Identification of HGD and GSTZ1 as Biomarkers Involved Metabolic Reprogramming in Kidney Renal Clear Cell Carcinoma
Abstract
1. Introduction
2. Results
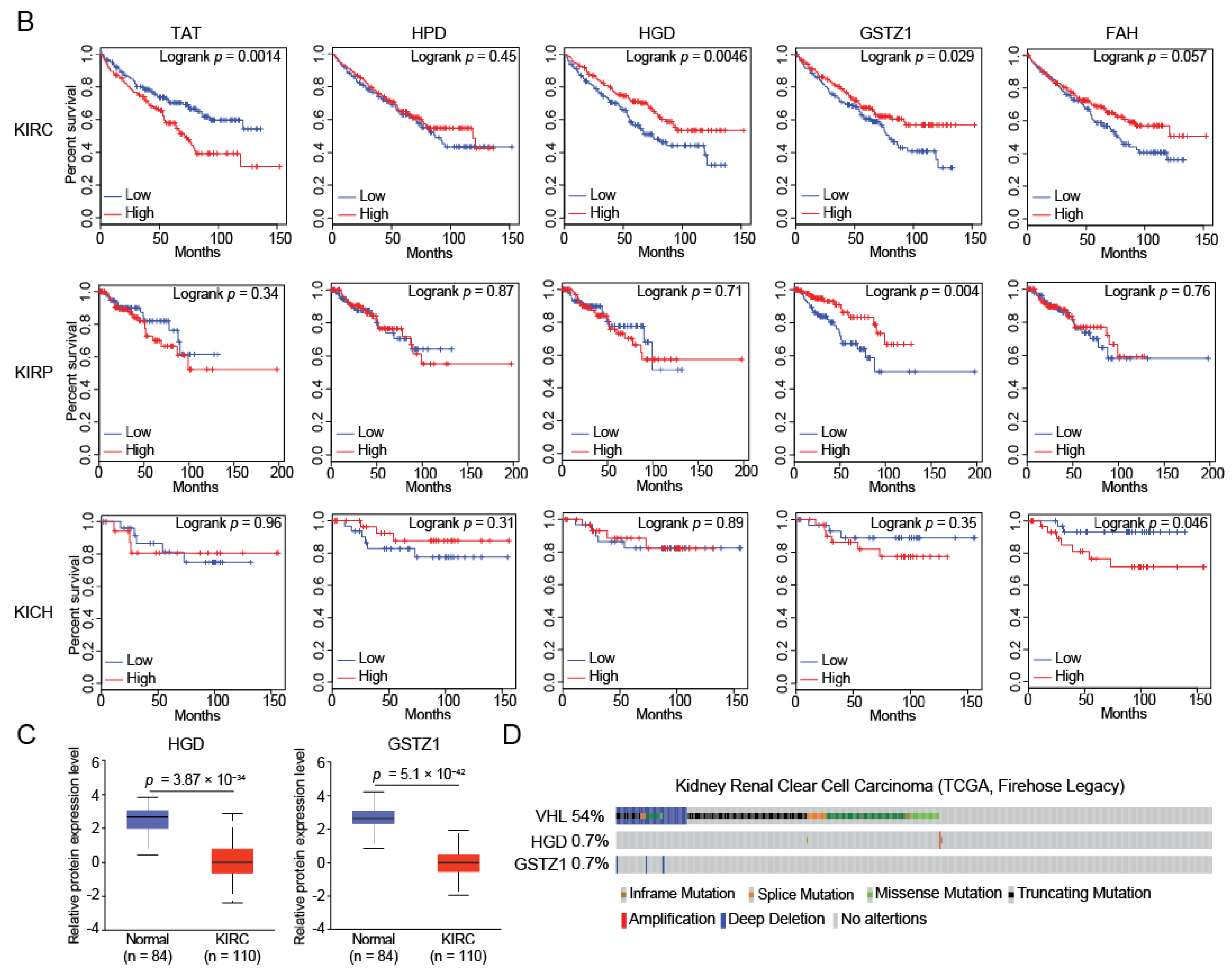
2.1. HGD and GSTZ1 Were Down-Regulated and Associated with Prognosis in KIRC
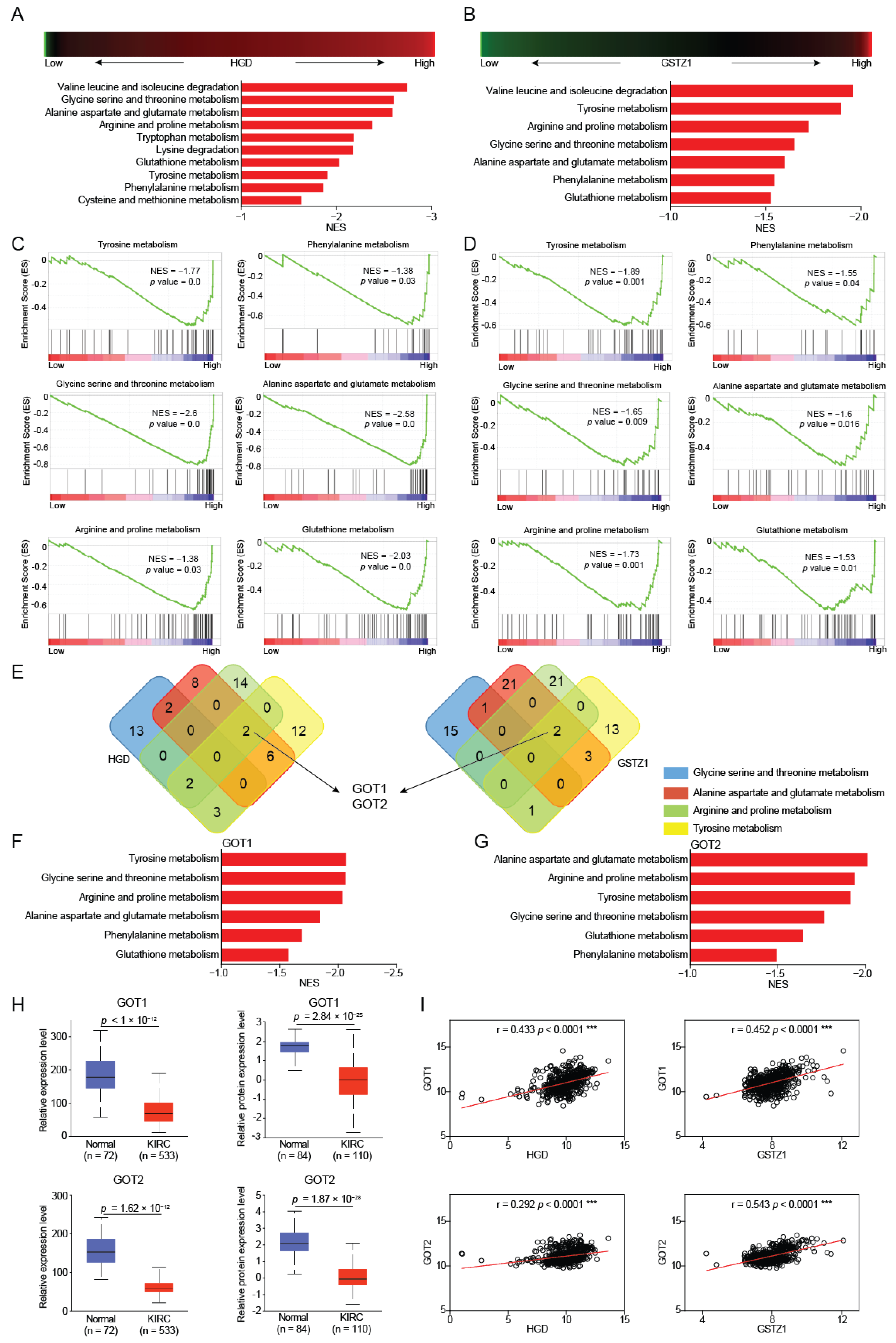
2.2. GOT1/2 Was Required for Amino Acid Metabolism Regulated by HGD/GSTZ1
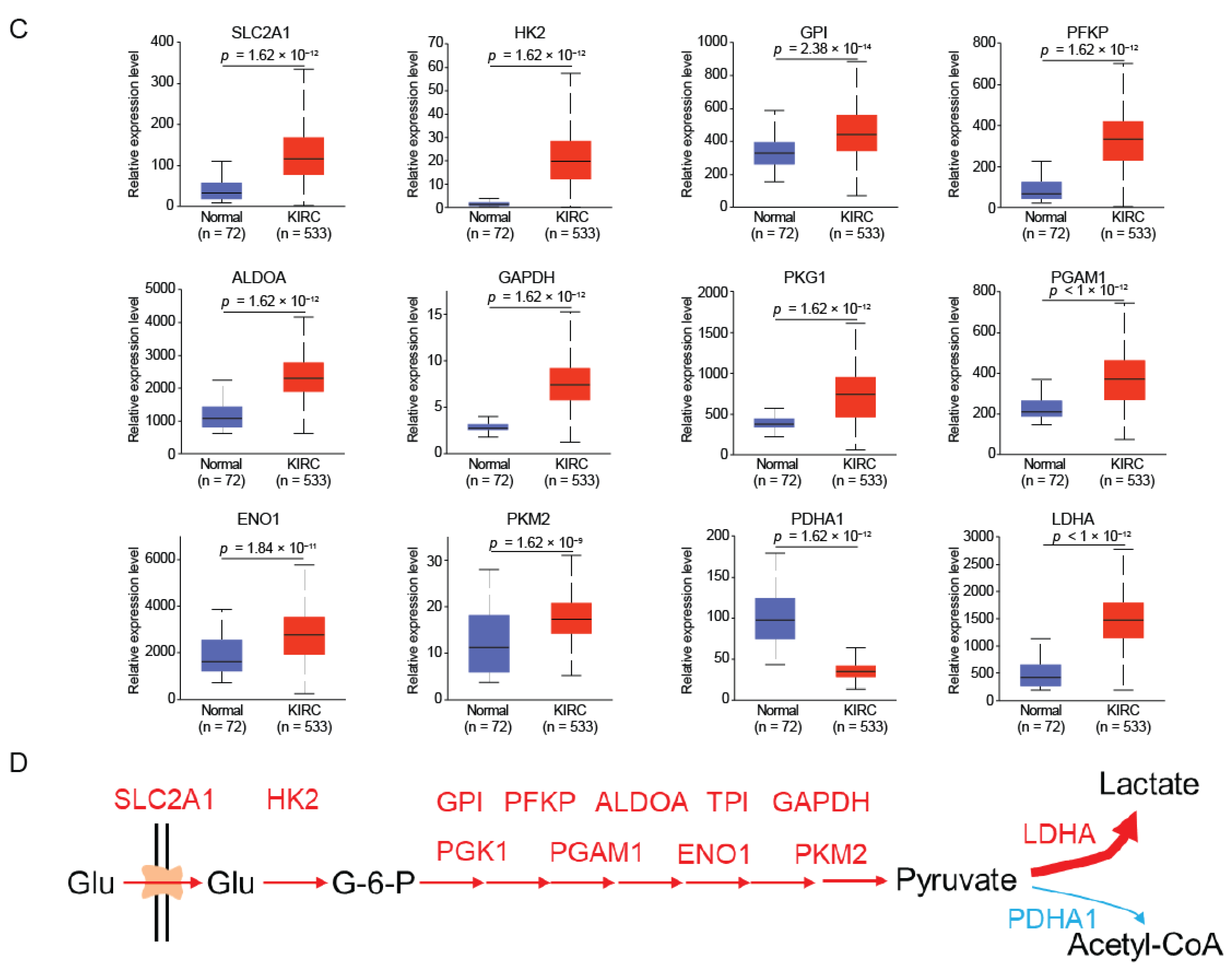
2.3. HGD and GSTZ1 Promoted the Conversion of Glucose to Lactate in KIRC
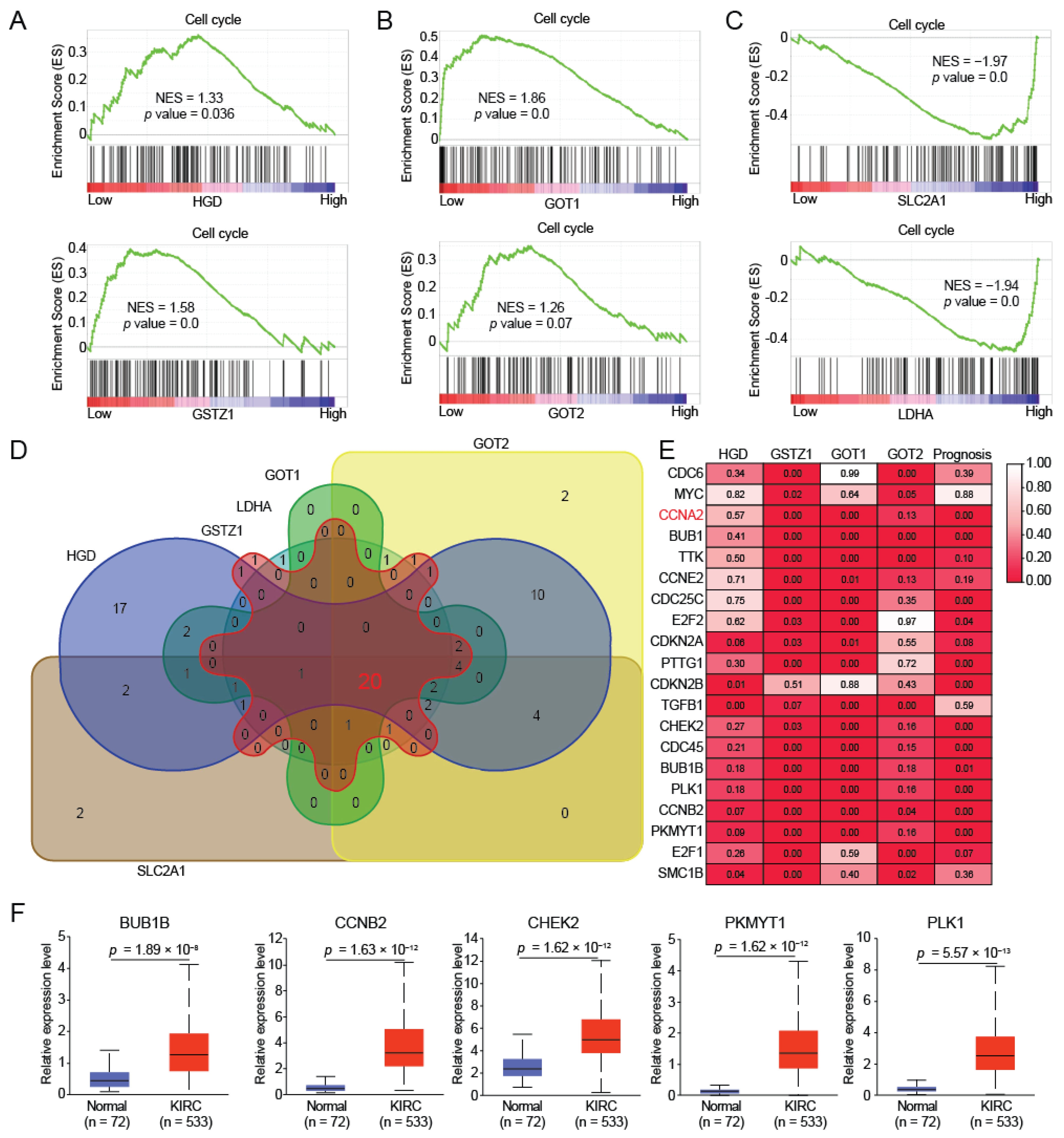
2.4. HGD/GSTZ1 Promoted Cell Cycle and Tumor Progression
3. Discussion
4. Materials and Methods
4.1. The Analysis for Differential Gene Expression and Mutation
4.2. Survival Prognostic Analysis
4.3. Gene Set Enrichment Analysis (GSEA)
4.4. Correlation Analysis
4.5. Target Overlap Analysis
4.6. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Makhov, P.; Joshi, S.; Ghatalia, P.; Kutikov, A.; Uzzo, R.G.; Kolenko, V.M. Resistance to Systemic Therapies in Clear Cell Renal Cell Carcinoma: Mechanisms and Management Strategies. Mol. Cancer Ther. 2018, 17, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Seliger, B.; Jasinski, S.; Dressler, S.P.; Marincola, F.M.; Recktenwald, C.V.; Wang, E.; Lichtenfels, R. Linkage of microRNA and proteome-based profiling data sets: A perspective for the priorization of candidate biomarkers in renal cell carcinoma? J. Proteome Res. 2011, 10, 191–199. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Staehler, M.; Schuler, T.; Spek, A.; Rodler, S.; Tamalunas, A.; Fürweger, C.; Muacevic, A. Propensity Score-Matched Analysis of Single Fraction Robotic Radiosurgery Versus Open Partial Nephrectomy in Renal Cell Carcinoma: Oncological Outcomes. Cureus 2022, 14, e21623. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Wettersten, H.I.; Hakimi, A.A.; Morin, D.; Bianchi, C.; Johnstone, M.E.; Donohoe, D.R.; Trott, J.F.; Aboud, O.A.; Stirdivant, S.; Neri, B.; et al. Grade-Dependent Metabolic Reprogramming in Kidney Cancer Revealed by Combined Proteomics and Metabolomics Analysis. Cancer Res. 2015, 75, 2541–2552. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Reznik, E.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef]
- Nizioł, J.; Copié, V.; Tripet, B.P.; Nogueira, L.B.; Nogueira, K.; Ossoliński, K.; Arendowski, A.; Ruman, T. Metabolomic and elemental profiling of human tissue in kidney cancer. Metabolomics 2021, 17, 30. [Google Scholar] [CrossRef]
- Linehan, W.M.; Schmidt, L.S.; Crooks, D.R.; Wei, D.; Srinivasan, R.; Lang, M. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019, 9, 1006–1021. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qiao, Y.; Sun, H.; Chang, H.; Zhao, H.; Zhang, S.; Shan, C. Decreased SLC27A5 Suppresses Lipid Synthesis and Tyrosine Metabolism to Activate the Cell Cycle in Hepatocellular Carcinoma. Biomedicines 2022, 10, 234. [Google Scholar] [CrossRef] [PubMed]
- Lubensky, I.A.; Gnarra, J.R.; Bertheau, P.; Walther, M.M.; Linehan, W.M.; Zhuang, Z. Allelic deletions of the VHL gene detected in multiple microscopic clear cell renal lesions in von Hippel-Lindau disease patients. Am. J. Pathol. 1996, 149, 2089–2094. [Google Scholar] [PubMed]
- Lonser, R.R.; Glenn, G.M.; Walther, M.; Chew, E.Y.; Libutti, S.K.; Linehan, W.M.; Oldfield, E.H. Von Hippel-Lindau disease. Lancet 2003, 361, 2059–2067. [Google Scholar] [CrossRef]
- Choo, D.; Shotland, L.; Mastroianni, M.; Glenn, G.; van Waes, C.; Linehan, W.M.; Oldfield, E.H. Endolymphatic sac tumors in von Hippel-Lindau disease. J. Neurosurg. 2004, 100, 480–487. [Google Scholar] [CrossRef]
- Lubensky, I.A.; Pack, S.; Ault, D.; Vortmeyer, A.O.; Libutti, S.K.; Choyke, P.L.; Walther, M.M.; Linehan, W.M.; Zhuang, Z. Multiple neuroendocrine tumors of the pancreas in von Hippel-Lindau disease patients: Histopathological and molecular genetic analysis. Am. J. Pathol. 1998, 153, 223–231. [Google Scholar] [CrossRef]
