Moderate-Risk Genes for Hereditary Ovarian Cancers Involved in the Homologous Recombination Repair Pathway
Abstract
1. Introduction
2. Predisposition Genes Included in This Study
3. ATM (Ataxia–Telangiectasia Mutated) Gene
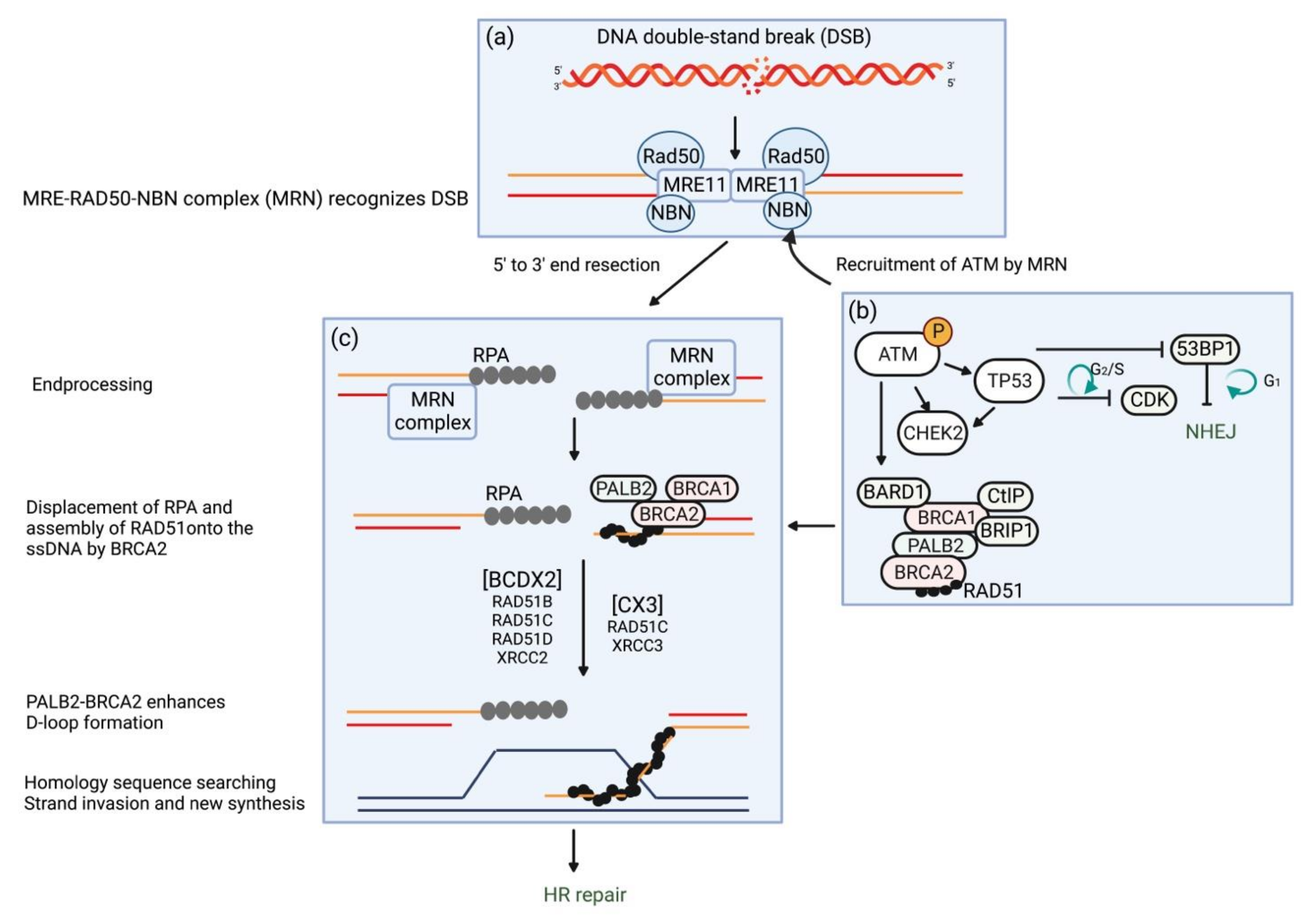
3.1. Molecular Function in the Response to DSBs
3.2. Prevalence and Risk of Developing EOC
3.3. Medical Management for the Prevention of EOC
4. BRIP1 (BRCA1 Interacting Helicase 1) Gene
4.1. Molecular Function in the Response to DSBs
4.2. Prevalence and Risk of Developing EOC
4.3. Medical Management for the Prevention of EOC
5. NBN (Nibrin) Gene
5.1. Molecular Function in the Response to DSBs
5.2. Prevalence and Risk of Developing EOC
5.3. Medical Management for the Prevention of EOC
6. PALB2 (Partner and Localizer of BRCA2) Gene
6.1. Molecular Function in the Response to DSBs
6.2. Prevalence and Risk of Developing EOC
6.3. Medical Management for the Prevention of EOC
7. RAD51C/RAD51D Gene
7.1. Molecular Function in the Response to DSBs
7.2. Prevalence and Risk of Developing EOC
7.3. Medical Management for the Prevention of EOC
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. (Eds.) SEER Cancer Statistics [Review]; National Cancer Institute: Bethesda, MD, USA. Available online: https://seer.cancer.gov/archive/csr/1975_2017/ (accessed on 12 May 2022).
