The First Molecular Detection of Aedes albopictus in Sudan Associates with Increased Outbreaks of Chikungunya and Dengue
Abstract
1. Introduction
2. Results
2.1. Phylogenetic Analysis
2.2. Haplotype Analysis and Global Network
2.3. Population Diversity and Evolutionary Characteristics
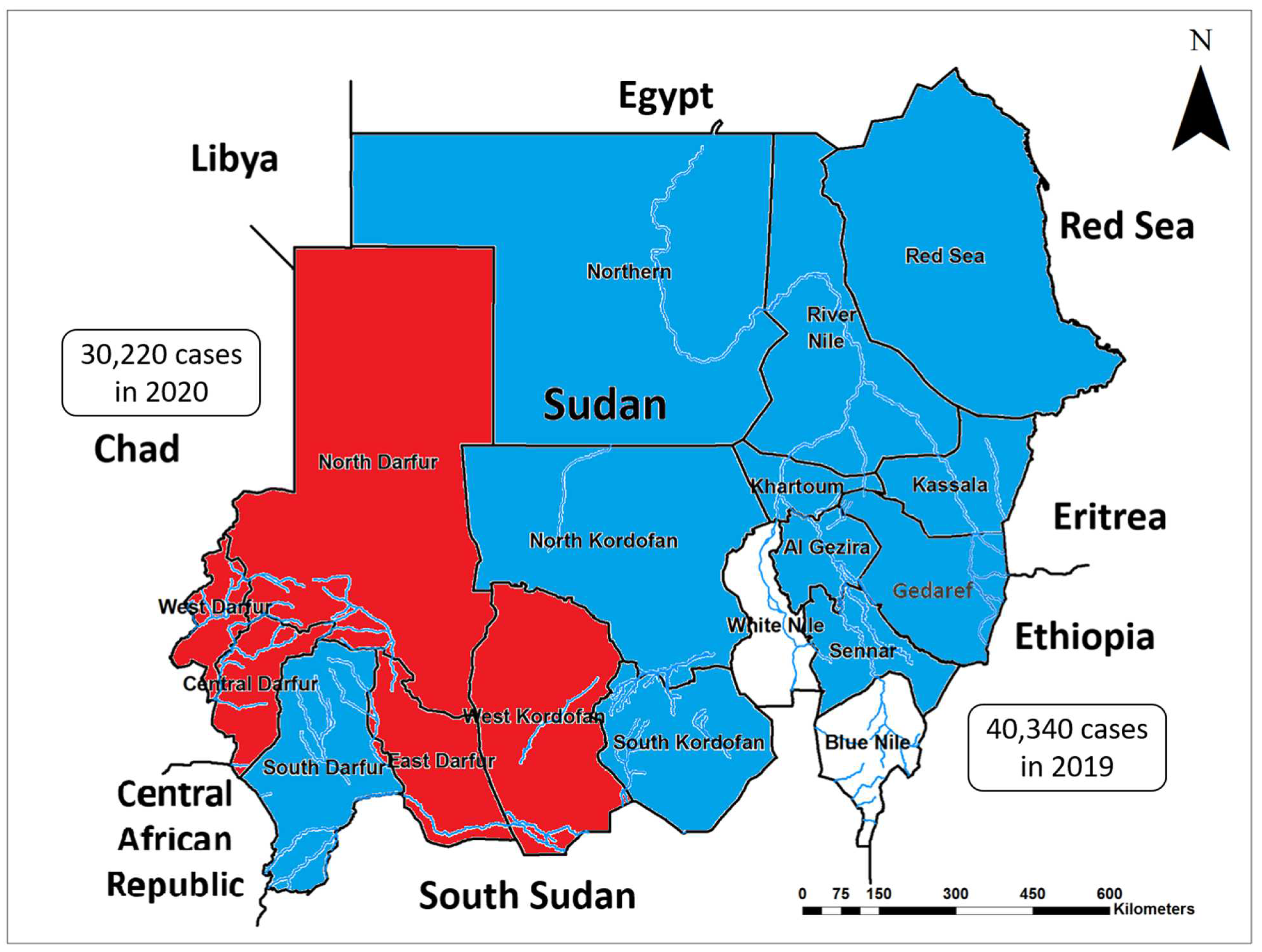
2.4. Recent Increase in the Burden and Spread of CHIKV in Sudan and the Neighboring Countries
3. Discussion
4. Materials and Methods
4.1. Exploratory Surveys
4.2. Mosquito Samples Collection and Morphological Identification
4.3. DNA Extraction from Mosquito and Polymerase Chain Reaction
4.4. PCR Amplicons Sequencing and Sequences’ Identity Confirmation
4.5. Bioinformatics and Phylogenetic Analysis
4.6. Exploring the Potential Health Impacts of the Emergence of This Invasive Vector in the Area
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kraemer, M.U.G.; Reiner, R.C., Jr.; Brady, O.J.; Messina, J.P.; Gilbert, M.; Pigott, D.M.; Yi, D.; Johnson, K.; Earl, L.; Marczak, L.B.; et al. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat. Microbiol. 2019, 4, 854–863. [Google Scholar] [CrossRef]
- Weaver, S.C.; Barrett, A.D.T. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2004, 2, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Cleton, N.; Koopmans, M.; Reimerink, J.; Godeke, G.-J.; Reusken, C. Come fly with me: Review of clinically important arboviruses for global travelers. J. Clin. Virol. 2012, 55, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Ali, Y.; Salim, B.; Dietrich, I.; Zinsstag, J. Epidemics of Crimean-Congo Hemorrhagic Fever (CCHF) in Sudan between 2010 and 2020. Microorganisms 2022, 10, 928. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Ali, Y.; Elduma, A.; Eldigail, M.H.; Mhmoud, R.A.; Mohamed, N.S.; Ksiazek, T.G.; Dietrich, I.; Weaver, S.C. Unique Outbreak of Rift Valley Fever in Sudan, 2019. Emerg. Infect. Dis. 2020, 26, 3030–3033. [Google Scholar] [CrossRef]
- Zientara, S.; Ponsart, C. Viral emergence and consequences for reproductive performance in ruminants: Two recent examples (bluetongue and Schmallenberg viruses). Reprod. Fertil. Dev. 2015, 27, 63–71. [Google Scholar] [CrossRef]