- Walther, M.M.; Keiser, H.R.; Choyke, P.L.; Rayford, W.; Lyne, J.C.; Linehan, W.M. Management of hereditary pheochromocytoma in von Hippel-Lindau kindreds with partial adrenalectomy. J. Urol. 1999, 161, 395–398. [Google Scholar] [CrossRef]
- Laschi, M.; Tinti, L.; Braconi, D.; Millucci, L.; Ghezzi, L.; Amato, L.; Selvi, E.; Spreafico, A.; Bernardini, G.; Santucci, A. Homogentisate 1,2 dioxygenase is expressed in human osteoarticular cells: Implications in alkaptonuria. J. Cell Physiol. 2012, 227, 3254–3257. [Google Scholar] [CrossRef]
- Zatková, A.; Polaková, H.; Micutková, L.; Zvarík, M.; Bosák, V.; Feráková, E.; Matusek, J.; Ferák, V.; Kádasi, L. Novel mutations in the homogentisate-1,2-dioxygenase gene identified in Slovak patients with alkaptonuria. J. Med. Genet. 2000, 37, 539–542. [Google Scholar] [CrossRef]
- Ascher, D.B.; Spiga, O.; Sekelska, M.; Pires, D.E.V.; Bernini, A.; Tiezzi, M.; Kralovicova, J.; Borovska, I.; Soltysova, A.; Olsson, B.; et al. Homogentisate 1,2-dioxygenase (HGD) gene variants, their analysis and genotype-phenotype correlations in the largest cohort of patients with AKU. Eur. J. Hum. Genet. 2019, 27, 888–902. [Google Scholar] [CrossRef]
