A New Phospholipase D from Moritella sp. JT01: Biochemical Characterization, Crystallization and Application in the Synthesis of Phosphatidic Acid
Abstract
:1. Introduction
2. Results
2.1. Bioinformatic Analysis of MsPLD
2.2. Recombinant Expression and Purification of Wild-Type MsPLD
2.3. Enzyme Characterization of MsPLD
2.3.1. Optimum Temperature and Thermostability of MsPLD
2.3.2. Optimum pH and pH Stability of the MsPLD
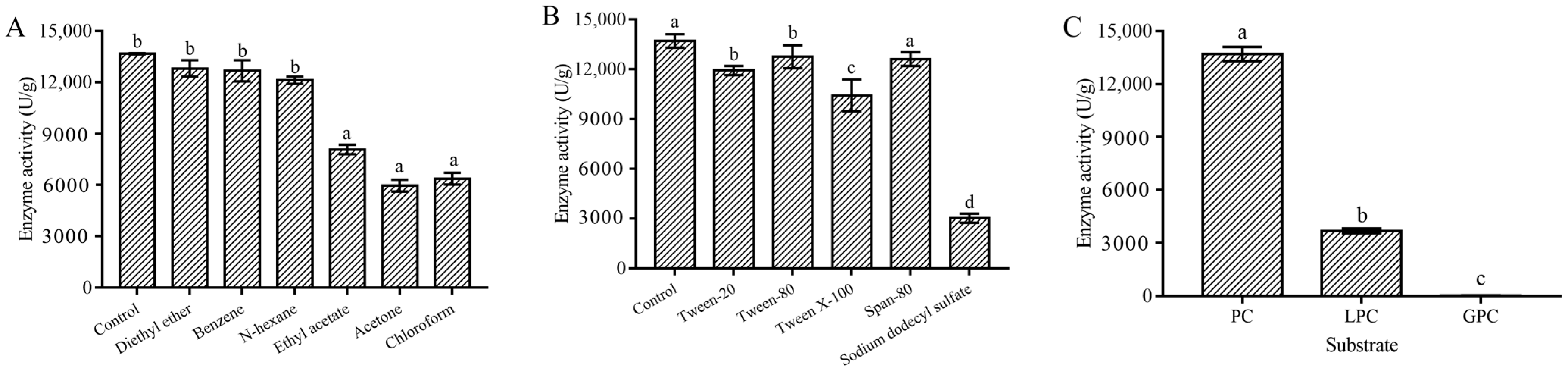
2.3.3. Metal Ion, Organic Solvent, and Surfactant Stability of the MsPLD
2.3.4. Substrate Selectivity and Kinetic Parameters of MsPLD
2.4. Crystallization and Structural Analysis of the MsPLD
2.4.1. Overall Structure
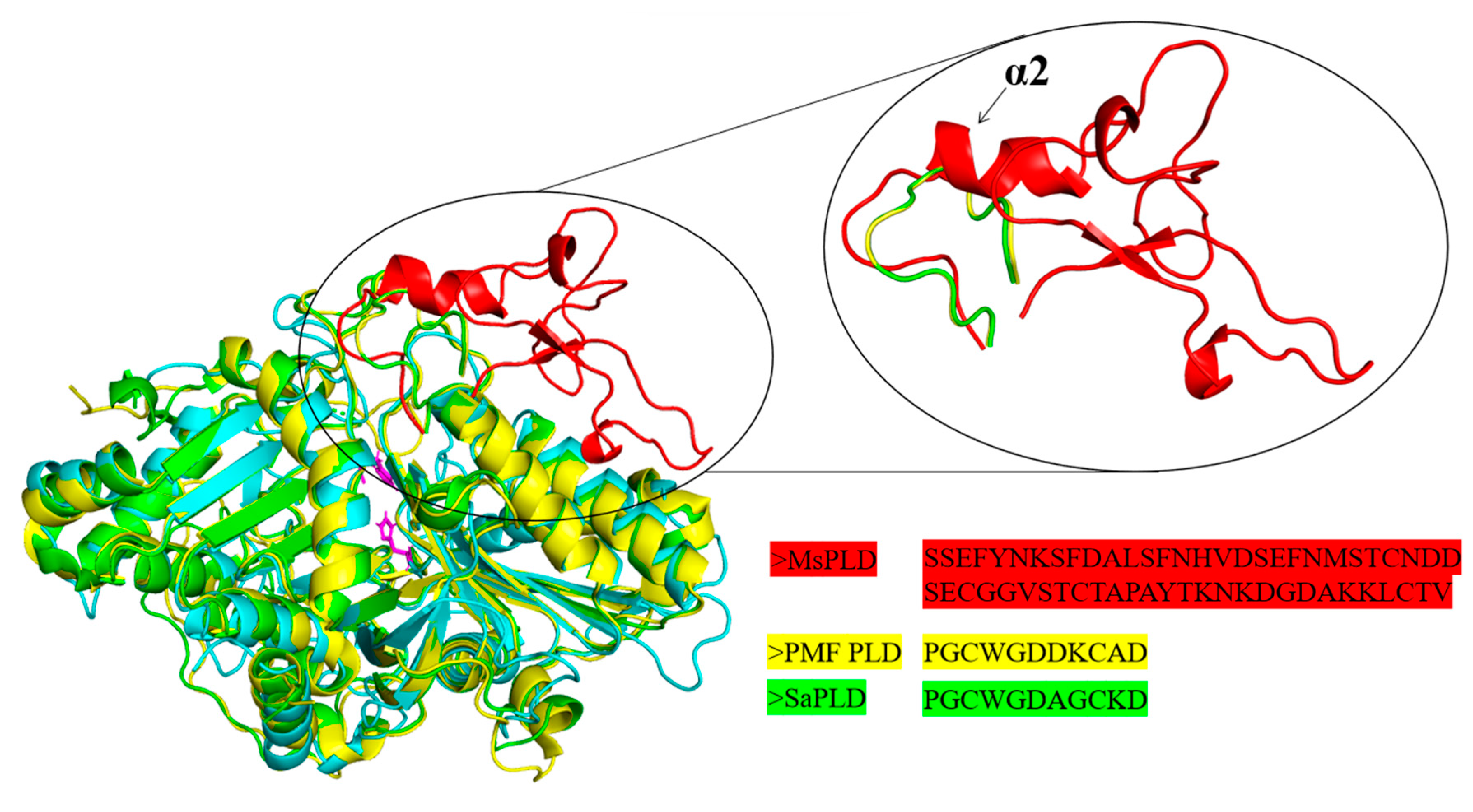
2.4.2. An Extra Loop Segment Was Found in MsPLD Compared to Other Reported PLDs
2.4.3. Deletion or Fasting of the Extra Loop Segment Resulted in Complete Loss of Its Hydrolysis Activity
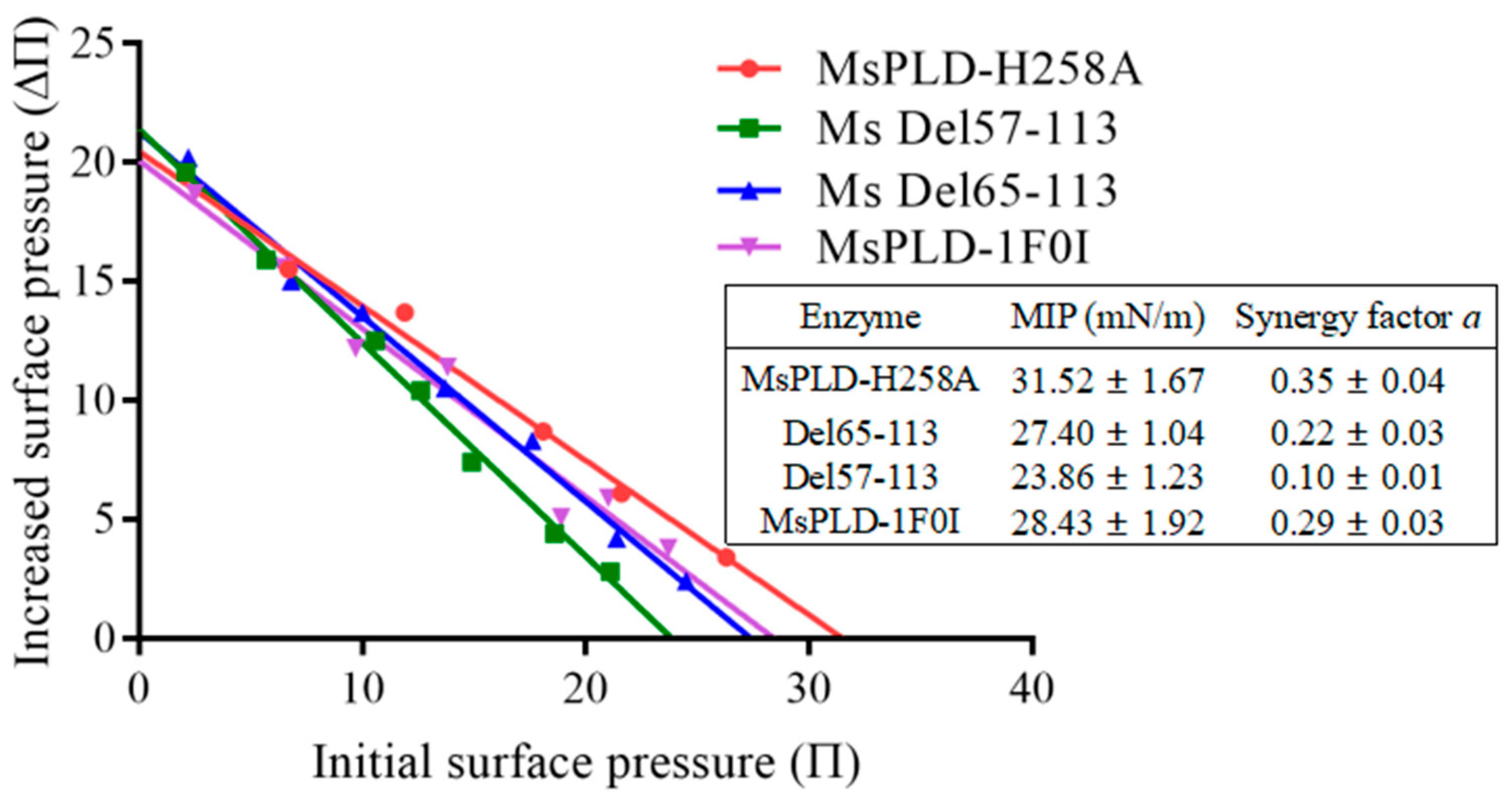
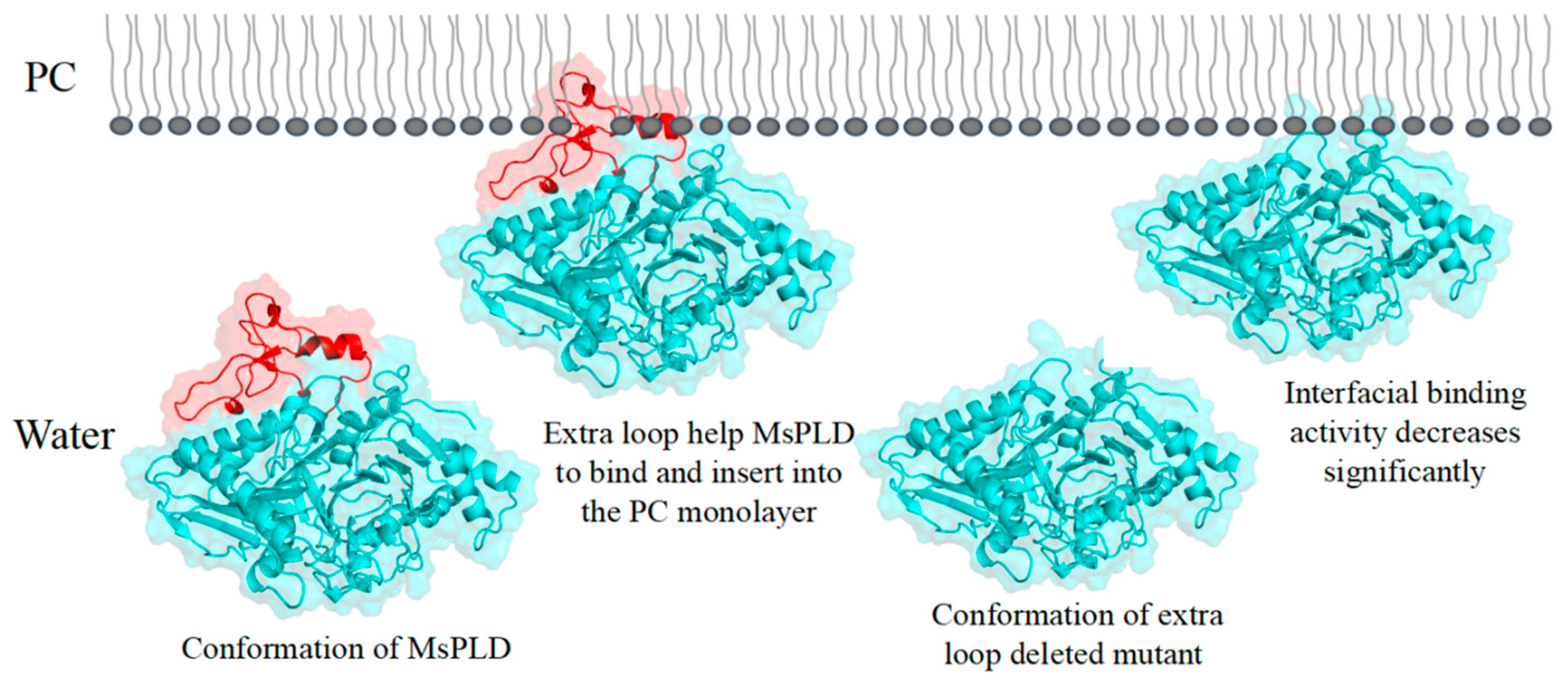
2.4.4. Deletion of the Extra Loop Segment Significantly Decreased the Binding Properties of MsPLD to PC Monolayer
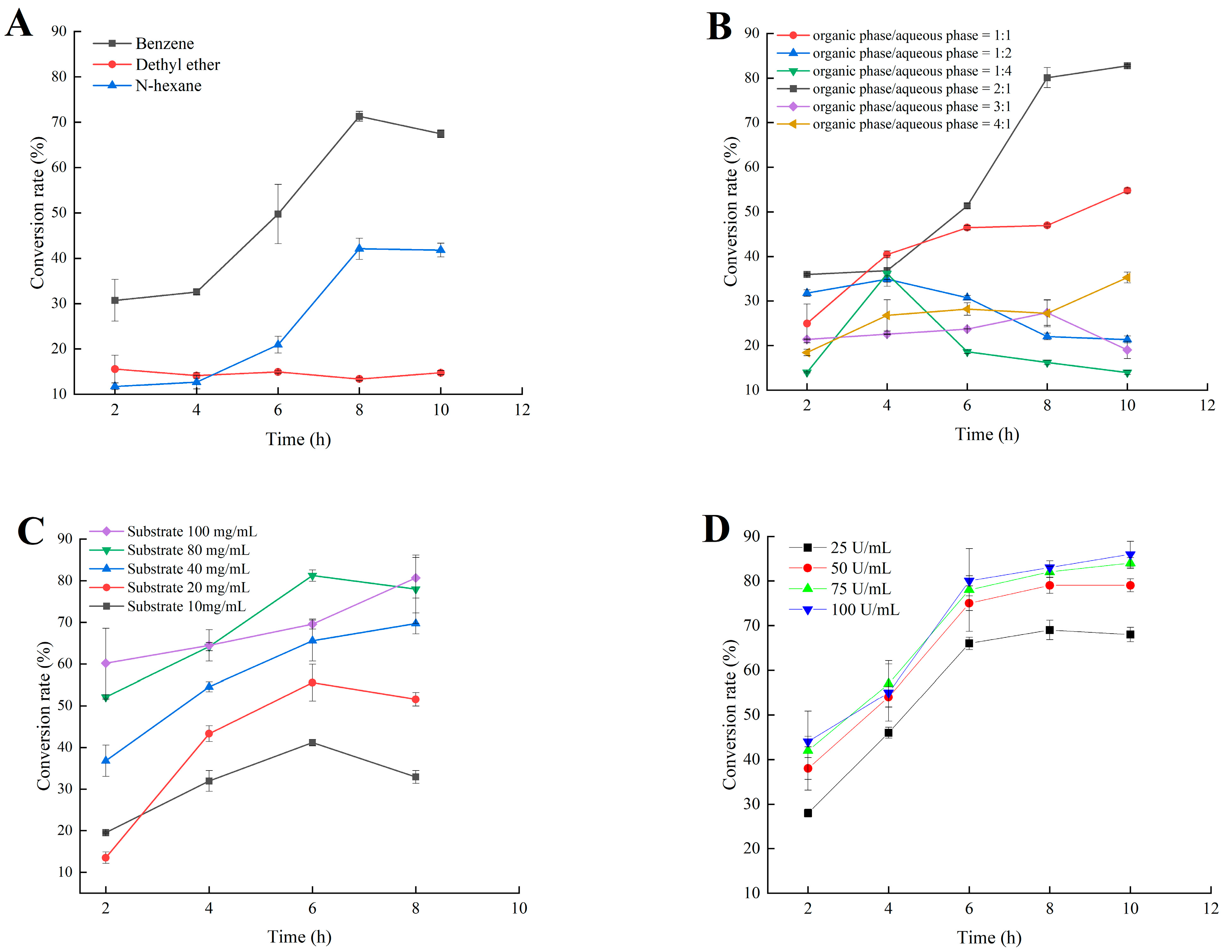
2.5. Application of MsPLD for the Synthesis of PA by Using PC as Substrate
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Bioinformatic Analysis of MsPLD
4.3. Recombinant Expression of MsPLD in Escherichia coli
4.4. Enzyme Characterization
4.4.1. MsPLD Hydrolysis Activity Assays
4.4.2. Effects of Temperature and pH on Enzyme Activity
4.4.3. Metal Ion, Organic Solvent, and Surfactant Stability of MsPLD
