. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R. Mutations causing neurodegenerative tauopathies. Biochim. et Biophys. Acta (BBA)-Mol. Basis Dis. 2005, 1739, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Arriagada, P.V.; Growdon, J.H.; Hedley-Whyte, E.T.; Hyman, B.T. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology 1992, 42, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Staging of alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging 1995, 16, 271–278; discussion 278–284. [Google Scholar] [CrossRef]
- Schöll, M.; Lockhart, S.N.; Schonhaut, D.R.; O’Neil, J.P.; Janabi, M.; Ossenkoppele, R.; Baker, S.L.; Vogel, J.W.; Faria, J.; Schwimmer, H.D.; et al. PET Imaging of Tau Deposition in the Aging Human Brain. Neuron 2016, 89, 971–982. [Google Scholar] [CrossRef]
- Frost, B.; Jacks, R.L.; Diamond, M.I. Propagation of Tau Misfolding from the Outside to the Inside of a Cell. J. Biol. Chem. 2009, 284, 12845–12852. [Google Scholar] [CrossRef]
- Clavaguera, F.; Bolmont, T.; Crowther, R.A.; Abramowski, D.; Frank, S.; Probst, A.; Fraser, G.; Stalder, A.K.; Beibel, M.; Staufenbiel, M.; et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 2009, 11, 909–913. [Google Scholar] [CrossRef]
- Franzmeier, N.; Brendel, M.; Beyer, L.; Slemann, L.; Kovacs, G.G.; Arzberger, T.; Kurz, C.; Respondek, G.; Lukic, M.J.; Biel, D.; et al. Tau deposition patterns are associated with functional connectivity in primary tauopathies. Nat. Commun. 2022, 13, 1362. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Avila, J. Tau protein, the main component of paired helical filaments. J. Alzheimer’s Dis. 2006, 9, 171–175. [Google Scholar] [CrossRef]
- Avila, J.; Jiménez, J.S.; Sayas, C.L.; Bolós, M.; Zabala, J.C.; Rivas, G.; Hernández, F. Tau Structures. Front. Aging Neurosci. 2016, 8, 262. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G.; Ghetti, B.; Goedert, M. Classification of diseases with accumulation of Tau protein. Neuropathol. Appl. Neurobiol. 2022, 48, e12792. [Google Scholar] [CrossRef]
- Schweers, O.; Schönbrunn-Hanebeck, E.; Marx, A.; Mandelkow, E. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for beta-structure. J. Biol. Chem. 1994, 269, 24290–24297. [Google Scholar] [CrossRef]
- Gong, C.-X.; Liu, F.; Grundke-Iqbal, I.; Iqbal, K. Post-translational modifications of tau protein in Alzheimer’s disease. J. Neural Transm. 2004, 112, 813–838. [Google Scholar] [CrossRef]
- Martin, L.; Latypova, X.; Terro, F. Post-translational modifications of tau protein: Implications for Alzheimer’s disease. Neurochem. Int. 2011, 58, 458–471. [Google Scholar] [CrossRef]
- Medina, M.; Hernández, F.; Avila, J. New Features about Tau Function and Dysfunction. Biomolecules 2016, 6, 21. [Google Scholar] [CrossRef]
- Hoover, B.R.; Reed, M.N.; Su, J.; Penrod, R.D.; Kotilinek, L.A.; Grant, M.K.; Pitstick, R.; Carlson, G.A.; Lanier, L.M.; Yuan, L.-L.; et al. Tau Mislocalization to Dendritic Spines Mediates Synaptic Dysfunction Independently of Neurodegeneration. Neuron 2010, 68, 1067–1081. [Google Scholar] [CrossRef]
- Avila, J. Tau phosphorylation and aggregation in Alzheimer’s disease pathology. FEBS Lett. 2006, 580, 2922–2927. [Google Scholar] [CrossRef] [PubMed]
- Mudher, A.; Colin, M.; Dujardin, S.; Medina, M.; Dewachter, I.; Alavi Naini, S.M.; Mandelkow, E.-M.; Mandelkow, E.; Buee, L.; Goedert, M.; et al. What is the evidence that tau pathology spreads through prion-like propagation? Acta Neuropathol. Commun. 2017, 5, 99. [Google Scholar] [CrossRef] [PubMed]
- Bancher, C.; Braak, H.; Fischer, P.; Jellinger, K.A. Neuropathological staging of Alzheimer lesions and intellectual status in Alzheimer’s and Parkinson’s disease patients. Neurosci. Lett. 1993, 162, 179–182. [Google Scholar] [CrossRef]
- Braak, H.; Zetterberg, H.; Del Tredici, K.; Blennow, K. Intraneuronal tau aggregation precedes diffuse plaque deposition, but amyloid-β changes occur before increases of tau in cerebrospinal fluid. Acta Neuropathol. 2013, 126, 631–641. [Google Scholar] [CrossRef]
- Fu, H.; Hardy, J.; Duff, K.E. Selective vulnerability in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Possenti, A.; Freer, R.; Nakano, Y.; Villegas, N.C.H.; Tang, M.; Cauhy, P.V.M.; Lassus, B.A.; Chen, S.; Fowler, S.L.; et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 2018, 22, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Rüb, U.; Schultz, C.; Del Tredici, K. Vulnerability of cortical neurons to Alzheimer’s and Parkinson’s diseases. J. Alzheimer’s Dis. 2006, 9, 35–44. [Google Scholar] [CrossRef]
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef]
- Prusiner, S. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef]
- del Río, J.A.; Ferrer, I.; Gavín, R. Role of cellular prion protein in interneuronal amyloid transmission. Prog. Neurobiol. 2018, 165–167, 87–102. [Google Scholar] [CrossRef]
