Putative Transcription Factor Genes Associated with Regulation of Carotenoid Biosynthesis in Chili Pepper Fruits Revealed by RNA-Seq Coexpression Analysis
Abstract
1. Introduction
2. Results
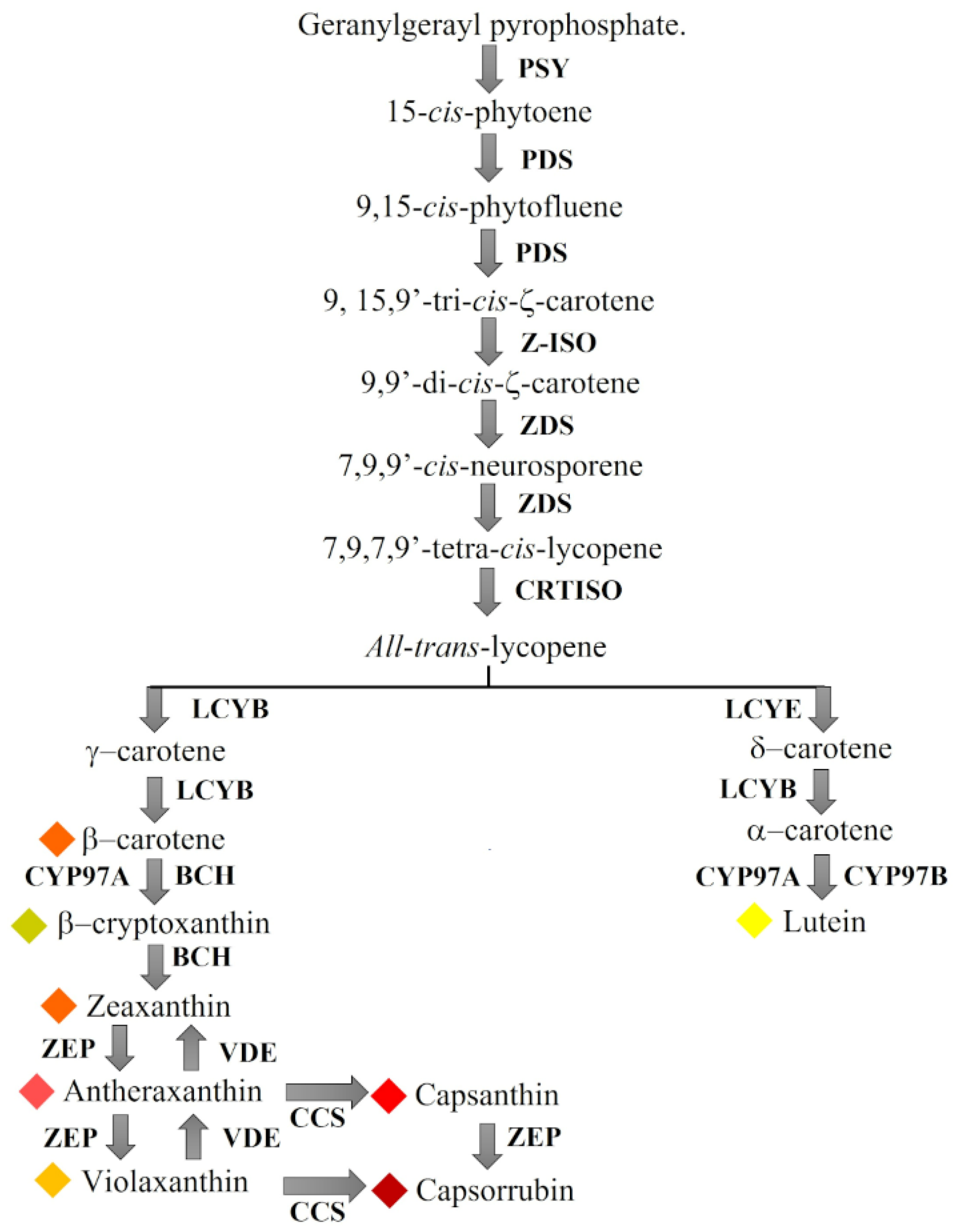
2.1. Expression Profiles of Carotenoid-Biosynthesis-Related Genes
2.2. Coexpression Analysis of Carotenoid-Biosynthesis-Related Genes and TF Genes
3. Discussion
4. Materials and Methods
4.1. Biological Materials, Sequencing, and RNA-Seq Data Analysis
4.2. Selection of TF Candidates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gómez-García, M.d.R.; Ochoa-Alejo, N. Biochemistry and molecular biology of carotenoid biosynthesis in chili peppers (Capsicum spp.). Int. J. Mol. Sci. 2013, 14, 19025. [Google Scholar] [CrossRef] [PubMed]
- Villa-Rivera, M.G.; Ochoa-Alejo, N. Chili pepper carotenoids: Nutraceutical properties and mechanisms of action. Molecules 2020, 25, 5573. [Google Scholar] [CrossRef] [PubMed]
- Villa-Rivera, M.G.; Ochoa-Alejo, N. Transcriptional regulation of ripening in chili pepper fruits (Capsicum spp.). Int. J. Mol. Sci. 2021, 22, 12151. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.; Fray, R.; Marshall, A.; Jayasuta, S.; Baxter, C.; van Wijk, R.; et al. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef]
- Song, J.; Chen, C.; Zhang, S.; Wang, J.; Huang, Z.; Chen, M.; Cao, B.; Zhu, Z.; Lei, J. Systematic analysis of the Capsicum ERF transcription factor family: Identification of regulatory factors involved in the regulation of species-specific metabolites. BMC Genom. 2020, 21, 573. [Google Scholar] [CrossRef]
- Arce-Rodríguez, M.L.; Martinez, O.; Ochoa-Alejo, N. Genome-wide identification and analysis of the MYB transcription factor gene family in chili pepper (Capsicum spp.). Int. J. Mol. Sci. 2021, 22, 2229. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.H.; Yang, S.P.; Yu, Y.N.; Khan, A.; Feng, P.L.; Ali, M.; Shao, D.K.; Wang, Y.Y.; Zhang, R.X.; Gai, W.X.; et al. Comprehensive transcriptome-based characterization of differentially expressed genes involved in carotenoid biosynthesis of different ripening stages of Capsicum. Sci. Hortic. 2021, 288, 110311. [Google Scholar] [CrossRef]
- Ma, X.; Yu, Y.-N.; Jia, J.-H.; Li, Q.-H.; Gong, Z.-H. The pepper MYB transcription factor CaMYB306 accelerates fruit coloration and negatively regulates cold resistance. Sci. Hortic. 2022, 295, 110892. [Google Scholar] [CrossRef]
- Welsch, R.; Maass, D.; Voegel, T.; Dellapenna, D.; Beyer, P. Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007, 145, 1073–1085. [Google Scholar] [CrossRef]
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The tomato MADS-Box transcription factor ripening inhibitor interacts with promoters involved in numerous ripening processes in a colorless nonripening-dependent manner. Plant Physiol. 2011, 157, 1568–1579. [Google Scholar] [CrossRef]