4.4.4. Substrate Selectivity and Kinetic Parameters
4.5. Crystallization and Structure Determination
4.6. Molecular Dynamic Simulations of MsPLD
4.7. Structure-Based Mutants Design, Construction, and Expression
4.8. Interfacial Adsorption Measurements by Using Monolayer Technology
4.9. Enzymatic Synthesis of PA by MsPLD
4.9.1. Reaction System Optimization
4.9.2. Analysis of PA by High-Performance Liquid Chromatography
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kooijman, E.E.; Burger, K.N. Biophysics and function of phosphatidic acid: A molecular perspective. Biochim. Et Biophys. Acta 2009, 1791, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Wang, X. Phosphatidic acid: An emerging versatile class of cellular mediators. Essays Biochem. 2020, 64, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Devaiah, S.P.; Zhang, W.; Welti, R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006, 45, 250–278. [Google Scholar] [CrossRef] [PubMed]
- Athenstaedt, K. Phosphatidic acid biosynthesis in the model organism yeast Saccharomyces cerevisiae—A survey. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158907. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, X. Phospholipase D and phosphatidic acid in plant immunity. Plant Sci. Int. J. Exp. Plant Biol. 2019, 279, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Raben, D.M.; Barber, C.N. Phosphatidic acid and neurotransmission. Adv. Biol. Regul. 