- Pereira De Oliveira, R.; Hutet, E.; Lancelot, R.; Paboeuf, F.; Duhayon, M.; Boinas, F.; Pérez de León, A.A.; Filatov, S.; Le Potier, M.F.; Vial, L. Differential vector competence of Ornithodoros soft ticks for African swine fever virus: What if it involves more than just crossing organic barriers in ticks? Parasites Vectors 2020, 13, 618. [Google Scholar] [CrossRef]
- Blahove, M.R.; Carter, J.R. Flavivirus persistence in wildlife populations. Viruses 2021, 13, 2099. [Google Scholar] [CrossRef]
- Braack, L.; Gouveia de Almeida, A.P.; Cornel, A.J.; Swanepoel, R.; de Jager, C. Mosquito-borne arboviruses of African origin: Review of key viruses and vectors. Parasites Vectors 2018, 11, 29. [Google Scholar] [CrossRef]
- Weetman, D.; Kamgang, B.; Badolo, A.; Moyes, C.L.; Shearer, F.M.; Coulibaly, M.; Pinto, J.; Lambrechts, L.; McCall, P.J. Aedes Mosquitoes and Aedes-Borne Arboviruses in Africa: Current and Future Threats. Int. J. Environ. Res. Public Health 2018, 15, 220. [Google Scholar] [CrossRef]
- Whiteman, A.; Loaiza, J.R.; Yee, D.A.; Poh, K.C.; Watkins, A.S.; Lucas, K.J.; Rapp, T.J.; Kline, L.; Ahmed, A.; Chen, S.; et al. Do socioeconomic factors drive Aedes mosquito vectors and their arboviral diseases? A systematic review of dengue, chikungunya, yellow fever, and Zika Virus. One Health 2020, 11, 100188. [Google Scholar] [CrossRef]
- Elaagip, A.; Alsedig, K.; Altahir, O.; Ageep, T.; Ahmed, A.; Siam, H.A.; Samy, A.M.; Mohamed, W.; Khalid, F.; Gumaa, S.; et al. Seroprevalence and associated risk factors of Dengue fever in Kassala state, eastern Sudan. PLoS Negl. Trop. Dis. 2020, 14, e0008918. [Google Scholar] [CrossRef]
- Müller, P.; Engeler, L.; Vavassori, L.; Suter, T.; Guidi, V.; Gschwind, M.; Tonolla, M.; Flacio, E. Surveillance of invasive Aedes mosquitoes along Swiss traffic axes reveals different dispersal modes for Aedes albopictus and Ae. japonicus. PLoS Negl. Trop. Dis. 2020, 14, e0008705. [Google Scholar] [CrossRef]
- Flacio, E.; Engeler, L.; Tonolla, M.; Lüthy, P.; Patocchi, N. Strategies of a thirteen year surveillance programme on Aedes albopictus (Stegomyia albopicta) in southern Switzerland. Parasites Vectors 2015, 8, 208. [Google Scholar] [CrossRef]
- Carrieri, M.; Albieri, A.; Angelini, P.; Baldacchini, F.; Venturelli, C.; Zeo, S.M.; Bellini, R. Surveillance of the chikungunya vector Aedes albopictus (Skuse) in Emilia-Romagna (northern Italy): Organizational and technical aspects of a large scale monitoring system. J. Vector Ecol. 2011, 36, 108–116. [Google Scholar] [CrossRef]