- Mohanty, B.; Lakshmanan, M.; Lim, S.-H.; Kim, J.K.; Ha, S.-H.; Lee, D.-Y. Light-specific transcriptional regulation of the accumulation of carotenoids and phenolic compounds in rice leaves. Plant Signal. Behav. 2016, 11, e1184808. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.C.; Han, Y.C.; Kuang, J.F.; Chen, J.Y.; Lu, W.J. Papaya CpEIN3a and CpNAC2 co-operatively regulate carotenoid biosynthesis-related genes CpPDS2/4, CpLCY-e and CpCHY-b during fruit ripening. Plant Cell Physiol. 2017, 58, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, Y.; Su, D.; Lu, W.; Li, Z. SlGRAS4 accelerates fruit ripening by regulating ethylene biosynthesis genes and SlMADS1 in tomato. Hortic. Res. 2021, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Ríos, G.; Naranjo, M.A.; Rodrigo, M.-J.; Alós, E.; Zacarías, L.; Cercós, M.; Talón, M. Identification of a GCC transcription factor responding to fruit colour change events in citrus through the transcriptomic analyses of two mutants. BMC Plant Biol. 2010, 10, 276. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Trapero-Mozos, A.; Gómez, M.D.; Rubio-Moraga, A.; Ahrazem, O. Identification and possible role of a MYB transcription factor from saffron (Crocus sativus). J. Plant Physiol. 2012, 169, 509–515. [Google Scholar] [CrossRef]
- Grassi, S.; Piro, G.; Lee, J.M.; Zheng, Y.; Fei, Z.; Dalessandro, G.; Giovannoni, J.J.; Lenucci, M.S. Comparative genomics reveals candidate carotenoid pathway regulators of ripening watermelon fruit. BMC Genom. 2013, 14, 781. [Google Scholar] [CrossRef]
- Ye, J.; Hu, T.; Yang, C.; Li, H.; Yang, M.; Ijaz, R.; Ye, Z.; Zhang, Y. Transcriptome profiling of tomato fruit development reveals transcription factors associated with ascorbic acid, carotenoid and flavonoid biosynthesis. PLoS ONE 2015, 10, e0130885. [Google Scholar] [CrossRef]
- Jiang, C.C.; Zhang, Y.F.; Lin, Y.J.; Chen, Y.; Lu, X.K. Illumina® sequencing reveals candidate genes of carotenoid metabolism in three pummelo cultivars (Citrus maxima) with different pulp color. Int. J. Mol. Sci. 2019, 20, 2246. [Google Scholar] [CrossRef]
- Lu, C.; Pu, Y.; Liu, Y.; Li, Y.; Qu, J.; Huang, H.; Dai, S. Comparative transcriptomics and weighted gene co-expression correlation network analysis (WGCNA) reveal potential regulation mechanism of carotenoid accumulation in Chrysanthemum × morifolium. Plant Physiol. Biochem. 2019, 142, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Martínez, O.; Arce-Rodríguez, M.L.; Hernández-Godinez, F.; Escoto-Sandoval, C.; Cervantes-Hernández, F.; Hayano-Kanashiro, C.; Ordaz-Ortíz, J.J.; Reyes-Valdes, M.H.; Razo-Mendivil, F.G.; Garces-Claver, A.; et al. Transcriptome analyses throughout chili pepper fruit development reveal novel insights into the domestication process. Plants 2021, 10, 585. [Google Scholar] [CrossRef] [PubMed]
- Escoto-Sandoval, C.; Flores-Diaz, A.; Reyes-Valdes, M.H.; Ochoa-Alejo, N.; Martínez, O. A method to analyze time expression profiles demonstrated in a database of chili pepper fruit development. Sci. Rep. 2021, 11, 13181. [Google Scholar] [CrossRef] [PubMed]
- Martínez-López, L.A.; Ochoa-Alejo, N.; Martínez, O. Dynamics of the chili pepper transcriptome during fruit development. BMC Genom. 2014, 15, 143. [Google Scholar] [CrossRef]
