Homozygous CRISPR/Cas9 Knockout Generated a Novel Functionally Active Exon 1 Skipping XPA Variant in Melanoma Cells
Abstract
1. Introduction
2. Results
2.1. CRISPR/Cas9 Application
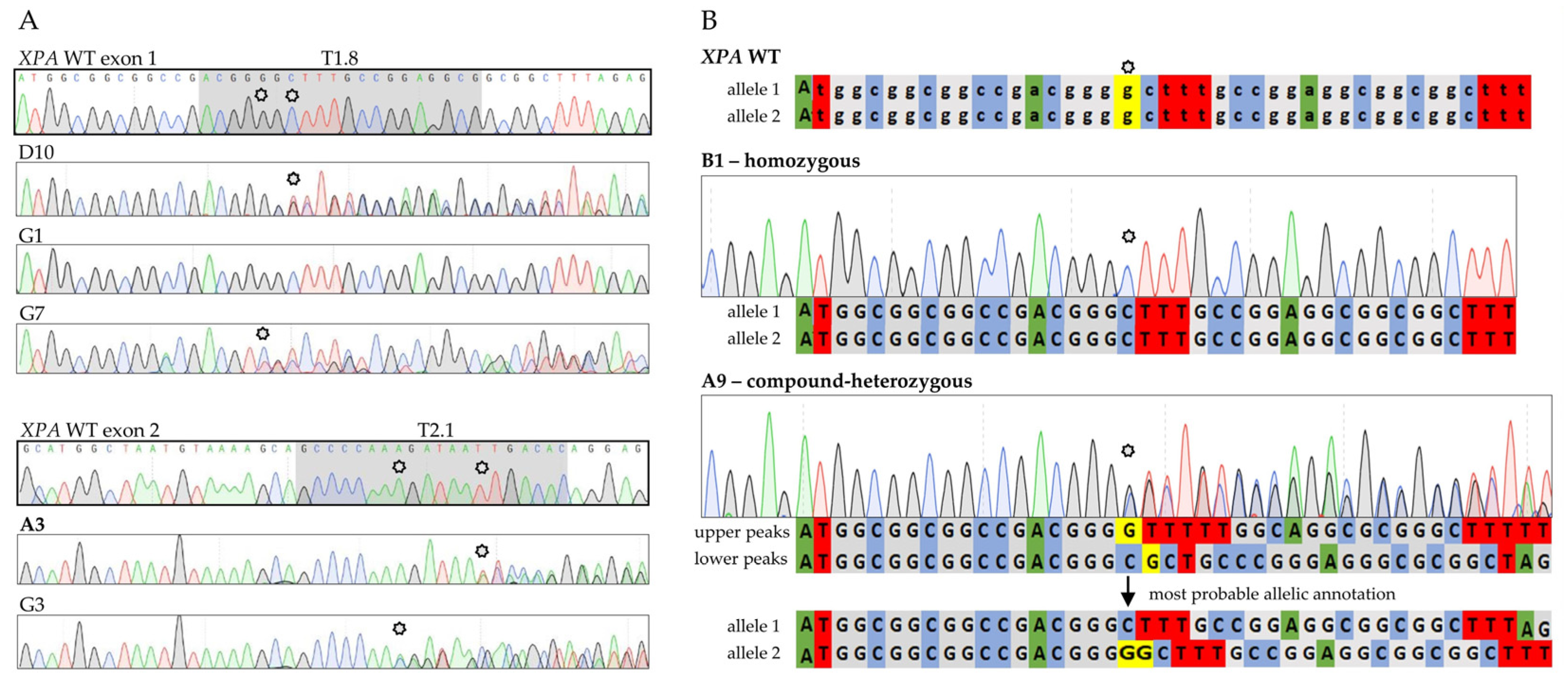
2.2. Establishment of Stable XPA-Mutated Cell Lines
2.3. XPA Expression
2.4. UV-Induced DNA Damage and Its Repair
2.5. Post-UVC Metabolic Activity Indicating Cell Survival
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Generation of sgRNA-PX459 Plasmids
4.3. Generation of XPA-Mutated A375 Subclonal Cell Lines
4.4. Immunofluorescence
4.5. Immunoblotting
4.6. Host Cell Reactivation (HCR) Assay
4.7. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide Tetrazolium (MTT) Assay
4.8. RNA Extraction
4.9. Two-Step Quantitative Reverse Transcription PCR (RT-qPCR)
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stewart, R.A.; Pilié, P.G.; Yap, T.A. Development of PARP and Immune-Checkpoint Inhibitor Combinations. Cancer Res. 2018, 78, 6717–6725. [Google Scholar] [CrossRef] [PubMed]
- Mouw, K.W.; Goldberg, M.S.; Konstantinopoulos, P.A.; D’Andrea, A.D. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov. 2017, 7, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Siddiqui, B.A.; Anandhan, S.; Yadav, S.S.; Subudhi, S.K.; Gao, J.; Goswami, S.; Allison, J.P. The Next Decade of Immune Checkpoint Therapy. Cancer Discov. 2021, 11, 838–857. [Google Scholar] [CrossRef] [PubMed]
- Marcus, L.; Lemery, S.J.; Keegan, P.; Pazdur, R. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clin. Cancer Res. 2019, 25, 3753–3758. [Google Scholar] [CrossRef] [PubMed]
- Chambon, F.; Osdoit, S.; Bagny, K.; Moro, A.; Nguyen, J.; Réguerre, Y. Dramatic response to nivolumab in xeroderma pigmentosum skin tumor. Pediatr. Blood Cancer 2018, 65, e26837. [Google Scholar] [CrossRef]
- Deinlein, T.; Lax, S.F.; Schwarz, T.; Giuffrida, R.; Schmid-Zalaudek, K.; Zalaudek, I. Rapid response of metastatic cutaneous squamous cell carcinoma to pembrolizumab in a patient with xeroderma pigmentosum: Case report and review of the literature. Eur. J. Cancer 2017, 83, 99–102. [Google Scholar] [CrossRef]
- Hauschild, A.; Eichstaedt, J.; Möbus, L.; Kähler, K.; Weichenthal, M.; Schwarz, T.; Weidinger, S. Regression of melanoma metastases and multiple non-melanoma skin cancers in xeroderma pigmentosum by the PD1-antibody pembrolizumab. Eur. J. Cancer 2017, 77, 84–87. [Google Scholar] [CrossRef]
- Salomon, G.; Maza, A.; Boulinguez, S.; Paul, C.; Lamant, L.; Tournier, E.; Mazereeuw-Hautier, J.; Meyer, N. Efficacy of anti-programmed cell death-1 immunotherapy for skin carcinomas and melanoma metastases in a patient with xeroderma pigmentosum. Br. J. Dermatol. 2018, 178, 1199–1203. [Google Scholar] [CrossRef]
- Steineck, A.; Krumm, N.; Sarthy, J.F.; Pritchard, C.C.; Chapman, T.; Stacey, A.W.; Vitanza, N.A.; Cole, B. Response to Pembrolizumab in a Patient With Xeroderma Pigmentosum and Advanced Squamous Cell Carcinoma. JCO Precis. Oncol. 2019, 3, PO.19.00028. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Permata, T.B.M.; Hagiwara, Y.; Sato, H.; Yasuhara, T.; Oike, T.; Gondhowiardjo, S.; Held, K.D.; Nakano, T.; Shibata, A. Base excision repair regulates PD-L1 expression in cancer cells. Oncogene 2019, 38, 4452–4466. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Hamed, M.; Emmert, S.; Wolkenhauer, O.; Fuellen, G.; Thiem, A. The Prognostic and Predictive Role of Xeroderma Pigmentosum Gene Expression in Melanoma. Front. Oncol. 2022, 12, 810058. [Google Scholar] [CrossRef] [PubMed]