- Canali, M.; Rivas-Morales, S.; Beutels, P.; Venturelli, C. The Cost of Arbovirus Disease Prevention in Europe: Area-Wide Integrated Control of Tiger Mosquito, Aedes albopictus, in Emilia-Romagna, Northern Italy. Int. J. Environ. Res. Public Health 2017, 14, 444. [Google Scholar] [CrossRef]
- Ahmed, A.; Ali, Y.; Mohamed, N.S. Arboviral diseases: The emergence of a major yet ignored public health threat in Africa. Lancet Planet Health 2020, 4, e555. [Google Scholar] [CrossRef]
- de Santi, V.P.; Khaireh, B.A.; Chiniard, T.; Pradines, B.; Taudon, N.; Larréché, S.; Mohamed, A.B.; de Laval, F.; Berger, F.; Gala, F.; et al. Role of Anopheles stephensi Mosquitoes in Malaria Outbreak, Djibouti, 2019. Emerg. Infect. Dis. 2021, 27, 1697–1700. [Google Scholar] [CrossRef]
- Faulde, M.K.; Rueda, L.M.; Khaireh, B.A. First record of the Asian malaria vector Anopheles stephensi and its possible role in the resurgence of malaria in Djibouti, Horn of Africa. Acta Trop. 2014, 139, 39–43. [Google Scholar] [CrossRef]
- Ahmed, A.; Pignatelli, P.; Elaagip, A.; Hamid, M.M.A.; Alrahman, O.F.; Weetman, D. Invasive Malaria Vector Anopheles stephensi Mosquitoes in Sudan, 2016–2018. Emerg. Infect. Dis. J.-CDC 2021, 27, 2952–2954. [Google Scholar] [CrossRef]
- Ahmed, A.; Dietrich, I.; LaBeaud, A.D.; Lindsay, S.W.; Musa, A.; Weaver, S.C. Risks and Challenges of Arboviral Diseases in Sudan: The Urgent Need for Actions. Viruses 2020, 12, 81. [Google Scholar] [CrossRef]
- Ahmed, A.; Ali, Y.; Elmagboul, B.; Mohamed, O.; Elduma, A.; Bashab, H.; Mahamoud, A.; Khogali, H.; Elaagip, A.; Higazi, T. Dengue Fever in the Darfur Area, Western Sudan. Emerg. Infect. Dis. 2019, 25, 2126. [Google Scholar] [CrossRef]
- Ahmed, A.; Eldigail, M.; Elduma, A.; Breima, T.; Dietrich, I.; Ali, Y.; Weaver, S.C. First report of epidemic dengue fever and malaria co-infections among internally displaced persons in humanitarian camps of North Darfur, Sudan. Int. J. Infect. Dis. 2021, 108, 513–516. [Google Scholar] [CrossRef]
- Ahmed, A.; Elduma, A.; Magboul, B.; Higazi, T.; Ali, Y. The First Outbreak of Dengue Fever in Greater Darfur, Western Sudan. Trop. Med. Infect. Dis. 2019, 4, 43. [Google Scholar] [CrossRef]