- Vascellari, S.; Manzin, A. Parkinson’s Disease: A Prionopathy? Int. J. Mol. Sci. 2021, 22, 8022. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Tau proteinopathies and the prion concept. Prog. Mol. Biol. Transl. Sci. 2020, 175, 239–259. [Google Scholar] [CrossRef] [PubMed]
- Coca, J.R.; Eraña, H.; Castilla, J. Biosemiotics comprehension of PrP code and prion disease. Biosystems 2021, 210, 104542. [Google Scholar] [CrossRef] [PubMed]
- Jarrett, J.T.; Lansbury, P.T., Jr. Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer’s disease and scrapie? Cell 1993, 73, 1055–1058. [Google Scholar] [CrossRef]
- Brunello, C.A.; Merezhko, M.; Uronen-Mattila, R.-L.; Huttunen, H.J. Mechanisms of secretion and spreading of pathological tau protein. Cell. Mol. Life Sci. 2020, 77, 1721–1744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cao, Y.; Ma, L.; Wei, Y.; Li, H. Possible Mechanisms of Tau Spread and Toxicity in Alzheimer’s Disease. Front. Cell Dev. Biol. 2021, 9, 707268. [Google Scholar] [CrossRef]
- De La-Rocque, S.; Moretto, E.; Butnaru, I.; Schiavo, G. Knockin’ on heaven’s door: Molecular mechanisms of neuronal tau uptake. J. Neurochem. 2020, 156, 563–588. [Google Scholar] [CrossRef]
- Mirbaha, H.; Holmes, B.; Sanders, D.; Bieschke, J.; Diamond, M.I. Tau Trimers Are the Minimal Propagation Unit Spontaneously Internalized to Seed Intracellular Aggregation. J. Biol. Chem. 2015, 290, 14893–14903. [Google Scholar] [CrossRef]
- Sharma, A.M.; Thomas, T.L.; Woodard, D.R.; Kashmer, O.M.; Diamond, M.I. Tau monomer encodes strains. eLife 2018, 7, e37813. [Google Scholar] [CrossRef]
- Kim, D.; Lim, S.; Haque, M.; Ryoo, N.; Hong, H.S.; Rhim, H.; Lee, D.-E.; Chang, Y.-T.; Lee, J.-S.; Cheong, E.; et al. Identification of disulfide cross-linked tau dimer responsible for tau propagation. Sci. Rep. 2015, 5, 15231. [Google Scholar] [CrossRef]
- Vaquer-Alicea, J.; Diamond, M.I.; Joachimiak, L.A. Tau strains shape disease. Acta Neuropathol. 2021, 142, 57–71. [Google Scholar] [CrossRef]
- Taniguchi-Watanabe, S.; Arai, T.; Kametani, F.; Nonaka, T.; Masuda-Suzukake, M.; Tarutani, A.; Murayama, S.; Saito, Y.; Arima, K.; Yoshida, M.; et al. Biochemical classification of tauopathies by immunoblot, protein sequence and mass spectrometric analyses of sarkosyl-insoluble and trypsin-resistant tau. Acta Neuropathol. 2015, 131, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Clavaguera, F.; Akatsu, H.; Fraser, G.; Crowther, R.A.; Frank, S.; Hench, J.; Probst, A.; Winkler, D.T.; Reichwald, J.; Staufenbiel, M.; et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. USA 2013, 110, 9535–9540. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Andrés-Benito, P.; Zelaya, M.V.; Aguirre, M.E.E.; Carmona, M.; Ausín, K.; Lachén-Montes, M.; Fernández-Irigoyen, J.; Santamaría, E.; del Rio, J.A. Familial globular glial tauopathy linked to MAPT mutations: Molecular neuropathology and seeding capacity of a prototypical mixed neuronal and glial tauopathy. Acta Neuropathol. 2020, 139, 735–771. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Cryo-EM structures of τ filaments from human brain. Essays Biochem. 2021, 65, 949–959. [Google Scholar] [CrossRef]
- Falcon, B.; Zhang, W.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Ghetti, B.; Scheres, S.H.W.; Goedert, M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 2018, 561, 137–140. [Google Scholar] [CrossRef]
- Han, Z.Z.; Kang, S.G.; Arce, L.; Westaway, D. Prion-like strain effects in tauopathies. Cell Tissue Res. 2022. [Google Scholar] [CrossRef]
- Robert, A.; Schöll, M.; Vogels, T. Tau Seeding Mouse Models with Patient Brain-Derived Aggregates. Int. J. Mol. Sci. 2021, 22, 6132. [Google Scholar] [CrossRef] [PubMed]
- Dujardin, S.; Colin, M.; Buée, L. Invited review: Animal models of tauopathies and their implications for research/translation into the clinic. Neuropathol. Appl. Neurobiol. 2015, 41, 59–80. [Google Scholar] [CrossRef]
- Götz, J.; Bodea, L.-G.; Goedert, M. Rodent models for Alzheimer disease. Nat. Rev. Neurosci. 2018, 19, 583–598. [Google Scholar] [CrossRef]
- Yoshiyama, Y.; Higuchi, M.; Zhang, B.; Huang, S.-M.; Iwata, N.; Saido, T.C.; Maeda, J.; Suhara, T.; Trojanowski, J.Q.; Lee, V.M.-Y. Synapse Loss and Microglial Activation Precede Tangles in a P301S Tauopathy Mouse Model. Neuron 2007, 53, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Allen, B.; Ingram, E.; Takao, M.; Smith, M.J.; Jakes, R.; Virdee, K.; Yoshida, H.; Holzer, M.; Craxton, M.; Emson, P.C.; et al. Abundant Tau Filaments and Nonapoptotic Neurodegeneration in Transgenic Mice Expressing Human P301S Tau Protein. J. Neurosci. 2002, 22, 9340–9351. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.-L.; Chen, X.; Kazim, S.F.; Liu, F.; Gong, C.-X.; Grundke-Iqbal, I.; Iqbal, K. Passive immunization targeting the N-terminal projection domain of tau decreases tau pathology and improves cognition in a transgenic mouse model of Alzheimer disease and tauopathies. J. Neural Transm. 2014, 122, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.-L.; Hu, W.; Tung, Y.C.; Liu, F.; Gong, C.-X.; Iqbal, K. Tau passive immunization blocks seeding and spread of Alzheimer hyperphosphorylated Tau-induced pathology in 3 × Tg-AD mice. Alzheimer’s Res. Ther. 2018, 10, 13. [Google Scholar] [CrossRef]