- Borszéková Pulzová, L.; Ward, T.A.; Chovanec, M. XPA: DNA Repair Protein of Significant Clinical Importance. Int. J. Mol. Sci 2020, 21, 2182. [Google Scholar] [CrossRef] [PubMed]
- Fadda, E. Role of the XPA protein in the NER pathway: A perspective on the function of structural disorder in macromolecular assembly. Comput. Struct. Biotechnol. J. 2016, 14, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef]
- Sugitani, N.; Sivley, R.M.; Perry, K.E.; Capra, J.A.; Chazin, W.J. XPA: A key scaffold for human nucleotide excision repair. DNA Repair 2016, 44, 123–135. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Jansen, R.; Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, C.J.; Lin, A.; Girish, V.; Sheltzer, J.M. Generating Single Cell-Derived Knockout Clones in Mammalian Cells with CRISPR/Cas9. Curr. Protoc. Mol. Biol. 2019, 128, e100. [Google Scholar] [CrossRef] [PubMed]
- Tuladhar, R.; Yeu, Y.; Tyler Piazza, J.; Tan, Z.; Rene Clemenceau, J.; Wu, X.; Barrett, Q.; Herbert, J.; Mathews, D.H.; Kim, J.; et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 2019, 10, 4056. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Lindeboom, R.G.H.; Vermeulen, M.; Lehner, B.; Supek, F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat. Genet. 2019, 51, 1645–1651. [Google Scholar] [CrossRef]
- Popp, M.W.; Maquat, L.E. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell 2016, 165, 1319–1322. [Google Scholar] [CrossRef]
- Liu, Y.; Peterson, D.A.; Kimura, H.; Schubert, D. Mechanism of cellular 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. J. Neurochem. 1997, 69, 581–593. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Ghasemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 12827. [Google Scholar] [CrossRef]
- Śliwka, L.; Wiktorska, K.; Suchocki, P.; Milczarek, M.; Mielczarek, S.; Lubelska, K.; Cierpiał, T.; Łyżwa, P.; Kiełbasiński, P.; Jaromin, A.; et al. The Comparison of MTT and CVS Assays for the Assessment of Anticancer Agent Interactions. PLoS ONE 2016, 11, e0155772. [Google Scholar] [CrossRef] [PubMed]
- van Tonder, A.; Joubert, A.M.; Cromarty, A.D. Limitations of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay when compared to three commonly used cell enumeration assays. BMC Res. Notes 2015, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.; Rieper, P.; Ohlenbusch, A.; Seebode, C.; Lehmann, J.; Gratchev, A.; Emmert, S. A unique chromosomal in-frame deletion identified among seven XP-C patients. Photodermatol. Photoimmunol. Photomed. 2016, 32, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Bartels, C.L.; Lambert, M.W. Domains in the XPA protein important in its role as a processivity factor. Biochem. Biophys. Res. Commun. 2007, 356, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Fagbemi, A.F.; Orelli, B.; Schärer, O.D. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair 2011, 10, 722–729. [Google Scholar] [CrossRef]