- Ahmed, A.; Mahmoud, I.; Eldigail, M.; Elhassan, R.M.; Weaver, S.C. The Emergence of Rift Valley Fever in Gedaref State Urges the Need for a Cross-Border One Health Strategy and Enforcement of the International Health Regulations. Pathogens 2021, 10, 885. [Google Scholar] [CrossRef]
- Hamid, Z.; Hamid, T.; Alsedig, K.; Abdallah, T.; Elaagip, A.; Ahmed, A.; Khalid, F.; Abdel Hamid, M. Molecular Investigation of Dengue virus serotype 2 Circulation in Kassala State, Sudan. Jpn. J. Infect. Dis. 2019, 72, 58–61. [Google Scholar] [CrossRef]
- Lewis, D.J. The Aēdes mosquitoes of the Sudan. Ann. Trop. Med. Parasitol. 1955, 49, 164–173. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- World Health Organization. Regional Office for Africa. Weekly Bulletin on Outbreak and Other Emergencies: Week 38: 16–22 September 2019. 2019. Available online: https://apps.who.int/iris/handle/10665/327836 (accessed on 27 August 2022).
- World Health Organization. Regional Office for Africa, Health Emergencies Programme. Weekly Bulletin on Outbreaks and other Emergencies: Week 02: 06–12 January 2018. Health Emergency Information and Risk Assessment. 2018. Available online: https://apps.who.int/iris/handle/10665/259850 (accessed on 24 August 2022).
- World Health Organization. Regional Office for Africa. Weekly Bulletin on Outbreak and other Emergencies: Week 39: 21–27 September 2020. 2020. Available online: https://apps.who.int/iris/handle/10665/335723 (accessed on 21 August 2022).
- World Health Organization. Regional Office for Africa. Weekly Bulletin on Outbreak and other Emergencies: Week 32: 05–11 August 2019. 2019. Available online: https://apps.who.int/iris/handle/10665/326304 (accessed on 24 August 2022).
- Ahmed, A.; Mohamed, N.S.; Siddig, E.E.; Algaily, T.; Sulaiman, S.; Ali, Y. The impacts of climate change on displaced populations: A call for actions. J. Clim. Change Health 2021, 3, 100057. [Google Scholar] [CrossRef]
- Walter Reed Biosystematics Unit Website. Aedes Aegypti Species Page. Available online: http://wrbu.si.edu/vectorspecies/mosquitoes/aegypti (accessed on 27 August 2022).
- Walter Reed Biosystematics Unit Website. Aedes Africanus Species Page. Available online: http://wrbu.si.edu/vectorspecies/mosquitoes/africanus (accessed on 7 August 2022).
- Walter Reed Biosystematics Unit Website. Aedes Luteocephalus Species Page. Available online: http://wrbu.si.edu/vectorspecies/mosquitoes/luteocephalus (accessed on 15 August 2022).