- Medina, M.; Avila, J. The need for better AD animal models. Front. Pharmacol. 2014, 5, 227. [Google Scholar] [CrossRef][Green Version]
- Saito, T.; Mihira, N.; Matsuba, Y.; Sasaguri, H.; Hashimoto, S.; Narasimhan, S.; Zhang, B.; Murayama, S.; Higuchi, M.; Lee, V.M.; et al. Humanization of the entire murine Mapt gene provides a murine model of pathological human tau propagation. J. Biol. Chem. 2019, 294, 12754–12765. [Google Scholar] [CrossRef]
- Andorfer, C.; Kress, Y.; Espinoza, M.; De Silva, R.; Tucker, K.L.; Barde, Y.A.; Duff, K.; Davies, P. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J. Neurochem. 2003, 86, 582–590. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, X.; Tung, Y.C.; Xie, S.; Liu, F.; Iqbal, K. Hyperphosphorylation determines both the spread and the morphology of tau pathology. Alzheimer’s Dement. 2016, 12, 1066–1077. [Google Scholar] [CrossRef]
- Harris, J.A.; Koyama, A.; Maeda, S.; Ho, K.; Devidze, N.; Dubal, D.B.; Yu, G.-Q.; Masliah, E.; Mucke, L. Human P301L-Mutant Tau Expression in Mouse Entorhinal-Hippocampal Network Causes Tau Aggregation and Presynaptic Pathology but No Cognitive Deficits. PLoS ONE 2012, 7, e45881. [Google Scholar] [CrossRef]
- Liu, L.; Drouet, V.; Wu, J.W.; Witter, M.P.; Small, S.A.; Clelland, C.; Duff, K. Trans-Synaptic Spread of Tau Pathology In Vivo. PLoS ONE 2012, 7, e31302. [Google Scholar] [CrossRef]
- Caillierez, R.; Bégard, S.; Lécolle, K.; Deramecourt, V.; Zommer, N.; Dujardin, S.; Loyens, A.; Dufour, N.; Aurégan, G.; Winderickx, J.; et al. Lentiviral Delivery of the Human Wild-type Tau Protein Mediates a Slow and Progressive Neurodegenerative Tau Pathology in the Rat Brain. Mol. Ther. 2013, 21, 1358–1368. [Google Scholar] [CrossRef] [PubMed]
- Osinde, M.; Clavaguera, F.; May-Nass, R.; Tolnay, M.; Dev, K.K. Lentivirus Tau (P301S) expression in adult amyloid precursor protein (APP)-transgenic mice leads to tangle formation. Neuropathol. Appl. Neurobiol. 2008, 34, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Kirk, R.G.W. Recovering The Principles of Humane Experimental Technique. Sci. Technol. Hum. Values 2018, 43, 622–648. [Google Scholar] [CrossRef] [PubMed]
- Vanhelmont, T.; Vandebroek, T.; De Vos, A.; Terwel, D.; Lemaire, K.; Anandhakumar, J.; Franssens, V.; Swinnen, E.; Van Leuven, F.; Winderickx, J. Serine-409 phosphorylation and oxidative damage define aggregation of human protein tau in yeast. FEMS Yeast Res. 2010, 10, 992–1005. [Google Scholar] [CrossRef]
- Porzoor, A.; Macreadie, I.G. Application of Yeast to Study the Tau and Amyloid-β Abnormalities of Alzheimer’s Disease. J. Alzheimer’s Dis. 2013, 35, 217–225. [Google Scholar] [CrossRef]
- Guo, J.L.; Lee, V.M.-Y. Seeding of Normal Tau by Pathological Tau Conformers Drives Pathogenesis of Alzheimer-like Tangles. J. Biol. Chem. 2011, 286, 15317–15331. [Google Scholar] [CrossRef]
- Calafate, S.; Buist, A.; Miskiewicz, K.; Vijayan, V.; Daneels, G.; de Strooper, B.; de Wit, J.; Verstreken, P.; Moechars, D. Synaptic Contacts Enhance Cell-to-Cell Tau Pathology Propagation. Cell Rep. 2015, 11, 1176–1183. [Google Scholar] [CrossRef]
- Wu, J.W.; Herman, M.; Liu, L.; Simoes, S.; Acker, C.M.; Figueroa, H.; Steinberg, J.I.; Margittai, M.; Kayed, R.; Zurzolo, C.; et al. Small Misfolded Tau Species Are Internalized via Bulk Endocytosis and Anterogradely and Retrogradely Transported in Neurons. J. Biol. Chem. 2013, 288, 1856–1870. [Google Scholar] [CrossRef]
- Gonzalez, C.; Armijo, E.; Bravo-Alegria, J.; Becerra-Calixto, A.; Mays, C.E.; Soto, C. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry 2018, 23, 2363–2374. [Google Scholar] [CrossRef]
- Oakley, D.H.; Klickstein, N.; Commins, C.; Chung, M.; Dujardin, S.; Bennett, R.E.; Hyman, B.T.; Frosch, M.P. Continuous Monitoring of Tau-Induced Neurotoxicity in Patient-Derived iPSC-Neurons. J. Neurosci. 2021, 41, 4335–4348. [Google Scholar] [CrossRef]
- Usenovic, M.; Niroomand, S.; Drolet, R.E.; Yao, L.; Gaspar, R.C.; Hatcher, N.G.; Schachter, J.; Renger, J.J.; Parmentier-Batteur, S. Internalized Tau Oligomers Cause Neurodegeneration by Inducing Accumulation of Pathogenic Tau in Human Neurons Derived from Induced Pluripotent Stem Cells. J. Neurosci. 2015, 35, 14234–14250. [Google Scholar] [CrossRef] [PubMed]
- Khlistunova, I.; Biernat, J.; Wang, Y.; Pickhardt, M.; von Bergen, M.; Gazova, Z.; Mandelkow, E.; Mandelkow, E.-M. Inducible Expression of Tau Repeat Domain in Cell Models of Tauopathy. J. Biol. Chem. 2006, 281, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, B.; Li, G.; Yin, H.; Kuret, J. Tau Aggregation and Toxicity in a Cell Culture Model of Tauopathy. J. Biol. Chem. 2007, 282, 16454–16464. [Google Scholar] [CrossRef]