- Feltes, B.C. Every protagonist has a sidekick: Structural aspects of human xeroderma pigmentosum-binding proteins in nucleotide excision repair. Protein Sci. 2021, 30, 2187–2205. [Google Scholar] [CrossRef]
- Krasikova, Y.S.; Rechkunova, N.I.; Maltseva, E.A.; Lavrik, O.I. RPA and XPA interaction with DNA structures mimicking intermediates of the late stages in nucleotide excision repair. PLoS ONE 2018, 13, e0190782. [Google Scholar] [CrossRef]
- Volker, M.; Moné, M.J.; Karmakar, P.; van Hoffen, A.; Schul, W.; Vermeulen, W.; Hoeijmakers, J.H.; van Driel, R.; van Zeeland, A.A.; Mullenders, L.H. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 2001, 8, 213–224. [Google Scholar] [CrossRef]
- Touat, M.; Sourisseau, T.; Dorvault, N.; Chabanon, R.M.; Garrido, M.; Morel, D.; Krastev, D.B.; Bigot, L.; Adam, J.; Frankum, J.R.; et al. DNA repair deficiency sensitizes lung cancer cells to NAD+ biosynthesis blockade. J. Clin. Investig. 2018, 128, 1671–1687. [Google Scholar] [CrossRef]
- Martens, M.C.; Edelkamp, J.; Seebode, C.; Schafer, M.; Stahlke, S.; Krohn, S.; Jung, O.; Murua Escobar, H.; Emmert, S.; Boeckmann, L. Generation and Characterization of a CRISPR/Cas9-Mediated SNAP29 Knockout in Human Fibroblasts. Int. J. Mol. Sci. 2021, 22, 5293. [Google Scholar] [CrossRef]
- Ribas, A. Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov. 2015, 5, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Thiem, A.; Hesbacher, S.; Kneitz, H.; di Primio, T.; Heppt, M.V.; Hermanns, H.M.; Goebeler, M.; Meierjohann, S.; Houben, R.; Schrama, D. IFN-gamma-induced PD-L1 expression in melanoma depends on p53 expression. J. Exp. Clin. Cancer Res. 2019, 38, 397. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banicka, V.; Martens, M.C.; Panzer, R.; Schrama, D.; Emmert, S.; Boeckmann, L.; Thiem, A. Homozygous CRISPR/Cas9 Knockout Generated a Novel Functionally Active Exon 1 Skipping XPA Variant in Melanoma Cells. Int. J. Mol. Sci. 2022, 23, 11649. https://doi.org/10.3390/ijms231911649
Banicka V, Martens MC, Panzer R, Schrama D, Emmert S, Boeckmann L, Thiem A. Homozygous CRISPR/Cas9 Knockout Generated a Novel Functionally Active Exon 1 Skipping XPA Variant in Melanoma Cells. International Journal of Molecular Sciences. 2022; 23(19):11649. https://doi.org/10.3390/ijms231911649
Chicago/Turabian StyleBanicka, Veronika, Marie Christine Martens, Rüdiger Panzer, David Schrama, Steffen Emmert, Lars Boeckmann, and Alexander Thiem. 2022. "Homozygous CRISPR/Cas9 Knockout Generated a Novel Functionally Active Exon 1 Skipping XPA Variant in Melanoma Cells" International Journal of Molecular Sciences 23, no. 19: 11649. https://doi.org/10.3390/ijms231911649
APA StyleBanicka, V., Martens, M. C., Panzer, R., Schrama, D., Emmert, S., Boeckmann, L., & Thiem, A. (2022). Homozygous CRISPR/Cas9 Knockout Generated a Novel Functionally Active Exon 1 Skipping XPA Variant in Melanoma Cells. International Journal of Molecular Sciences, 23(19), 11649. https://doi.org/10.3390/ijms231911649