- Lheureux, S.; Gourley, C.; Vergote, I.; Oza, A.M. Epithelial ovarian cancer. Lancet 2019, 393, 1240–1253. [Google Scholar] [CrossRef]
- Henderson, J.T.; Webber, E.M.; Sawaya, G.F. Screening for ovarian cancer: Updated evidence report and systematic review for the US Preventive Services Task Force. JAMA 2018, 319, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Prat, J. Ovarian carcinomas: Five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012, 460, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Cabasag, C.J.; Arnold, M.; Butler, J.; Inoue, M.; Trabert, B.; Webb, P.M.; Bray, F.; Soerjomataram, I. The influence of birth cohort and calendar period on global trends in ovarian cancer incidence. Int. J. Cancer 2020, 146, 749–758. [Google Scholar] [CrossRef]
- Moyer, V.A.; U.S. Preventive Services Task Force. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2014, 160, 271–281. [Google Scholar] [CrossRef]
- Tanha, K.; Mottaghi, A.; Nojomi, M.; Moradi, M.; Rajabzadeh, R.; Lotfi, S.; Janani, L. Investigation on factors associated with ovarian cancer: An umbrella review of systematic review and meta-analyses. J. Ovarian Res. 2021, 14, 153. [Google Scholar] [CrossRef]
- Abdulaziz, G.; Welc, N.A.; Gąsiorowska, E.; Nowak-Markwitz, E. Assessment of gynecological and lifestyle-related risk factors of ovarian cancer. Prz. Menopauzalny 2021, 20, 184–192. [Google Scholar] [CrossRef]
- Ferguson, L.R. Chronic inflammation and mutagenesis. Mutat. Res. 2010, 690, 3–11. [Google Scholar] [CrossRef]
- Kawanishi, S.; Ohnishi, S.; Ma, N.; Hiraku, Y.; Murata, M. Crosstalk between DNA damage and inflammation in the multiple steps of carcinogenesis. Int. J. Mol. Sci. 2017, 18, 1808. [Google Scholar] [CrossRef]
- Zhou, Z.; Zeng, F.; Yuan, J.; Tang, J.; Colditz, G.A.; Tworoger, S.S.; Trabert, B.; Su, X. Pelvic inflammatory disease and the risk of ovarian cancer: A meta-analysis. Cancer Causes Control 2017, 28, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Trabert, B.; Ness, R.B.; lo Ciganic, W.-H.; Murphy, M.A.; Goode, E.L.; Poole, E.M.; Brinton, L.A.; Webb, P.M.; Nagle, C.M.; Jordan, S.J.; et al. Aspirin, nonaspirin nonsteroidal anti-inflammatory drug, and acetaminophen use and risk of invasive epithelial ovarian cancer: A pooled analysis in the Ovarian Cancer Association Consortium. J. Natl. Cancer Inst. 2014, 106, djt431. [Google Scholar] [CrossRef] [PubMed]
- Risch, H.A.; Howe, G.R. Pelvic inflammatory disease and the risk of epithelial ovarian cancer. Cancer Epidemiol. Biomark. Prev. 1995, 4, 447–451. [Google Scholar]
- Ness, R.B.; Grisso, J.A.; Cottreau, C.; Klapper, J.; Vergona, R.; Wheeler, J.E.; Morgan, M.; Schlesselman, J.J. Factors related to inflammation of the ovarian epithelium and risk of ovarian cancer. Epidemiology 2000, 11, 111–117. [Google Scholar] [CrossRef]
- Brilhante, A.V.M.; Augusto, K.L.; Portela, M.C.; Sucupira, L.C.G.; Oliveira, L.A.F.; Pouchaim, A.J.M.V.; Nobrega, L.R.M.; Magalhaes, T.F.M.; Sobreira, L.R.P. Endometriosis and ovarian cancer: An integrative review (endometriosis and ovarian cancer). Asian Pac. J. Cancer Prev. 2017, 18, 11–16. [Google Scholar] [CrossRef]
- Pennington, K.P.; Walsh, T.; Harrell, M.I.; Lee, M.K.; Pennil, C.C.; Rendi, M.H.; Thornton, A.; Norquist, B.M.; Casadei, S.; Nord, A.S.; et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, Fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 2014, 20, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.; Burger, R.A.; et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016, 2, 482–490. [Google Scholar] [CrossRef]