- Chun, W.; Johnson, G.V.W. Activation of Glycogen Synthase Kinase 3β Promotes the Intermolecular Association of Tau. J. Biol. Chem. 2007, 282, 23410–23417. [Google Scholar] [CrossRef]
- Gómez-Ramos, A.; Diaz-Hernandez, M.; Cuadros, R.; Hernandez, F.; Avila, J. Extracellular tau is toxic to neuronal cells. FEBS Lett. 2006, 580, 4842–4850. [Google Scholar] [CrossRef] [PubMed]
- Friedhoff, P.; Schneider, A.; Mandelkow, E.-M.; Mandelkow, E. Rapid Assembly of Alzheimer-like Paired Helical Filaments from Microtubule-Associated Protein Tau Monitored by Fluorescence in Solution. Biochemistry 1998, 37, 10223–10230. [Google Scholar] [CrossRef]
- Terwel, D.; Lasrado, R.; Snauwaert, J.; Vandeweert, E.; Van Haesendonck, C.; Borghgraef, P.; Van Leuven, F. Changed Conformation of Mutant Tau-P301L Underlies the Moribund Tauopathy, Absent in Progressive, Nonlethal Axonopathy of Tau-4R/2N Transgenic Mice. J. Biol. Chem. 2005, 280, 3963–3973. [Google Scholar] [CrossRef]
- Kanai, Y.; Takemura, R.; Oshima, T.; Mori, H.; Ihara, Y.; Yanagisawa, M.; Masaki, T.; Hirokawa, N. Expression of multiple tau isoforms and microtubule bundle formation in fibroblasts transfected with a single tau cDNA. J. Cell Biol. 1989, 109, 1173–1184. [Google Scholar] [CrossRef]
- Vogelsberg-Ragaglia, V.; Bruce, J.; Richter-Landsberg, C.; Zhang, B.; Hong, M.; Trojanowski, J.Q.; Lee, V.M.-Y. Distinct FTDP-17 Missense Mutations in Tau Produce Tau Aggregates and Other Pathological Phenotypes in Transfected CHO Cells. Mol. Biol. Cell 2000, 11, 4093–4104. [Google Scholar] [CrossRef]
- Guo, J.L.; Buist, A.; Soares, A.; Callaerts, K.; Calafate, S.; Stevenaert, F.; Daniels, J.P.; Zoll, B.E.; Crowe, A.; Brunden, K.R.; et al. The Dynamics and Turnover of Tau Aggregates in Cultured Cells. J. Biol. Chem. 2016, 291, 13175–13193. [Google Scholar] [CrossRef]
- Pickhardt, M.; Biernat, J.; Hübschmann, S.; Dennissen, F.; Timm, T.; Aho, A.; Mandelkow, E.-M.; Mandelkow, E. Time course of Tau toxicity and pharmacologic prevention in a cell model of Tauopathy. Neurobiol. Aging 2017, 57, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Chun, W.; Waldo, G.S.; Johnson, G.V.W. Split GFP complementation assay: A novel approach to quantitatively measure aggregation of tau in situ: Effects of GSK3β activation and caspase 3 cleavage. J. Neurochem. 2007, 103, 2529–2539. [Google Scholar] [CrossRef] [PubMed]
- Tak, H.; Haque, M.; Kim, M.J.; Lee, J.H.; Baik, J.-H.; Kim, Y.; Kim, D.J.; Grailhe, R.; Kim, Y.K. Bimolecular Fluorescence Complementation; Lighting-Up Tau-Tau Interaction in Living Cells. PLoS ONE 2013, 8, e81682. [Google Scholar] [CrossRef] [PubMed]
- Kfoury, N.; Holmes, B.B.; Jiang, H.; Holtzman, D.M.; Diamond, M.I. Trans-cellular Propagation of Tau Aggregation by Fibrillar Species. J. Biol. Chem. 2012, 287, 19440–19451. [Google Scholar] [CrossRef]
- Holmes, B.B.; Furman, J.L.; Mahan, T.E.; Yamasaki, T.R.; Mirbaha, H.; Eades, W.C.; Belaygorod, L.; Cairns, N.J.; Holtzman, D.M.; Diamond, M.I. Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, E4376–E4385. [Google Scholar] [CrossRef]
- Lo, C.H.; Lim, C.K.-W.; Ding, Z.; Wickramasinghe, S.P.; Braun, A.R.; Ashe, K.H.; Rhoades, E.; Thomas, D.D.; Sachs, J.N. Targeting the ensemble of heterogeneous tau oligomers in cells: A novel small molecule screening platform for tauopathies. Alzheimer’s Dement. 2019, 15, 1489–1502. [Google Scholar] [CrossRef]
- Hitt, B.D.; Vaquer-Alicea, J.; Manon, V.A.; Beaver, J.D.; Kashmer, O.M.; Garcia, J.N.; Diamond, M.I. Ultrasensitive tau biosensor cells detect no seeding in Alzheimer’s disease CSF. Acta Neuropathol. Commun. 2021, 9, 99. [Google Scholar] [CrossRef]
- Chen, J.J.; Nathaniel, D.L.; Raghavan, P.; Nelson, M.; Tian, R.; Tse, E.; Hong, J.Y.; See, S.K.; Mok, S.-A.; Hein, M.Y.; et al. Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J. Biol. Chem. 2019, 294, 18952–18966. [Google Scholar] [CrossRef]
- Han, D.H.; Na, H.-K.; Choi, W.H.; Lee, J.H.; Kim, Y.K.; Won, C.; Lee, S.-H.; Kim, K.P.; Kuret, J.; Min, D.-H.; et al. Direct cellular delivery of human proteasomes to delay tau aggregation. Nat. Commun. 2014, 5, 5633. [Google Scholar] [CrossRef]
- Heo, C.E.; Han, J.Y.; Lim, S.; Lee, J.; Im, D.; Lee, M.J.; Kim, Y.K.; Kim, H.I. ATP Kinetically Modulates Pathogenic Tau Fibrillations. ACS Chem. Neurosci. 2020, 11, 3144–3152. [Google Scholar] [CrossRef]
- Nonaka, T.; Watanabe, S.T.; Iwatsubo, T.; Hasegawa, M. Seeded Aggregation and Toxicity of α-Synuclein and Tau: Cellular models of neurodegenerative diseases. J. Biol. Chem. 2010, 285, 34885–34898. [Google Scholar] [CrossRef] [PubMed]