- Escoto-Sandoval, C.; Flores-Díaz, A.; Reyes-Valdés, M.H.; Ochoa-Alejo, N.; Martínez, O. An R Package for Data Mining Chili Pepper Fruit Transcriptomes; Research Square: Durham, CA, USA, 2020. [Google Scholar]
- Maoka, T. Carotenoids as natural functional pigments. J. Nat. Med. 2020, 74, 1–16. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Llorente, B.; Martinez-Garcia, J.F.; Stange, C.; Rodriguez-Concepcion, M. Illuminating colors: Regulation of carotenoid biosynthesis and accumulation by light. Curr. Opin. Plant Biol. 2017, 37, 49–55. [Google Scholar] [CrossRef]
- Sathasivam, R.; Radhakrishnan, R.; Kim, J.K.; Park, S.U. An update on biosynthesis and regulation of carotenoids in plants. S. Af. J. Bot. 2021, 140, 290–302. [Google Scholar] [CrossRef]
- Tian, S.L.; Li, L.; Shah, S.N.M.; Gong, Z.H. The relationship between red fruit colour formation and key genes of capsanthin biosynthesis pathway in Capsicum annuum. Biol. Plant. 2015, 59, 507–513. [Google Scholar] [CrossRef]
- Liu, R.; Song, J.; Liu, S.; Chen, C.; Zhang, S.; Wang, J.; Xiao, Y.; Cao, B.; Lei, J.; Zhu, Z. Genome-wide identification of the Capsicum bHLH transcription factor family: Discovery of a candidate regulator involved in the regulation of species-specific bioactive metabolites. BMC Plant Biol. 2021, 21, 262. [Google Scholar] [CrossRef]
- Chen, R.; Ma, J.; Luo, D.; Hou, X.; Ma, F.; Zhang, Y.; Meng, Y.; Zhang, H.; Guo, W. CaMADS, a MADS-box transcription factor from pepper, plays an important role in the response to cold, salt, and osmotic stress. Plant Sci. 2019, 280, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Soon-Kee, S.; Yong-Hwan, M.; Jae-Eun, C.; Sook-Yi, L.; HyoGuen, P.; Gynheung, A. Characterization of MADS box genes from hot pepper. Mol. Cells 2001, 11, 352–359. [Google Scholar]
- Dong, T.; Chen, G.; Tian, S.; Xie, Q.; Yin, W.; Zhang, Y.; Hu, Z. A non-climacteric fruit gene CaMADS-RIN regulates fruit ripening and ethylene biosynthesis in climacteric fruit. PLoS ONE 2014, 9, e95559. [Google Scholar] [CrossRef] [PubMed]
- Dubey, M.; Jaiswal, V.; Rawoof, A.; Kumar, A.; Nitin, M.; Chhapekar, S.S.; Kumar, N.; Ahmad, I.; Islam, K.; Brahma, V.; et al. Identification of genes involved in fruit development/ripening in Capsicum and development of functional markers. Genomics 2019, 111, 1913–1922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, Z.; Yao, Q.; Guo, X.; Nguyen, V.; Li, F.; Chen, G. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 2018, 8, 3413. [Google Scholar] [CrossRef]
- Li, C.X.; Hou, X.M.; Qi, N.A.; Liu, H.W.; Li, Y.H.; Huang, D.J.; Wang, C.L.; Liao, W.B. Insight into ripening-associated transcription factors in tomato: A review. Sci. Hortic. 2021, 288, 110363. [Google Scholar] [CrossRef]
- Li, J.G.; Li, H.L.; Peng, S.Q. Three R2R3 MYB transcription factor genes from Capsicum annuum showing differential expression during fruit ripening. Afr. J. Biotechnol. 2011, 10, 8267–8274. [Google Scholar] [CrossRef]
- Arce-Rodríguez, M.L.; Ochoa-Alejo, N. An R2R3-MYB transcription factor regulates capsaicinoid biosynthesis. Plant Physiol. 2017, 174, 1359–1370. [Google Scholar] [CrossRef]