- Calaf, G.M.; Urzua, U.; Termini, L.; Aguayo, F. Oxidative stress in female cancers. Oncotarget 2018, 9, 23824–23842. [Google Scholar] [CrossRef]
- Savage, K.I.; Matchett, K.B.; Barros, E.M.; Cooper, K.M.; Irwin, G.W.; Gorski, J.J.; Orr, K.S.; Vohhodina, J.; Kavanagh, J.N.; Madden, A.F.; et al. BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res. 2014, 74, 2773–2784. [Google Scholar] [CrossRef]
- Alayev, A.; Salamon, R.S.; Manna, S.; Schwartz, N.S.; Berman, A.Y.; Holz, M.K. Estrogen induces RAD 51C expression and localization to sites of DNA damage. Cell Cycle 2016, 15, 3230–3239. [Google Scholar] [CrossRef]
- Zach, L.; Yedidia-Aryeh, L.; Goldberg, M. Estrogen and DNA damage modulate mRNA levels of genes involved in homologous recombination repair in estrogen-deprived cells. J. Trans. Genet. Genom. 2022, 6, 266–280. [Google Scholar] [CrossRef]
- Yamamoto, H.; Hirasawa, A. Homologous recombination deficiencies and hereditary tumors. Int. J. Mol. Sci. 2022, 23, 348. [Google Scholar] [CrossRef]
- Ueki, A.; Hirasawa, A. Molecular features and clinical management of hereditary gynecological cancers. Int. J. Mol. Sci. 2020, 21, 9504. [Google Scholar] [CrossRef] [PubMed]
- Bono, M.; Fanale, D.; Incorvaia, L.; Cancellierando, D.; Fiorino, A.; Calo, V.; Dimino, A.; Filorizzo, C.; Corsini, L.R.; Brando, C.; et al. Impact of deleterious variants in other genes beyond BRCA1/2 detected in breast/ovarian and pancreatic cancer patients by NGS-based multi-gene panel testing: Looking over the hedge. ESMO Open 2021, 6, 100235. [Google Scholar] [CrossRef] [PubMed]
- Domchek, S.M.; Robson, M.E. Update on genetic testing in gynecologic cancer. J. Clin. Oncol. 2019, 37, 2501–2510. [Google Scholar] [CrossRef] [PubMed]
- Tsaousis, G.N.; Papadopoulou, E.; Apessos, A.; Agiannitopoulos, K.; Pepe, G.; Kampouri, S.; Diamatopoulos, N.; Floros, T.; Iosifidou, R.; Katopodi, O.; et al. Analysis of hereditary cancer syndromes by using a panel of genes: Novel and multiple pathogenic mutations. BMC Cancer 2019, 19, 535. [Google Scholar] [CrossRef]
- Samadder, N.J.; Riegert-Johnson, D.; Boardman, L.; Rhodes, D.; Wick, M.; Okuno, S.; Kunze, K.L.; Golafshar, M.; Uson, P.L.S.; Mountjoy, L.; et al. Comparison of universal genetic testing vs guideline-directed targeted testing for patients with hereditary cancer syndrome. JAMA Oncol. 2021, 7, 230–237. [Google Scholar] [CrossRef]
- Vietri, M.T.; D’Elia, G.; Caliendo, G.; Casamassimi, A.; Federico, A.; Passariello, L.; Cioffi, M.; Molinari, A.M. Prevalence of mutations in BRCA and MMR genes in patients affected with hereditary endometrial cancer. Med. Oncol. 2021, 38, 13. [Google Scholar] [CrossRef]
- Liu, Y.L.; Breen, K.; Catchings, A.; Ranganathan, M.; Latham, A.; Goldfrank, D.J.; Grisham, R.N.; Long Roche, K.; Frey, M.K.; Chi, D.S.; et al. Risk-reducing bilateral salpingo-oophorectomy for ovarian cancer: A review and clinical guide for hereditary predisposition genes. JCO Oncol. Pract. 2022, 18, 201–209. [Google Scholar] [CrossRef]
- Phelan, C.M.; Kuchenbaecker, K.B.; Tyrer, J.P.; Kar, S.P.; Lawrenson, K.; Winham, S.J.; Dennis, J.; Pirie, A.; Riggan, M.J.; Chornokur, G.; et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017, 49, 680–691. [Google Scholar] [CrossRef]
- Kar, S.P.; Berchuck, A.; Gayther, S.A.; Goode, E.L.; Moysich, K.B.; Pearce, C.L.; Ramus, S.J.; Schildkraut, J.M.; Sellers, T.A.; Pharoah, P.D.P. Common genetic variation and susceptibility to ovarian cancer: Current insights and future directions. Cancer Epidemiol. Biomark. Prev. 2018, 27, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Eleje, G.U.; Eke, A.C.; Ezebialu, I.U.; Ikechebelu, J.I.; Ugwu, E.O.; Okonkwo, O.O. Risk-reducing bilateral salpingo-oophorectomy in women with BRCA1 or BRCA2 mutations. Cochrane Database Syst. Rev. 2018, 8, CD012464. [Google Scholar] [CrossRef] [PubMed]