- Yang, Y.J.; Guo, J.H.; Chen, W.J.; Zhao, R.; Tang, J.S.; Meng, X.H.; Zhao, L.; Tu, M.; He, X.Y.; Wu, L.Q.; et al. First report of HGD mutations in a Chinese with alkaptonuria. Gene 2013, 518, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Bin, C.; Xue, Q.; Gao, Q.; Huang, A. GSTZ1 sensitizes hepatocellular carcinoma cells to sorafenib-induced ferroptosis via inhibition of NRF2/GPX4 axis. Cell Death Dis. 2021, 12, 426. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Wang, Q.; Tang, N.; Wang, K. GSTZ1-1 downregulates Wnt/β-catenin signalling in hepatocellular carcinoma cells. FEBS Open Bio 2020, 10, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Q.; Yang, Y.; Lei, C.; Yang, F.; Liang, L.; Chen, C.; Xia, J.; Wang, K.; Tang, N. GSTZ1 deficiency promotes hepatocellular carcinoma proliferation via activation of the KEAP1/NRF2 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 438. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, J.; Deng, H.; Wang, Y.; Lei, C.; Wang, Q.; Xiang, J.; Liang, L.; Xia, J.; Pan, X.; et al. GSTZ1-1 Deficiency Activates NRF2/IGF1R Axis in HCC via Accumulation of Oncometabolite Succinylacetone. EMBO J. 2019, 38, e101964. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019, 10, 5679. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Chang, H.; Su, M.; Qiao, Y.; Sun, H.; Zhao, Y.; Zhang, S.; Shan, C. Identification of HGD and GSTZ1 as Biomarkers Involved Metabolic Reprogramming in Kidney Renal Clear Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 4583. https://doi.org/10.3390/ijms23094583
Wang J, Chang H, Su M, Qiao Y, Sun H, Zhao Y, Zhang S, Shan C. Identification of HGD and GSTZ1 as Biomarkers Involved Metabolic Reprogramming in Kidney Renal Clear Cell Carcinoma. International Journal of Molecular Sciences. 2022; 23(9):4583. https://doi.org/10.3390/ijms23094583
Chicago/Turabian StyleWang, Jiyan, Hongkai Chang, Meng Su, Yaya Qiao, Huanran Sun, Yongshan Zhao, Shuai Zhang, and Changliang Shan. 2022. "Identification of HGD and GSTZ1 as Biomarkers Involved Metabolic Reprogramming in Kidney Renal Clear Cell Carcinoma" International Journal of Molecular Sciences 23, no. 9: 4583. https://doi.org/10.3390/ijms23094583
APA StyleWang, J., Chang, H., Su, M., Qiao, Y., Sun, H., Zhao, Y., Zhang, S., & Shan, C. (2022). Identification of HGD and GSTZ1 as Biomarkers Involved Metabolic Reprogramming in Kidney Renal Clear Cell Carcinoma. International Journal of Molecular Sciences, 23(9), 4583. https://doi.org/10.3390/ijms23094583