2017, 63, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.Y.; Xue, H.W. Phosphatidic acid plays key roles regulating plant development and stress responses. J. Integr. Plant Biol. 2018, 60, 851–863. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Fisher, J.; Ingham, E. Joint Lubricant. EP1204433A1, 15 May 2002. [Google Scholar]
- Ruenberg, D. Anti-Depressant, Stress Suppressor and Mood Improver. US6410522B1, 25 June 2002. [Google Scholar]
- Shenfeld, A. Compositions and Methods for Smoking Cessation. EP1791548A1, 6 June 2007. [Google Scholar]
- Shinitzky, M.; Shenfeld, A. Lipid-Based Immune Modulator Composition. CN1264304A, 29 December 2004. [Google Scholar]
- Hoffman, J.R.; Stout, J.R.; Williams, D.R.; Wells, A.J.; Fragala, M.S.; Mangine, G.T.; Gonzalez, A.M.; Emerson, N.S.; McCormack, W.P.; Scanlon, T.C.; et al. Efficacy of phosphatidic acid ingestion on lean body mass, muscle thickness and strength gains in resistance-trained men. J. Int. Soc. Sports Nutr. 2012, 9, 47. [Google Scholar] [CrossRef] [Green Version]
- Toldra, F.; Gallego, M.; Reig, M.; Aristoy, M.-C.; Mora, L. Recent progress in enzymatic release of peptides in foods of animal origin and assessment of bioactivity. J. Agric. Food Chem. 2020, 68, 12842–12855. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Czarna, A.; Chen, W.; Yang, B.; Wang, Y. Function of C-terminal peptides on enzymatic and interfacial adsorption properties of lipase from Gibberella zeae. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2018, 1862, 2623–2631. [Google Scholar] [CrossRef]