- Walter Reed Biosystematics Unit Website. Aedes Vittatus Species Page. Available online: http://wrbu.si.edu/vectorspecies/mosquitoes/ae_vittatus (accessed on 8 August 2022).
- Walter Reed Biosystematics Unit Website. Aedes Vexans Species Page. Available online: http://wrbu.si.edu/vectorspecies/mosquitoes/vexans (accessed on 8 August 2022).
- Himeidan, Y.E.; Kweka, E.J.; Mahgoub, M.M.; El Rayah, E.A.; Ouma, J.O. Recent Outbreaks of Rift Valley Fever in East Africa and the Middle East. Front. Public Health 2014, 2, 169. [Google Scholar] [CrossRef]
- Davies, F.G.; Linthicum, K.J.; James, A.D. Rainfall and epizootic Rift Valley fever. Bull. World Health Organ. 1985, 63, 941–943. [Google Scholar] [PubMed]
- Gerdes, G.H. Rift Valley fever. Rev. Sci. Tech. 2004, 23, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A. Urgent call for a global enforcement of the public sharing of health emergencies data: Lesson learned from serious arboviral disease epidemics in Sudan. Int. Health 2020, 12, 238–240. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A. Current status of Mosquito-borne Arboviruses in Sudan, and challenges of surveillance and responses. In Proceedings of the Mosquito-Borne Arboviruses: The Rising Global Threat, Malaria Consortium Webinar, Online, 10 February 2021. [Google Scholar] [CrossRef]
- Elagali, A.; Ahmed, A.; Makki, N.; Ismail, H.; Ajak, M.; Alene, K.A.; Weiss, D.J.; Mohammed, A.A.; Abubakr, M.; Cameron, E.; et al. Spatiotemporal mapping of malaria incidence in Sudan using routine surveillance data. Sci. Rep. 2022, 12, 14114. [Google Scholar] [CrossRef]
- Mohamed, N.S.; Ali, Y.; Muneer, M.S.; Siddig, E.E.; Sibley, C.H.; Ahmed, A. Malaria epidemic in humanitarian crisis settings the case of South Kordofan state, Sudan. J. Infect. Dev. Ctries. 2021, 15, 168–171. [Google Scholar] [CrossRef]
- Franklinos, L.H.V.; Jones, K.E.; Redding, D.W.; Abubakar, I. The effect of global change on mosquito-borne disease. Lancet Infect. Dis. 2019, 19, e302–e312. [Google Scholar] [CrossRef]
- Caminade, C.; McIntyre, K.M.; Jones, A.E. Impact of recent and future climate change on vector-borne diseases. Ann. N. Y. Acad. Sci. 2019, 1436, 157–173. [Google Scholar] [CrossRef]
- Crowl, T.A.; Crist, T.O.; Parmenter, R.R.; Belovsky, G.; Lugo, A.E. The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 2008, 6, 238–246. [Google Scholar] [CrossRef]
- Gubler, D.J.; Reiter, P.; Ebi, K.L.; Yap, W.; Nasci, R.; Patz, J.A. Climate variability and change in the United States: Potential impacts on vector- and rodent-borne diseases. Environ. Health Perspect. 2001, 109, 223–233. [Google Scholar] [CrossRef]
- Ahmed, A.; Abubakr, M.; Ali, Y.; Siddig, E.E.; Mohamed, N.S. Vector control strategy for Anopheles stephensi in Africa. Lancet Microbe 2022, 3, e403. [Google Scholar] [CrossRef]
- Ahmed, A.; Mohamed, N.S.; EL-Sadig, S.M.; Fahal, L.A.; Abelrahim, Z.B.; Ahmed, E.S.; Siddig, E.E. COVID-19 in Sudan. J. Infect. Dev. Ctries. 2021, 15, 204–208. [Google Scholar] [CrossRef]