- Rebbeck, T.R.; Lynch, H.T.; Neuhausen, S.L.; Narod, S.A.; Van’t Veer, L.; Garber, J.E.; Evans, G.; Isaacs, C.; Daly, M.B.; Matloff, E.; et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N. Engl. J. Med. 2002, 346, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Kauff, N.D.; Satagopan, J.M.; Robson, M.E.; Scheuer, L.; Hensley, M.; Hudis, C.A.; Ellis, N.A.; Boyd, J.; Borgen, P.I.; Barakat, R.R.; et al. Risk-reducing salpingo-oophorectomy in Women with a BRCA1 or BRCA2 Mutation. N. Engl. J. Med. 2002, 346, 1609–1615. [Google Scholar] [CrossRef]
- Neff, R.T.; Senter, L.; Salani, R. BRCA mutation in ovarian cancer: Testing, implications and treatment considerations. Ther. Adv. Med. Oncol. 2017, 9, 519–531. [Google Scholar] [CrossRef]
- Tung, N.; Domchek, S.M.; Stadler, Z.; Nathanson, K.L.; Couch, F.; Garber, J.E.; Offit, K.; Robson, M.E. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat. Rev. Clin. Oncol. 2016, 13, 581–588. [Google Scholar] [CrossRef]
- Samuel, D.; Diaz-Barbe, A.; Pinto, A.; Schlumbrecht, M.; George, S. Hereditary ovarian carcinoma: Cancer pathogenesis looking beyond BRCA1 and BRCA2. Cells 2022, 11, 539. [Google Scholar] [CrossRef]
- Leblanc, E.; Narducci, F.; Hudry, D.; Bresson, L.; Charvolin, J.Y.; Ferron, G.; Guyon, F.; Fourchotte, V.; Lambaudie, E.; Baron, M.; et al. First results of a prospective national controlled study: Prophylactic radical fimbriectomy (NCT01608074), in women with a hereditary familial risk of breast/ovarian cancer—Tolerance and pathological findings. J. Clin. Oncol. 2018, 36, 5574. [Google Scholar] [CrossRef]
- Gaba, F.; Robbani, S.; Singh, N.; McCluggage, W.G.; Wilkinson, N.; Ganesan, R.; Bryson, G.; Rowlands, G.; Tyson, C.; Arora, R.; et al. Preventing ovarian cancer through early excision of tubes and late ovarian removal (PROTECTOR): Protocol for a prospective non-randomised multi-center trial. Int. J. Gynecol. Cancer 2021, 31, 286–291. [Google Scholar] [CrossRef]
- Steenbeek, M.P.; Bommel, M.V.; Swisher, E.; Lu, K.; Hermens, R.; Hullu, J.D. Tubectomy with delayed oophorectomy in high risk women to assess the safety of prevention (TUBA-WISP-II). Int. J. Gynecol. Cancer 2021, 31, A314. [Google Scholar] [CrossRef]
- Pavanello, M.; Chan, I.H.; Ariff, A.; Pharoah, P.D.; Gayther, S.A.; Ramus, S.J. Rare germline genetic variants and the risks of epithelial ovarian cancer. Cancers 2020, 12, 3046. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Soeda, S.; Endo, Y.; Okabe, C.; Sato, T.; Kamo, N.; Ueda, M.; Kojima, M.; Furukawa, S.; Nishigori, H.; et al. Rare hereditary gynecological cancer syndromes. Int. J. Mol. Sci. 2022, 23, 1563. [Google Scholar] [CrossRef] [PubMed]
- Pietragalla, A.; Arcieri, M.; Marchetti, C.; Fagotti, A. Ovarian cancer predisposition beyond BRCA1 and BRCA2 genes. Int. J. Gynecol. Cancer 2020, 30, 1803–1810. [Google Scholar] [CrossRef] [PubMed]
- Suszynska, M.; Klonowska, K.; Jasinska, A.J.; Kozlowski, P. Large-scale meta-analysis of mutations identified in panels of breast/ovarian cancer-related genes—Providing evidence of cancer predisposition genes. Gynecol. Oncol. 2019, 153, 452–462. [Google Scholar] [CrossRef] [PubMed]
- National Comprehensive Cancer Network (NCCN) in Oncology. Genetic/Familial High-Risk Assessment: Breast, Ovarian and Pancreatic. Version 2. 2022. Available online: https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf (accessed on 12 May 2022).