- Woerman, A.L.; Aoyagi, A.; Patel, S.; Kazmi, S.A.; Lobach, I.; Grinberg, L.T.; McKee, A.C.; Seeley, W.W.; Olson, S.H.; Prusiner, S.B. Tau prions from Alzheimer’s disease and chronic traumatic encephalopathy patients propagate in cultured cells. Proc. Natl. Acad. Sci. USA 2016, 113, E8187–E8196. [Google Scholar] [CrossRef] [PubMed]
- Crowe, A.; Henderson, M.J.; Anderson, J.; Titus, S.A.; Zakharov, A.; Simeonov, A.; Buist, A.; Delay, C.; Moechars, D.; Trojanowski, J.Q.; et al. Compound screening in cell-based models of tau inclusion formation: Comparison of primary neuron and HEK293 cell assays. J. Biol. Chem. 2020, 295, 4001–4013. [Google Scholar] [CrossRef] [PubMed]
- Holzer, M.; Schade, N.; Opitz, A.; Hilbrich, I.; Stieler, J.; Vogel, T.; Neukel, V.; Oberstadt, M.; Totzke, F.; Schächtele, C.; et al. Novel Protein Kinase Inhibitors Related to Tau Pathology Modulate Tau Protein-Self Interaction Using a Luciferase Complementation Assay. Molecules 2018, 23, 2335. [Google Scholar] [CrossRef]
- Wegmann, S.; Nicholls, S.; Takeda, S.; Fan, Z.; Hyman, B.T. Formation, release, and internalization of stable tau oligomers in cells. J. Neurochem. 2016, 139, 1163–1174. [Google Scholar] [CrossRef]
- Kampers, T.; Friedhoff, P.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 1996, 399, 344–349. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R.; Spillantini, M.G.; Hasegawa, M.; Smith, M.J.; Crowther, R.A. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 1996, 383, 550–553. [Google Scholar] [CrossRef]
- Zhang, W.; Falcon, B.; Murzin, A.G.; Fan, J.; Crowther, R.A.; Goedert, M.; Scheres, S.H. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife 2019, 8. [Google Scholar] [CrossRef]
- Takeda, S.; Bs, C.C.; DeVos, S.L.; Nobuhara, C.K.; Wegmann, S.; Bs, A.D.R.; Ba, I.C.; Fan, Z.; Nicholls, S.B.; Sherman, A.E.; et al. Seed-competent high-molecular-weight tau species accumulates in the cerebrospinal fluid of Alzheimer’s disease mouse model and human patients. Ann. Neurol. 2016, 80, 355–367. [Google Scholar] [CrossRef]
- Seidler, P.M.; Boyer, D.R.; Murray, K.A.; Yang, T.P.; Bentzel, M.; Sawaya, M.R.; Rosenberg, G.; Cascio, D.; Williams, C.K.; Newell, K.L.; et al. Structure-based inhibitors halt prion-like seeding by Alzheimer’s disease–and tauopathy–derived brain tissue samples. J. Biol. Chem. 2019, 294, 16451–16464. [Google Scholar] [CrossRef]
- Siano, G.; Caiazza, M.C.; Ollà, I.; Varisco, M.; Madaro, G.; Quercioli, V.; Calvello, M.; Cattaneo, A.; Di Primio, C. Identification of an ERK Inhibitor as a Therapeutic Drug Against Tau Aggregation in a New Cell-Based Assay. Front. Cell. Neurosci. 2019, 13, 386. [Google Scholar] [CrossRef] [PubMed]
- Holmes, B.B.; Devos, S.L.; Kfoury, N.; Li, M.; Jacks, R.; Yanamandra, K.; Ouidja, M.O.; Brodsky, F.M.; Marasa, J.; Bagchi, D.P.; et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. USA 2013, 110, E3138–E3147. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ramos, A.; Diaz-Hernandez, M.; Rubio, A.; Miras-Portugal, M.T.; Avila, J. Extracellular tau promotes intracellular calcium increase through M1 and M3 muscarinic receptors in neuronal cells. Mol. Cell. Neurosci. 2008, 37, 673–681. [Google Scholar] [CrossRef] [PubMed]
- del Rio, J.A.; Ferrer, I. Potential of Microfluidics and Lab-on-Chip Platforms to Improve Understanding of “prion-like” Protein Assembly and Behavior. Front. Bioeng. Biotechnol. 2020, 8, 570692. [Google Scholar] [CrossRef] [PubMed]
- Neto, E.; Leitão, L.; Sousa, D.M.; Alves, C.J.; Alencastre, I.S.; Aguiar, P.; Lamghari, M. Compartmentalized Microfluidic Platforms: The Unrivaled Breakthrough of In Vitro Tools for Neurobiological Research. J. Neurosci. 2016, 36, 11573–11584. [Google Scholar] [CrossRef]
- Taylor, A.M.; Jeon, N.L. Micro-scale and microfluidic devices for neurobiology. Curr. Opin. Neurobiol. 2010, 20, 640–647. [Google Scholar] [CrossRef]
- Wu, J.W.; Hussaini, S.A.; Bastille, I.M.; Rodriguez, G.; Mrejeru, A.; Rilett, K.; Sanders, D.; Cook, C.; Fu, H.; Boonen, R.A.; et al. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci. 2016, 19, 1085–1092. [Google Scholar] [CrossRef]
- Takeda, S.; Wegmann, S.; Cho, H.; Devos, S.L.; Commins, C.; Roe, A.D.; Nicholls, S.B.; Carlson, G.A.; Pitstick, R.; Nobuhara, C.K.; et al. Neuronal uptake and propagation of a rare phosphorylated high-molecular-weight tau derived from Alzheimer’s disease brain. Nat. Commun. 2015, 6, 8490. [Google Scholar] [CrossRef]
- Dujardin, S.; Commins, C.; Lathuiliere, A.; Beerepoot, P.; Fernandes, A.R.; Kamath, T.V.; De Los Santos, M.B.; Klickstein, N.; Corjuc, D.L.; Corjuc, B.T.; et al. Author Correction: Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat. Med. 2021, 27, 356. [Google Scholar] [CrossRef]
- de Wit, J.; Sylwestrak, E.; O’Sullivan, M.L.; Otto, S.; Tiglio, K.; Savas, J.N.; Yates, J.R., 3rd; Comoletti, D.; Taylor, P.; Ghosh, A. LRRTM2 Interacts with Neurexin1 and Regulates Excitatory Synapse Formation. Neuron 2009, 64, 799–806. [Google Scholar] [CrossRef]