- Sun, B.; Zhu, Z.; Chen, C.; Chen, G.; Cao, B.; Chen, C.; Lei, J. Jasmonate-inducible R2R3-MYB transcription factor regulates capsaicinoid biosynthesis and stamen development in Capsicum. J. Agric. Food Chem. 2019, 67, 10891–10903. [Google Scholar] [CrossRef]
- Zhang, R.-X.; Cheng, G.-X.; Liu, G.-T.; Chen, S.-Y.; Haq, S.u.; Khan, A.; Li, Q.-H.; Gong, Z.-H. Assessing the functional role of color-related CaMYB gene under cold stress using virus-induced gene silencing in the fruit of pepper (Capsicum annuum L.). Sci. Hortic. 2020, 272, 109504. [Google Scholar] [CrossRef]
- Yu, C.; Cai, X.; Ye, Z.; Li, H. Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem. Biophys. Res. Commun. 2015, 468, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors—Light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y.; Inaba, T.; Furuhashi, H.; Sasaki, Y. Trihelix DNA-binding protein with specificities for two distinct cis-elements—Both important for light down-regulated and dark-inducible gene expression in higher plants. J. Biol. Chem. 2001, 276, 22238–22243. [Google Scholar] [CrossRef]
- Cheng, Y.; Ahammed, G.J.; Yu, J.; Yao, Z.; Ruan, M.; Ye, Q.; Li, Z.; Wang, R.; Feng, K.; Zhou, G.; et al. Putative WRKYs associated with regulation of fruit ripening revealed by detailed expression analysis of the WRKY gene family in pepper. Sci. Rep. 2016, 6, 39000. [Google Scholar] [CrossRef]
- Kondo, F.; Hatakeyama, K.; Sakai, A.; Minami, M.; Nemoto, K.; Matsushima, K. The pungent-variable sweet chili pepper ‘Shishito’ (Capsicum annuum) provides insights regarding the relationship between pungency, the number of seeds, and gene expression involving capsaicinoid biosynthesis. Mol. Genet. Genom. 2021, 296, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Li, X.; Weng, Y.; Liu, Z.; Ashraf, M.F.; Noman, A.; Yang, S.; Ifnan, M.; Qiu, S.; Yang, Y.; et al. CaWRKY22 acts as a positive regulator in pepper response to Ralstonia solanacearum by constituting networks with CaWRKY6, CaWRKY27, CaWRKY40, and CaWRKY58. Int. J. Mol. Sci. 2018, 19, 1426. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-K.; Lee, S.; Yu, S.H.; Choi, D. Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta 2005, 222, 876–887. [Google Scholar] [CrossRef]
- Guo, W.L.; Wang, S.B.; Chen, R.G.; Chen, B.H.; Du, X.H.; Yin, Y.X.; Gong, Z.H.; Zhang, Y.Y. Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 2015, 6, 755. [Google Scholar] [CrossRef]
- Hou, X.M.; Zhang, H.F.; Liu, S.Y.; Wang, X.K.; Zhang, Y.M.; Meng, Y.C.; Luo, D.; Chen, R.G. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef]
- Kong, X.M.; Zhou, Q.; Zhou, X.; Wei, B.D.; Ji, S.J. Transcription factor CaNAC1 regulates low-temperature-induced phospholipid degradation in green bell pepper. J. Exp. Bot. 2020, 71, 1078–1091. [Google Scholar] [CrossRef]
- Su, H.; Zhang, S.; Yin, Y.; Zhu, D.; Han, L. Genome-wide analysis of NAM-ATAF1,2-CUC2 transcription factor family in Solanum lycopersicum. J. Plant Biochem. Biotechnol. 2015, 24, 176–183. [Google Scholar] [CrossRef]