- National Comprehensive Cancer Network (NCCN) in Oncology. Genetic/Familial High-Risk Assessment: Colorectal. Version 2. 2021. Available online: https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf (accessed on 12 May 2022).
- Suszynska, M.; Ratajska, M.; Kozlowski, P. BRIP1, RAD51C, and RAD51D mutations are associated with high susceptibility to ovarian cancer: Mutation prevalence and precise risk estimates based on a pooled analysis of ~30,000 cases. J. Ovarian Res. 2020, 13, 50. [Google Scholar] [CrossRef] [PubMed]
- Momozawa, Y.; Sasai, R.; Usui, Y.; Shiraishi, K.; Iwasaki, Y.; Taniyama, Y.; Parsons, M.T.; Mizukami, K.; Sekine, Y.; Hirata, M.; et al. Expansion of Cancer Risk Profile for BRCA1 and BRCA2 Pathogenic Variants. JAMA Oncol. 2022, 8, 871–878. [Google Scholar] [CrossRef]
- Huertas, P. DNA resection in eukaryotes: Deciding how to fix the break. Nat. Struct. Mol. Biol. 2010, 17, 11–16. [Google Scholar] [CrossRef]
- Huang, R.X.; Zhou, P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef]
- Pardo, B.; Gómez-González, B.; Aguilera, A. DNA repair in mammalian cells: DNA double-strand break repair—How to fix a broken relationship. Cell. Mol. Life Sci. 2009, 66, 1039–1056. [Google Scholar] [CrossRef]
- Hartlerode, A.J.; Scully, R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem. J. 2009, 423, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in patients with newly diagnosed advanced ovarian cancer. N. Eng. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Matos-Rodrigues, G.; Guirouilh-Barbat, J.; Martini, E.; Lopez, B.S. Homologous recombination, cancer and the ‘RAD51 paradox’. NAR Cancer 2021, 3, zcab016. [Google Scholar] [CrossRef] [PubMed]
- Burkle, A. Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 2001, 163, 1–5. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Pioli, C. Multifaceted role of PARP-1 in DNA repair and inflammation: Pathological and therapeutic implications in cancer and non-cancer diseases. Cells 2020, 9, 41. [Google Scholar] [CrossRef]
- Haince, J.F.; McDonald, D.; Rodrigue, A.; Déry, U.; Masson, J.Y.; Hendze, M.J.; Poirier, G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008, 283, 1197–1208. [Google Scholar] [CrossRef]
- Griguolo, G.; Dieci, M.V.; Guarneri, V.; Conte, P. Olaparib for the treatment of breast cancer. Expert Rev. Anticancer Ther. 2018, 18, 519–530. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Curtin, N.J.; Szabo, C. Poly(ADP-ribose) polymerase inhibition: Past, present and future. Nat. Rev. Drug Discov. 2020, 19, 711–736. [Google Scholar] [CrossRef]
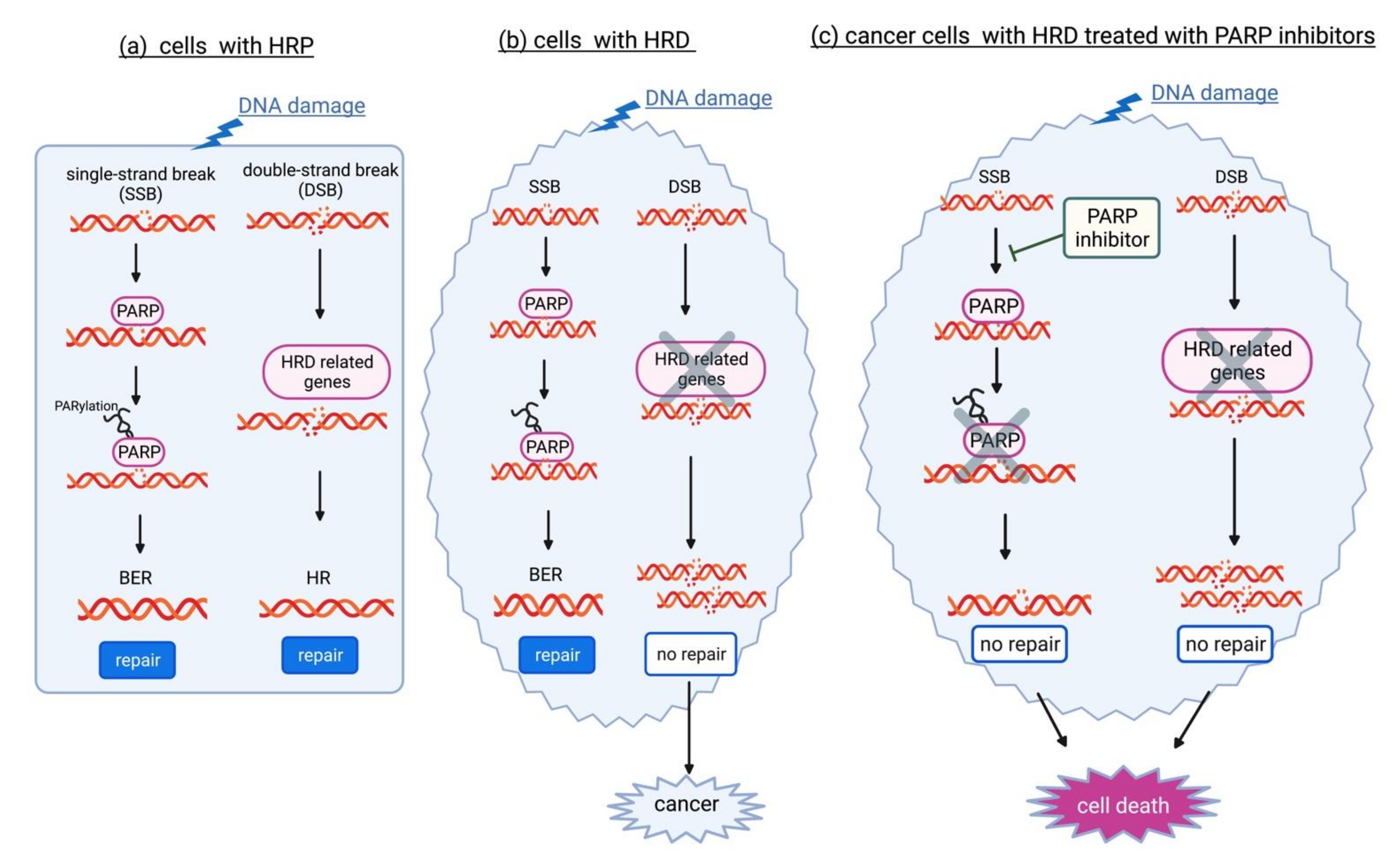
- Paulet, L.; Trecourt, A.; Leary, A.; Peron, J.; Descotes, F.; Devouassoux-Shisheboran, M.; Leroy, K.; You, B.; Lopez, J. Cracking the homologous recombination deficiency code: How to identify responders to PARP inhibitors. Eur. J. Cancer 2022, 166, 87–99. [Google Scholar] [CrossRef]
- Stucci, L.S.; Internò, V.; Tucci, M.; Perrone, M.; Mannavola, F.; Palmirotta, R.; Porta, C. The ATM gene in breast cancer: Its relevance in clinical practice. Genes 2021, 12, 727. [Google Scholar] [CrossRef]