- Scheiffele, P.; Fan, J.; Choih, J.; Fetter, R.; Serafini, T. Neuroligin Expressed in Nonneuronal Cells Triggers Presynaptic Development in Contacting Axons. Cell 2000, 101, 657–669. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Ting, H.-C.; Liu, C.-A.; Su, H.-L.; Chiou, T.-W.; Lin, S.-Z.; Harn, H.-J.; Ho, T.-J. Induced Pluripotent Stem Cell (iPSC)-Based Neurodegenerative Disease Models for Phenotype Recapitulation and Drug Screening. Molecules 2020, 25, 2000. [Google Scholar] [CrossRef] [PubMed]
- Slanzi, A.; Iannoto, G.; Rossi, B.; Zenaro, E.; Constantin, G. In vitro Models of Neurodegenerative Diseases. Front. Cell Dev. Biol. 2020, 8, 328. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Malboubi, M.; Li, Y.; George, J.; Jerusalem, A.; Szele, F.; Thompson, M.S.; Ye, H. Rapid and efficient differentiation of functional motor neurons from human iPSC for neural injury modelling. Stem Cell Res. 2018, 32, 126–134. [Google Scholar] [CrossRef]
- di Domenico, A.; Carola, G.; Calatayud, C.; Pons-Espinal, M.; Muñoz, J.P.; Richaud-Patin, Y.; Fernandez-Carasa, I.; Gut, M.; Faella, A.; Parameswaran, J.; et al. Patient-Specific iPSC-Derived Astrocytes Contribute to Non-Cell-Autonomous Neurodegeneration in Parkinson’s Disease. Stem Cell Rep. 2019, 12, 213–229. [Google Scholar] [CrossRef]
- Ehrlich, M.; Mozafari, S.; Glatza, M.; Starost, L.; Velychko, S.; Hallmann, A.-L.; Cui, Q.-L.; Schambach, A.; Kim, K.-P.; Bachelin, C.; et al. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc. Natl. Acad. Sci. USA 2017, 114, E2243–E2252. [Google Scholar] [CrossRef]
- Pomeshchik, Y.; Klementieva, O.; Gil, J.; Martinsson, I.; Hansen, M.G.; de Vries, T.; Sancho-Balsells, A.; Russ, K.; Savchenko, E.; Collin, A.; et al. Human iPSC-Derived Hippocampal Spheroids: An Innovative Tool for Stratifying Alzheimer Disease Patient-Specific Cellular Phenotypes and Developing Therapies. Stem Cell Rep. 2020, 15, 256–273. [Google Scholar] [CrossRef]
- Shi, Y.; Kirwan, P.; Livesey, F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012, 7, 1836–1846. [Google Scholar] [CrossRef]
- Swistowski, A.; Peng, J.; Liu, Q.; Mali, P.; Rao, M.S.; Cheng, L.; Zeng, X. Efficient Generation of Functional Dopaminergic Neurons from Human Induced Pluripotent Stem Cells Under Defined Conditions. Stem Cells 2010, 28, 1893–1904. [Google Scholar] [CrossRef]
- Israel, M.A.; Yuan, S.H.; Bardy, C.; Reyna, S.M.; Mu, Y.; Herrera, C.; Hefferan, M.P.; van Gorp, S.; Nazor, K.L.; Boscolo, F.S.; et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 2012, 482, 216–220. [Google Scholar] [CrossRef]
- Yagi, T.; Ito, D.; Okada, Y.; Akamatsu, W.; Nihei, Y.; Yoshizaki, T.; Yamanaka, S.; Okano, H.; Suzuki, N. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum. Mol. Genet. 2011, 20, 4530–4539. [Google Scholar] [CrossRef] [PubMed]
- Paquet, D.; Kwart, D.; Chen, A.; Sproul, A.; Jacob, S.; Teo, S.; Olsen, K.M.; Gregg, A.; Noggle, S.; Tessier-Lavigne, M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 2016, 533, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, A.; Diels, A.; Dijkmans, J.; Oyelami, T.; Meneghello, G.; Mertens, L.; Versweyveld, S.; Borgers, M.; Buist, A.; Peeters, P.; et al. Using Human iPSC-Derived Neurons to Model TAU Aggregation. PLoS ONE 2015, 10, e0146127. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, A.; Diels, A.; Reumers, J.; Van Hoorde, K.; Van den Wyngaert, I.; van Outryve d’Ydewalle, C.; De Bondt, A.; Kuijlaars, J.; De Muynck, L.; De Hoogt, R.; et al. Genetically Engineered iPSC-Derived FTDP-17 MAPT Neurons Display Mutation-Specific Neurodegenerative and Neurodevelopmental Phenotypes. Stem Cell Rep. 2018, 11, 363–379. [Google Scholar] [CrossRef]
- Manos, J.D.; Preiss, C.N.; Venkat, N.; Tamm, J.; Reinhardt, P.; Kwon, T.; Wu, J.; Winter, A.D.; Jahn, T.R.; Yanamandra, K.; et al. Uncovering specificity of endogenous TAU aggregation in a human iPSC-neuron TAU seeding model. iScience 2022, 25, 103658. [Google Scholar] [CrossRef]
- Choi, S.H.; Kim, Y.H.; Hebisch, M.; Sliwinski, C.; Lee, S.; D’Avanzo, C.; Chen, J.; Hooli, B.; Asselin, C.; Muffat, J.; et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014, 515, 274–278. [Google Scholar] [CrossRef]
- Raja, W.K.; Mungenast, A.E.; Lin, Y.T.; Ko, T.; Abdurrob, F.; Seo, J.; Tsai, L.H. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS ONE 2016, 11, e0161969. [Google Scholar] [CrossRef]
- Gerson, J.E.; Kayed, R. Formation and Propagation of Tau Oligomeric Seeds. Front. Neurol. 2013, 4, 93. [Google Scholar] [CrossRef]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.; Sengupta, U.; Guerrero-Munoz, M.J.; Kiritoshi, T.; Neugebauer, V.; Jackson, G.R.; Kayed, R. Alzheimer brain-derived tau oligomers propagate pathology from endogenous tau. Sci. Rep. 2012, 2, 700. [Google Scholar] [CrossRef]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.L.; Sengupta, U.; Clos, A.L.; Jackson, G.R.; Kayed, R. Tau oligomers impair memory and induce synaptic and mitochondrial dysfunction in wild-type mice. Mol. Neurodegener. 2011, 6, 39. [Google Scholar] [CrossRef]