- Ma, N.; Feng, H.; Meng, X.; Li, D.; Yang, D.; Wu, C.; Meng, Q. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014, 14, 351. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Chen, G.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.; Hu, Z. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2014, 55, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wei, W.; Zhao, X.; Tan, X.; Fan, Z.; Zhang, Y.; Jing, Y.; Meng, L.; Zhu, B.; Zhu, H.; et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 2018, 5, 75. [Google Scholar] [CrossRef]
- Noguero, M.; Atif, R.M.; Ochatt, S.; Thompson, R.D. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013, 209, 32–45. [Google Scholar] [CrossRef]
- Ruta, V.; Longo, C.; Lepri, A.; De Angelis, V.; Occhigrossi, S.; Costantino, P.; Vittorioso, P. The DOF transcription factors in seed and seedling development. Plants 2020, 9, 218. [Google Scholar] [CrossRef]
- Kim, S.; An, C.S.; Hong, Y.N.; Lee, K.W. Cold-inducible transcription factor, CaCBF, is associated with a homeodomain leucine zipper protein in hot pepper (Capsicum annuum L.). Mol. Cells 2004, 18, 300–308. [Google Scholar]
- Mou, S.; Liu, Z.; Gao, F.; Yang, S.; Su, M.; Shen, L.; Wu, Y.; He, S. CaHDZ27, a Homeodomain-Leucine Zipper I Protein, positively regulates the resistance to Ralstonia solanacearum infection in pepper. Mol. Plant Microbe Interact. 2017, 30, 960–973. [Google Scholar] [CrossRef]
- Lin, Z.; Hong, Y.; Yin, M.; Li, C.; Zhang, K.; Grierson, D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008, 55, 301–310. [Google Scholar] [CrossRef]
- Joo, H.; Lim, C.W.; Lee, S.C. Roles of pepper bZIP transcription factor CaATBZ1 and its interacting partner RING-type E3 ligase CaASRF1 in modulation of ABA signalling and drought tolerance. Plant J. 2019, 100, 399–410. [Google Scholar] [CrossRef]
- Lee, B.J.; Park, C.J.; Kim, S.K.; Kim, K.J.; Paek, K.H. In vivo binding of hot pepper bZIP transcription factor CabZIP1 to the G-box region of pathogenesis-related protein 1 promoter. Biochem. Biophys. Res. Commun. 2006, 344, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Lim, S.; Han, S.W.; Lee, S.C. Expression and functional roles of the pepper pathogen-induced bZIP transcription factor CabZIP2 in enhanced disease resistance to bacterial pathogen infection. Mol. Plant Microbe Interact. 2015, 28, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liu, Z.; Yang, S.; Yang, T.; Liang, J.; Wen, J.; Liu, Y.; Li, J.; Shi, L.; Tang, Q.; et al. Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature-high humidity challenge in a positive feedback loop with CaWRKY40. J. Exp. Bot. 2016, 67, 2439–2451. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.-N.; Lee, T.-Y.; Hung, Y.-C.; Li, G.-Z.; Tseng, K.-C.; Liu, Y.-H.; Kuo, P.-L.; Zheng, H.-Q.; Chang, W.-C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef]


| Accession Type | Accession Name | Abbreviation |
|---|---|---|
| D | Ancho San Luis | AS |
| D | Criollo de Morelos 334 (CM334) | CM |
| D | California Wonder | CW |
| D | Jalapeño Espinalteco | JE |
| D | Serrano Tampiqueño 74 | ST |