- Yu, W.; Lescale, C.; Babin, L.; Bedora-Faure, M.; Lenden-Hasse, H.; Baron, L.; Demangel, C.; Yelamos, J.; Brunet, E.; Deriano, L. Repair of G1 induced DNA double-strand breaks in S-G2/M by alternative NHEJ. Nat. Commun. 2020, 11, 5239. [Google Scholar] [CrossRef] [PubMed]
- Balmus, G.; Pilger, D.; Coates, J.; Demir, M.; Sczaniecka-Clift, M.; Barros, A.C.; Woods, M.; Fu, B.; Yang, F.; Chen, E.; et al. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 2019, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Britton, S.; Chanut, P.; Delteil, C.; Barboule, N.; Frit, P.; Calsou, P. ATM antagonizes NHEJ proteins assembly and DNA-ends synapsis at single-ended DNA double strand breaks. Nucleic Acids Res. 2020, 48, 9710–9723. [Google Scholar] [CrossRef]
- Ueno, S.; Sudo, T.; Hirasawa, A. ATM: Functions of ATM kinase and its relevance to hereditary tumors. Int. J. Mol. Sci. 2022, 23, 523. [Google Scholar] [CrossRef]
- Putti, S.; Giovinazzo, A.; Merolle, M.; Falchetti, M.L.; Pellegrini, M. ATM kinase dead: From ataxia telangiectasia syndrome to cancer. Cancers 2021, 13, 5498. [Google Scholar] [CrossRef] [PubMed]
- Ouhtit, A.; Gupta, I.; Shaikh, Z. BRIP1, a potential candidate gene in development of non-BRCA1/2 breast cancer. Front. Biosci. (Elite Ed.) 2016, 8, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Kurian, A.W.; Ward, K.C.; Howlader, N.; Deapen, D.; Hamilton, A.S.; Mariotto, A.; Miller, D.; Penberthy, L.S.; Katz, S.J. Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients. J. Clin. Oncol. 2019, 37, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Cummings, S.; Roman, S.S.; Saam, J.; Bernhisel, R.; Brown, K.; Lancaster, J.M.; Usha, L. Age of ovarian cancer diagnosis among BRIP1, RAD51C, and RAD51D mutation carriers identified through multi-gene panel testing. J. Ovarian Res. 2021, 14, 61. [Google Scholar] [CrossRef]
- Levitus, M.; Waisfisz, Q.; Godthelp, B.C.; de Vries, Y.; Hussain, S.; Wiegant, W.W.; Elghalbzouri-Maghrani, E.; Steltenpool, J.; Rooimans, M.A.; Pals, G.; et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J. Nat. Genet. 2005, 37, 934–935. [Google Scholar] [CrossRef]
- Wen, J.; Cerosaletti, K.; Schultz, K.J.; Wright, J.A.; Concannon, P. NBN phosphorylation regulates the accumulation of MRN and ATM at sites of DNA double-strand breaks. Oncogene 2013, 32, 4448–4456. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.; Tainer, J.A. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 2018, 87, 263–294. [Google Scholar] [CrossRef] [PubMed]
- Menolfi, D.; Zha, S. ATM, ATR and DNA-PKcs Kinases-the lessons from the mouse models: Inhibition ≠ deletion. Cell Biosci. 2020, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Chen, J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J. Biol. Chem. 2010, 285, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, J.; Lim, C.U.K. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 2006, 16, 45–54. [Google Scholar] [CrossRef]
- Chrzanowska, K.H.; Gregorek, H.; Dembowska-Bagińska, B.; Kalina, M.A.; Digweed, M. Nijmegen breakage syndrome (NBS). Orphanet J. Rare Dis. 2012, 7, 13. [Google Scholar] [CrossRef]