- Bright, J.; Hussain, S.; Dang, V.; Wright, S.; Cooper, B.; Byun, T.; Ramos, C.; Singh, A.; Parry, G.; Stagliano, N.; et al. Human secreted tau increases amyloid-beta production. Neurobiol. Aging 2015, 36, 693–709. [Google Scholar] [CrossRef] [PubMed]
- Bang, S.; Lee, S.; Choi, N.; Kim, H.N. Emerging Brain-Pathophysiology-Mimetic Platforms for Studying Neurodegenerative Diseases: Brain Organoids and Brains-on-a-Chip. Adv. Healthc. Mater. 2021, 10, 2002119. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Song, H.; Ming, G.-L. Brain organoids: Advances, applications and challenges. Development 2019, 146, dev166074. [Google Scholar] [CrossRef]
- Pellegrini, L.; Lancaster, M.A. Modeling neurodegeneration with mutant-tau organoids. Cell 2021, 184, 4377–4379. [Google Scholar] [CrossRef]
- Sun, X.-Y.; Ju, X.-C.; Li, Y.; Zeng, P.-M.; Wu, J.; Zhou, Y.-Y.; Shen, L.-B.; Dong, J.; Chen, Y.-J.; Luo, Z.-G. Generation of vascularized brain organoids to study neurovascular interactions. eLife 2022, 11. [Google Scholar] [CrossRef]
- Beutel, T.; Dzimiera, J.; Kapell, H.; Engelhardt, M.; Gass, A.; Schirmer, L. Cortical projection neurons as a therapeutic target in multiple sclerosis. Expert Opin. Ther. Targets 2020, 24, 1211–1224. [Google Scholar] [CrossRef]



| Tau Isoform | Variants | Host Cell | Expression | Aggregation Inducer | Detection Method | Reference |
|---|---|---|---|---|---|---|
| 4RD | Wild-type ΔK280 ΔK280/PP (I277P/I308P) | N2a | Stable doxycycline-inducible | Spontaneous formation | ThS ICC | [83] |
| 2N4R | Wild-type | HEK293 | Stable doxycycline-inducible | Congo red | ICC | [84] |
| 0N4R Tau1-421 | Wild-type | HEK293 | Transient expression | Spontaneous formation Phosphorylation | FRET (CFP/YFP) | [85] |
| 0N4R 4RD | ΔK280 I277P/I308P | HEK293 | Transient expression | Spontaneous formation Phosphorylation | BiFC (Split GFP) | [93] |
| 2N4R | Wild-Type | HEK293 | Stable expression | Phosphorylation | BiFC (Split Venus) | [94] |
| Tau Isoform | Variants | Host Cell | Expression | Extracellular Tau | Detection Method | Reference |
|---|---|---|---|---|---|---|
| 4RD Tau40 | Wild-type | HEK293 C17.2 | Transient expression | Tau PFFs | YFP ICC | [20] |
| 1N3R 1N4R | Wild-type | SH-SY5Y | Transient expression | Tau PFFs | GFP ICC | [102] |
| 3RD 4RD | L226V/V337M P301L/V337M | HEK293 | Transient expression | Crude brain homogenates from human AD, PiD, CTE, AGD, CBD, and PSP patients | YFP | [103] |
| Tau40 | Wild-type ΔK280 P301L R406W | QBI-293 | Transient expression | Tau PFFs | ICC | [77] |
| Tau40 | P301L | QBI-293 | Stable doxycycline-inducible | Tau PFFs | GFP ICC | [91,104] |
| 4RD | P301S | HEK293 | Stable expression | -Tau PFFs -Tau assemblies purified from AD patients | SLC (NLuc/CLuc) | [49] |
| 4RD | P301S | HEK293 | Stable expression | -Tau PFFs -AD brain lysate -P301S brain lysate | FRET (CFP/YFP) | [96] |
| 4RD | P301S | HEK293 | Stable expression | -Tau PFFs -AD brain lysate -AD CSF | FRET (Clo/Cler) | [98] |
| 4RD | ΔK280 P301L/V337M ΔK280/I22P /I308P | HEK293 | Transient expression | Tau PFFs | FRET (CFP/YFP) Antibody | [95] |
| Model | Cell Source | Type of Tau Seed | Detection Method | Reference |
|---|---|---|---|---|
| 2D | Neurons derived from wild-type hiPSCs with two MAPT mutations | K18 fibrils (P301L) | AlphaLISA | [135] |
| 2D | Wild-type hiPSC- neurons | Full-length human tau monomer and oligomer seeds | ICC ThS | [82] |
| 2D | Familial AD patient hiPSC- neurons expressing a tau aggregation biosensor | Tau seeds derived from mice carrying the MAPT P301L mutation (rTg4510) | in vitro longitudinal single-cell live-imaging system | [81] |
| 2D | -MAPT-wild-type hiPSC- neurons -MAPT-P301S/E10 + 16 hiPSC-neurons | -Sarkosyl-insoluble material from AD brains -Sarkosyl-insoluble material from healthy control brains | ICC HTRF | [136] |
| 3D (cerebral organoid) | -Wild-type hiPSC-neurons -Familial AD patient hiPSC-neurons | Spontaneous formation | ICC | [80] |
| 3D (cells in Matrigel) | ReNcell human neural stem cells with familial AD mutations APPSL and PS1ΔE9 | Spontaneous formation | ICC Modified Gallyas silver staining | [137] |
| 3D (cerebral organoids) | -Wild-type hiPSC-neurons -Familial AD patient hiPSC-neurons | Spontaneous formation | ICC ThS | [138] |
| Model | Advantages | Disadvantages |
|---|---|---|
| In vivo: rodent models of tauopathies | Transgenic models reproduce many of the tau pathologies seen in the brains of human patients | Most transgenic models do not entirely mimic the hallmarks of sporadic human tauopathies in terms of the morphology of tau aggregates and the affected cell types |