| D | Zunla-1 | ZU |
| W | Piquín Coahuila | CO |
| W | Piquín Querétaro | QU |
| W | Piquín Sonora Red | SR |
| W | Piquín Sonora Yellow | SY |
| C | F1: CM female x QU male | CQ |
| C | F1: QU female x CM male | QC |
| Salsa Id | Description | Abbreviation | Protein Id |
|---|---|---|---|
| 20260 | Bifunctional 15-cis-phytoene synthase, chromoplastic | PSY1 | XP_016570422.2 |
| 10326 | Phytoene synthase 2, chloroplastic | PSY2 | XP_016560212.1 |
| 9117 | Phytoene synthase 2, chloroplastic-like | PSY3 | XP_016576229.2 |
| 31467 | 15-cis-phytoene desaturase, chloroplastic/chromoplastic | PDS | XP_016562405.2 |
| 22592 | 15-cis-zeta-carotene isomerase, chloroplastic | Z-ISO | XP_016550148.1 |
| 5981 | Zeta-carotene desaturase, chloroplastic/chromoplastic | ZDS1 | NP_001311497.1 |
| 5979 | Zeta-carotene desaturase, chloroplastic/chromoplastic | ZDS2 | NP_001311497.1 |
| 11712 | Prolycopene isomerase, chloroplastic | CRTISO1 | XP_016548432.1 |
| 28615 | Prolycopene isomerase, chloroplastic | CRTISO2 | XP_016555023.1 |
| 2529 | Lycopene epsilon cyclase, chloroplastic isoform X2 | LCYE | XP_016540361.1 |
| 4235 | Lycopene beta cyclase, chloroplastic/chromoplastic | LCYB1 | XP_016571836.1 |
| 27689 | Lycopene beta cyclase, chloroplastic | LCYB2 | XP_016543793.2 |
| 20137 | Protein LUTEIN DEFICIENT 5, chloroplastic | CYP97A | XP_016551303.1 |
| 4572 | Beta-carotene hydroxylase 1, chloroplastic | BCH1 | NP_001311784.1 |
| 13055 | Beta-carotene hydroxylase 2, chloroplastic isoform X1 | BCH2 | NP_001385279.1 |
| 28396 | Zeaxanthin epoxidase, chloroplastic | ZEP | XP_016561102.1 |
| 16272 | Violaxanthin de-epoxidase, chloroplastic isoform X1 | VDE | XP_016550436.1 |
| 34360 | Capsanthin/capsorubin synthase, chromoplastic | CCS | NP_001311998.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villa-Rivera, M.G.; Martínez, O.; Ochoa-Alejo, N. Putative Transcription Factor Genes Associated with Regulation of Carotenoid Biosynthesis in Chili Pepper Fruits Revealed by RNA-Seq Coexpression Analysis. Int. J. Mol. Sci. 2022, 23, 11774. https://doi.org/10.3390/ijms231911774
Villa-Rivera MG, Martínez O, Ochoa-Alejo N. Putative Transcription Factor Genes Associated with Regulation of Carotenoid Biosynthesis in Chili Pepper Fruits Revealed by RNA-Seq Coexpression Analysis. International Journal of Molecular Sciences. 2022; 23(19):11774. https://doi.org/10.3390/ijms231911774
Chicago/Turabian StyleVilla-Rivera, Maria Guadalupe, Octavio Martínez, and Neftalí Ochoa-Alejo. 2022. "Putative Transcription Factor Genes Associated with Regulation of Carotenoid Biosynthesis in Chili Pepper Fruits Revealed by RNA-Seq Coexpression Analysis" International Journal of Molecular Sciences 23, no. 19: 11774. https://doi.org/10.3390/ijms231911774
APA StyleVilla-Rivera, M. G., Martínez, O., & Ochoa-Alejo, N. (2022). Putative Transcription Factor Genes Associated with Regulation of Carotenoid Biosynthesis in Chili Pepper Fruits Revealed by RNA-Seq Coexpression Analysis. International Journal of Molecular Sciences, 23(19), 11774. https://doi.org/10.3390/ijms231911774