- Damnjanović, J.; Iwasaki, Y. Phospholipase D as a catalyst: Application in phospholipid synthesis, molecular structure and protein engineering. J. Biosci. Bioeng. 2013, 116, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.J.; Gong, J.S.; Dong, Y.X.; Qin, J.; Li, H.; Li, H.; Lu, Z.M.; Zhang, X.M.; Xu, Z.H.; Shi, J.S. Phospholipase D engineering for improving the biocatalytic synthesis of phosphatidylserine. Bioprocess Biosyst. Eng. 2019, 42, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Leiros, I.; Secundo, F.; Zambonelli, C.; Servi, S.; Hough, E. The first crystal structure of a phospholipase D. Structure 2000, 8, 655–667. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, A.; Kakuno, K.; Iwasaki, Y.; Yamane, T.; Yamane, T. Crystallization and preliminary X-ray diffraction studies of phospholipase D from Streptomyces antibioticus. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 317–319. [Google Scholar] [CrossRef]
- Bowling, F.Z.; Salazar, C.M.; Bell, J.A.; Huq, T.S.; Frohman, M.A.; Airola, M.V. Crystal structure of human PLD1 provides insight into activation by PI(4,5)P(2) and RhoA. Nat. Chem. Biol. 2020, 16, 400–407. [Google Scholar] [CrossRef]
- Metrick, C.M.; Peterson, E.A.; Santoro, J.C.; Enyedy, I.J.; Murugan, P.; Chen, T.; Michelsen, K.; Cullivan, M.; Spilker, K.A.; Kumar, P.R.; et al. Human PLD structures enable drug design and characterization of isoenzyme selectivity. Nat. Chem. Biol. 2020, 16, 391–399. [Google Scholar] [CrossRef]