| The overexpression of mutated forms results in a rapid and robust tau pathology | Most transgenic models rely on the overexpression of mutant tau in virtually all brain cells, making tau spreading studies nearly impossible | |
| Allow for the evaluation of behavioral impairments | Time consuming and expensive | |
| Inoculation models of patient-derived material are highly translationally relevant models as they allow the investigation of tau spreading | Not suitable for high-throughput approaches | |
| They include the complexity of the nervous system, improving their translational value compared to other models | Difficult to monitor tau aggregation and spreading with high spatiotemporal resolution | |
| 2D mammalian immortalized cell lines | Rapid experimental turnaround time | Most models do not reproduce neuronal phenotypes |
| Easy to culture and transfect | Models that partially differentiate to neuronal phenotypes (i.e., SH-SY5Y) are cancer-derived cells | |
| Labeling techniques are easily introduced to monitor and track aggregate formation with spatiotemporal resolution | Most models are not complex enough to produce transnationally relevant results regarding tau spreading | |
| Excellent platforms for high-throughput approaches such as drug screening, especially in monoclonal cell lines | They do not reproduce the complexity of the nervous system | |
| Microfluidic devices: murine neural cells | Excellent platforms for the study of tau spreading as they allow to track the movement of tau aggregates across synapses | Laborious to prepare and maintain |
| Ideal platform for spatiotemporal separation of neuronal populations, allowing neural network modeling | High levels of variability between independent experiments (e.g., different litters) | |
| Small reaction volumes needed | Difficult to transfect | |
| 2D iPSC-derived neurons | Maintain the genetic information of donors and can replicate the disease phenotype of the donor in vitro | Lack of complexity |
| Easily gene-edited to express tau mutations | Neuronal immaturity | |
| Tau seeds can be easily introduced to the culture | In vitro differentiation induced heterogeneity | |
| Labeling techniques are easily introduced to monitor and track aggregate formation with spatiotemporal resolution | Labor- and time-intensive generation and characterization | |
| Excellent platforms for high-throughput approaches such as drug screening | Lack of intercellular communication between different cell types | |
| 3D Cerebral organoids | Maintain the genetic information of donors | Highly variable culture protocols, which can lead to varying outcomes between groups |
| Can replicate the disease phenotype without genetic manipulation i.e., spontaneous tau phosphorylation/aggregation | Lack of vasculature | |
| Closely recapitulate the laminar organization of the developing human cortex and thus can model tau spreading in a more physiologically relevant manner | High variability of tau expression between organoids | |
| Allow for interactions between different neural cell types | Oligodendrocytes and microglia are often not well formed | |
| Viable for much longer than neural cells in 2D-culture, allowing the study of long-term effects of tau pathology | More difficult to monitor and track aggregate formation with spatiotemporal resolution due to the dense 3D tissue | |
| Capable of mimicking perfusion and diffusion-based molecular transport | Can develop a necrotic core caused by lack of oxygen and nutrient diffusion into the inner-most layers | |
| Can be used to study endolysosomal trafficking abnormalities that affect tau pathology | Labor- and time-intensive generation and characterization |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sala-Jarque, J.; Zimkowska, K.; Ávila, J.; Ferrer, I.; del Río, J.A. Towards a Mechanistic Model of Tau-Mediated Pathology in Tauopathies: What Can We Learn from Cell-Based In Vitro Assays? Int. J. Mol. Sci. 2022, 23, 11527. https://doi.org/10.3390/ijms231911527
Sala-Jarque J, Zimkowska K, Ávila J, Ferrer I, del Río JA. Towards a Mechanistic Model of Tau-Mediated Pathology in Tauopathies: What Can We Learn from Cell-Based In Vitro Assays? International Journal of Molecular Sciences. 2022; 23(19):11527. https://doi.org/10.3390/ijms231911527
Chicago/Turabian StyleSala-Jarque, Julia, Karolina Zimkowska, Jesús Ávila, Isidro Ferrer, and José Antonio del Río. 2022. "Towards a Mechanistic Model of Tau-Mediated Pathology in Tauopathies: What Can We Learn from Cell-Based In Vitro Assays?" International Journal of Molecular Sciences 23, no. 19: 11527. https://doi.org/10.3390/ijms231911527
APA StyleSala-Jarque, J., Zimkowska, K., Ávila, J., Ferrer, I., & del Río, J. A. (2022). Towards a Mechanistic Model of Tau-Mediated Pathology in Tauopathies: What Can We Learn from Cell-Based In Vitro Assays? International Journal of Molecular Sciences, 23(19), 11527. https://doi.org/10.3390/ijms231911527