- Simhadri, S.; Vincelli, G.; Huo, Y.; Misenko, S.; Foo, T.K.; Ahlskog, J.; Sørensen, C.S.; Oakley, G.G.; Ganesan, S.; Bunting, S.F.; et al. PALB2 connects BRCA1 and BRCA2 in the G2/M checkpoint response. Oncogene 2019, 38, 1585–1596. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhang, J.; Willers, H.; Wang, H.; Chung, J.H.; van Gent, D.C.; Hallahan, D.E.; Powell, S.N.; Xia, F. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res. 2006, 66, 1401–1408. [Google Scholar] [CrossRef][Green Version]
- Yang, X.; Leslie, G.; Doroszuk, A.; Schneider, S.; Allen, J.; Decker, B.; Dunning, A.M.; Redman, J.; Scarth, J.; Plaskocinska, I.; et al. Cancer risks associated With germline PALB2 pathogenic variants: An international study of 524 families. J. Clin. Oncol. 2020, 38, 674–685. [Google Scholar] [CrossRef]
- Tischkowitz, M.; Balmana, J.; Foulkes, W.D.; James, P.; Ngeow, J.; Schmutzler, R.; Voian, N.; Wick, M.J.; Stewart, D.R.; Pal, T.; et al. Management of individuals with germline variants in PALB2: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 1416–1423. [Google Scholar] [CrossRef]
- Yang, X.; Song, H.; Leslie, G.; Engel, C.; Hahnen, E.; Auber, B.; Horváth, J.; Kast, K.; Niederacher, D.; Turnbull, C.; et al. Ovarian and breast cancer risks associated with pathogenic variants in RAD51C and RAD51D. J. Natl. Cancer Inst. 2020, 112, 1242–1250. [Google Scholar] [CrossRef]
- Min, A.; Im, S.A.; Yoon, Y.K.; Song, S.H.; Nam, H.J.; Hur, H.S.; Kim, H.P.; Lee, K.H.; Han, S.W.; Oh, D.Y.; et al. RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol. Cancer Ther. 2013, 12, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Loveday, C.; Turnbull, C.; Ramsay, E.; Hughes, D.; Ruark, E.; Frankum, J.R.; Bowden, G.; Kalmyrzaev, B.; Warren-Perry, M.; Snape, K.; et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat. Genet. 2011, 43, 879–882. [Google Scholar] [CrossRef]
- Gayarre, J.; Martín-Gimeno, P.; Osorio, A.; Paumard, B.; Barroso, A.; Fernández, V.; de La Hoya, M.; Rojo, A.; Caldés, T.; Palacios, J.; et al. Characterisation of the novel deleterious RAD51C p.Arg312Trp variant and prioritisation criteria for functional analysis of RAD51C missense changes. Br. J. Cancer 2017, 117, 1048–1062. [Google Scholar] [CrossRef] [PubMed]
- Slade, D. PARP and PARG inhibitors in cancer treatment. Genes Dev. 2020, 34, 360–394. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, D.R.; Dougherty, B.A.; Lai, Z.; Fielding, A.; Grinsted, L.; Spencer, S.; O’Connor, M.J.; Ho, T.W.; Robertson, J.A.; Lanchbury, J.S.; et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br. J. Cancer 2018, 119, 1191–1409. [Google Scholar] [CrossRef]
- Chian, Y.C.; Lin, P.H.; Chen, W.F. Homologous recombination deficiency assays in epithelial ovarian cancer: Current status and future direction. Front. Oncol. 2021, 11, 675972. [Google Scholar] [CrossRef]
- Yoshimura, A.; Imoto, I.; Iwata, H. Functions of breast cancer predisposition genes: Implications for clinical management. Int. J. Mol. Sci. 2022, 23, 7481. [Google Scholar] [CrossRef]