- Zinsstag, J.; Utzinger, J.; Probst-Hensch, N.; Shan, L.; Zhou, X.-N. Towards integrated surveillance-response systems for the prevention of future pandemics. Infect. Dis. Poverty 2020, 9, 87–92. [Google Scholar] [CrossRef]
- Ali, Y.; Ahmed, A.; Siddig, E.E.; Mohamed, N.S. The role of integrated programs in the prevention of COVID-19 in a humanitarian setting. Trans. R. Soc. Trop. Med. Hyg. 2021, 116, 193–196. [Google Scholar] [CrossRef]
- Zinsstag, J.; Hediger, K.; Osman, Y.M.; Abukhattab, S.; Crump, L.; Kaiser-Grolimund, A.; Mauti, S.; Ahmed, A.; Hattendorf, J.; Bonfoh, B.; et al. The Promotion and Development of One Health at Swiss TPH and Its Greater Potential. Diseases 2022, 10, 65. [Google Scholar] [CrossRef]
- Elduma, A.H.; LaBeaud, A.D.; Plante, J.A.; Plante, K.S.; Ahmed, A. High Seroprevalence of Dengue Virus Infection in Sudan: Systematic Review and Meta-Analysis. Trop. Med. Infect. Dis. 2020, 5, 120. [Google Scholar] [CrossRef]
- Markoff, L. Yellow Fever Outbreak in Sudan. N. Engl. J. Med. 2013, 368, 689–691. [Google Scholar] [CrossRef]
- Ahmed, A.; Ali, Y.; Siddig, E.E.; Hamed, J.; Mohamed, N.S.; Khairy, A.; Zinsstag, J. Hepatitis E Virus Outbreak among Tigray War Refugees from Ethiopia, Sudan. Emerg. Infect. Dis. 2022, 28, 1722. [Google Scholar] [CrossRef]
- Mordecai, E.A.; Ryan, S.J.; Caldwell, J.M.; Shah, M.M.; LaBeaud, A.D. Climate change could shift disease burden from malaria to arboviruses in Africa. Lancet Planet. Health 2020, 4, e416–e423. [Google Scholar] [CrossRef]
- Abubakr, M.; Sami, H.; Mahdi, I.; Altahir, O.; Abdelbagi, H.; Mohamed, N.S.; Ahmed, A. The Phylodynamic and Spread of the Invasive Asian Malaria Vectors, Anopheles stephensi, in Sudan. Biology 2022, 11, 409. [Google Scholar] [CrossRef]
- Ahmed, A.; Khogali, R.; Elnour, M.-A.B.; Nakao, R.; Salim, B. Emergence of the invasive malaria vector Anopheles stephensi in Khartoum State, Central Sudan. Parasites Vectors 2021, 14, 511. [Google Scholar] [CrossRef]
- Gubler, D.J. Aedes albopictus in Africa. Lancet Infect. Dis. 2003, 3, 751–752. [Google Scholar] [CrossRef]
- Ngoagouni, C.; Kamgang, B.; Nakouné, E.; Paupy, C.; Kazanji, M. Invasion of Aedes albopictus (Diptera: Culicidae) into central Africa: What consequences for emerging diseases? Parasites Vectors 2015, 8, 191. [Google Scholar] [CrossRef]
- Kamgang, B.; Wilson-Bahun, T.A.; Irving, H.; Kusimo, M.O.; Lenga, A.; Wondji, C.S. Geographical distribution of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) and genetic diversity of invading population of Ae. albopictus in the Republic of the Congo. Wellcome Open Res. 2018, 3, 79. [Google Scholar] [CrossRef]
- Kamgang, B.; Brengues, C.; Fontenille, D.; Njiokou, F.; Simard, F.; Paupy, C. Genetic structure of the tiger mosquito, Aedes albopictus, in Cameroon (Central Africa). PLoS ONE 2011, 6, e20257. [Google Scholar] [CrossRef]