- Li, J.; Yu, F.; Guo, H.; Xiong, R.; Zhang, W.; He, F.; Zhang, M.; Zhang, P. Crystal structure of plant PLDα1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D. Cell Res. 2020, 30, 61–69. [Google Scholar] [CrossRef]
- Wang, F.; Liu, S.; Mao, X.; Cui, R.; Yang, B.; Wang, Y. Crystal Structure of a Phospholipase D from the Plant-Associated Bacteria Serratia plymuthica Strain AS9 Reveals a Unique Arrangement of Catalytic Pocket. Int. J. Mol. Sci. 2021, 22, 3219. [Google Scholar] [CrossRef]
- Wang, F.; Wu, Z.; Abousalham, A.; Yang, B.; Wang, Y. Deletion the C-terminal peptides of Vibrio harveyi phospholipase D significantly improved its enzymatic properties. Int. J. Biol. Macromol. 2019, 129, 1140–1147. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Ertan, H.; Poljak, A.; Bridge, W.J. Evaluating Enzymatic Productivity-The Missing Link to Enzyme Utility. Int. J. Mol. Sci. 2022, 23, 6908. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Cui, R.; Tang, Q.; Lan, D.; Wang, F.; Wang, Y. Enhancement of Phospholipid Binding and Catalytic Efficiency of Streptomyces klenkii Phospholipase D by Increasing Hydrophobicity of the Active Site Loop. J. Agric. Food Chem. 2021, 69, 11110–11120. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wei, R.; Abousalham, A.; Chen, W.; Yang, B.; Wang, Y. Effect of N- and C-Terminal Amino Acids on the Interfacial Binding Properties of Phospholipase D from Vibrio parahaemolyticus. Int. J. Mol. Sci. 2018, 19, 2447. [Google Scholar] [CrossRef] [Green Version]