| Gene | Suszynska et al. [45] | NCCN Guidelines [46,47] | ||||
|---|---|---|---|---|---|---|
| Frequency of GPV in EOC Patients (%) | Relative Risk for EOC | Absolute Risk for EOC | Evidence for Association | Management for Risk Reduction | ||
| OR (95% CI) | p-Value | |||||
| BRCA1 | 8.607 | 35.26 (29.56–42.05) | <0.0001 | 39–58% | very strong | RRSO recommended for patients aged 35–40 yrs |
| BRCA2 | 4.520 | 11.91 (9.87–14.39) | <0.0001 | 13–29% | very strong | RRSO recommended for patients aged 40–45 yrs |
| BRIP1 | 1.057 | 4.88 (3.73–6.38) | <0.0001 | >10% | strong | RRSO considered for patients aged 45–50 yrs |
| CHEK2 | 0.703 | 0.43 (0.29–0.63) | <0.0001 | not established | not established | not established |
| ATM | 0.677 | 1.98 (1.33–2.94) | 0.001 | <3% | insufficient | manage based on family history |
| RAD51C | 0.554 | 4.24 (2.56–7.02) | <0.0001 | >10% | strong | RRSO considered for patients aged 45–50 yrs |
| RAD51D | 0.583 | 7.28 (4.03–13.14) | <0.0001 | >10% | strong | RRSO considered for patients aged 45–50 yrs |
| MSH6 | 0.444 | 4.08 (2.43–6.85) | <0.0001 | <13% | insufficient, mixed | - |
| PALB2 | 0.423 | 2.13 (1.42–3.21) | 0.0003 | 3–5% | insufficient | manage based on family history |
| TP53 | 0.294 | 5.05 (2.41–10.58) | <0.0001 | not established | not established | not established |
| NBN | 0.284 | 2.17 (1.35–3.49) | 0.0020 | insufficient data | limited | manage based on family history |
| MSH2 | 0.238 | 3.98 (1.18–8.69) | 0.0007 | >10% | strong | RRSO should be individualized after childbearing |
| PMS2 | 0.183 | 0.71 (0.29–1.72) | 0.5633 | <3% | limited | - |
| MLH1 | 0.104 | 1.44 (0.53–3.91) | 0.6815 | >10% | strong | RRSO should be individualized after childbearing |
| BARD1 | 0.142 | 1.41 (0.69–2.89) | 0.4706 | not established | not established | not established |
| PTEN | 0.063 | 5.47 (1.26–23.82) | 0.0799 | not established | not established | not established |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abe, A.; Imoto, I.; Ueki, A.; Nomura, H.; Kanao, H. Moderate-Risk Genes for Hereditary Ovarian Cancers Involved in the Homologous Recombination Repair Pathway. Int. J. Mol. Sci. 2022, 23, 11790. https://doi.org/10.3390/ijms231911790
Abe A, Imoto I, Ueki A, Nomura H, Kanao H. Moderate-Risk Genes for Hereditary Ovarian Cancers Involved in the Homologous Recombination Repair Pathway. International Journal of Molecular Sciences. 2022; 23(19):11790. https://doi.org/10.3390/ijms231911790
Chicago/Turabian StyleAbe, Akiko, Issei Imoto, Arisa Ueki, Hidetaka Nomura, and Hiroyuki Kanao. 2022. "Moderate-Risk Genes for Hereditary Ovarian Cancers Involved in the Homologous Recombination Repair Pathway" International Journal of Molecular Sciences 23, no. 19: 11790. https://doi.org/10.3390/ijms231911790
APA StyleAbe, A., Imoto, I., Ueki, A., Nomura, H., & Kanao, H. (2022). Moderate-Risk Genes for Hereditary Ovarian Cancers Involved in the Homologous Recombination Repair Pathway. International Journal of Molecular Sciences, 23(19), 11790. https://doi.org/10.3390/ijms231911790