- Pasker, B.; Sosada, M.; Fraś, P.; Boryczka, M.; Górecki, M.; Zych, M. Rapeseed Phosphatidylcholine Hydrolysis to Phosphatidic Acid Using Plant Extracts with Phopspholipase D. Acta Pol. Pharm. 2015, 72, 335–340. [Google Scholar] [PubMed]
- Ribitsch, D.; Karl, W.; Wehrschütz-Sigl, E.; Tutz, S.; Remler, P.; Weber, H.J.; Gruber, K.; Stehr, R.; Bessler, C.; Hoven, N.; et al. Heterologous expression and characterization of choline oxidase from the soil bacterium Arthrobacter nicotianae. Appl. Microbiol. Biotechnol. 2009, 81, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Shimbo, K.; Yano, H.; Miyamoto, Y.J.A. Purification and properties of phospholipase D from Streptomyces lydicus. Agric. Biol. Chem. 1990, 54, 1189–1193. [Google Scholar] [CrossRef] [Green Version]
- Powell, N.; Walker, A.W.; Stolarczyk, E.; Canavan, J.B.; Gökmen, M.R.; Marks, E.; Jackson, I.; Hashim, A.; Curtis, M.A.; Jenner, R.G. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity 2012, 37, 674–684. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Cui, R.; Lan, D.; Wang, F.; Wang, Y. Acyl Chain Specificity of Marine Streptomyces klenkii PhosPholipase D and Its Application in Enzymatic Preparation of Phosphatidylserine. Int. J. Mol. Sci. 2021, 22, 10580. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hough, M.A.; Wilson, K.S. From crystal to structure with CCP4. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laskowski, R.A.; Moss, D.S.; Thornton, J.M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993, 231, 1049–1067. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Bramucci, E.; Paiardini, A.; Bossa, F.; Pascarella, S. PyMod: Sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinform. 2012, 13 (Suppl. S4), S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Mao, X.; Deng, F.; Cui, R.; Li, L.; Liu, S.; Yang, B.; Lan, D.; Wang, Y. A New Phospholipase D from Moritella sp. JT01: Biochemical Characterization, Crystallization and Application in the Synthesis of Phosphatidic Acid. Int. J. Mol. Sci. 2022, 23, 11633. https://doi.org/10.3390/ijms231911633
Wang F, Mao X, Deng F, Cui R, Li L, Liu S, Yang B, Lan D, Wang Y. A New Phospholipase D from Moritella sp. JT01: Biochemical Characterization, Crystallization and Application in the Synthesis of Phosphatidic Acid. International Journal of Molecular Sciences. 2022; 23(19):11633. https://doi.org/10.3390/ijms231911633
Chicago/Turabian StyleWang, Fanghua, Xuejing Mao, Fuli Deng, Ruiguo Cui, Lilang Li, Siyu Liu, Bo Yang, Dongming Lan, and Yonghua Wang. 2022. "A New Phospholipase D from Moritella sp. JT01: Biochemical Characterization, Crystallization and Application in the Synthesis of Phosphatidic Acid" International Journal of Molecular Sciences 23, no. 19: 11633. https://doi.org/10.3390/ijms231911633
APA StyleWang, F., Mao, X., Deng, F., Cui, R., Li, L., Liu, S., Yang, B., Lan, D., & Wang, Y. (2022). A New Phospholipase D from Moritella sp. JT01: Biochemical Characterization, Crystallization and Application in the Synthesis of Phosphatidic Acid. International Journal of Molecular Sciences, 23(19), 11633. https://doi.org/10.3390/ijms231911633