- Tedjou, A.N.; Kamgang, B.; Yougang, A.P.; Njiokou, F.; Wondji, C.S. Update on the geographical distribution and prevalence of Aedes aegypti and Aedes albopictus (Diptera: Culicidae), two major arbovirus vectors in Cameroon. PLoS Negl. Trop. Dis. 2019, 13, e0007137. [Google Scholar] [CrossRef]
- Ruiling, Z.; Tongkai, L.; Dezhen, M.; Zhong, Z. Genetic characters of the globally spread tiger mosquito, Aedes albopictus (Diptera, Culicidae): Implications from mitochondrial gene COI. J. Vector Ecol. 2018, 43, 89–97. [Google Scholar] [CrossRef]
- Weaver, S.C. Prediction and prevention of urban arbovirus epidemics: A challenge for the global virology community. Antivir. Res. 2018, 156, 80–84. [Google Scholar] [CrossRef]
- Weaver, S.C.; Reisen, W.K. Present and future arboviral threats. Antivir. Res. 2010, 85, 328–345. [Google Scholar] [CrossRef]
- Zinsstag, J.; Schelling, E.; Crump, L.; Whittaker, M.; Tanner, M.; Stephen, C. One Health: The Theory and Practice of Integrated Health Approaches, 2nd ed.; CABI: Wallingford, UK, 2020; p. 459. [Google Scholar]
- Pigott, D.M.; Deshpande, A.; Letourneau, I.; Morozoff, C.; Reiner, R.C.; Kraemer, M.U.G.; Brent, S.E.; Bogoch, I.I.; Khan, K.; Biehl, M.H.; et al. Local, national, and regional viral haemorrhagic fever pandemic potential in Africa: A multistage analysis. Lancet 2017, 390, 2662–2672. [Google Scholar] [CrossRef]
- Fauver, J.R.; Gendernalik, A.; Weger-Lucarelli, J.; Grubaugh, N.D.; Brackney, D.E.; Foy, B.D.; Ebel, G.D. The Use of Xenosurveillance to Detect Human Bacteria, Parasites, and Viruses in Mosquito Bloodmeals. Am. J. Trop. Med. Hyg. 2017, 97, 324–329. [Google Scholar] [CrossRef]
- Brinkmann, A.; Nitsche, A.; Kohl, C. Viral Metagenomics on Blood-Feeding Arthropods as a Tool for Human Disease Surveillance. Int. J. Mol. Sci. 2016, 17, 1743. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Sharma, S.; Krajacich, B.J.; Iii, L.S.F.; Bolay, F.K.; Ii, J.W.D.; Johnson, W.E.; Ebel, G.D.; Foy, B.D.; Brackney, D.E. Xenosurveillance: A Novel Mosquito-Based Approach for Examining the Human-Pathogen Landscape. PLOS Negl. Trop. Dis. 2015, 9, e0003628. [Google Scholar] [CrossRef] [PubMed]
- Zinsstag, J.; Crump, L.; Schelling, E.; Hattendorf, J.; Maidane, Y.O.; Ali, K.O.; Muhummed, A.; Umer, A.A.; Aliyi, F.; Nooh, F. Climate change and one health. FEMS Microbiol. Lett. 2018, 365, fny085. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Irish, S.R.; Zohdy, S.; Yoshimizu, M.; Tadesse, F.G. Strategies for conducting Anopheles stephensi surveys in non-endemic areas. Acta Trop. 2022, 236, 106671. [Google Scholar] [CrossRef]
- Coetzee, M. Key to the females of Afrotropical Anopheles mosquitoes (Diptera: Culicidae). Malar. J. 2020, 19, 70. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Rozas, J.; Sánchez-DelBarrio, J.C.; Messeguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef]





| Populations * | N | S | H | Hd ± VarHd | Pi | Tajima’s D | Fu Li’s D | Fu Li’s F |
|---|---|---|---|---|---|---|---|---|
| MadagasCAR | 12 | 6 | 7 | 0.864 ± 0.00618 | 0.00321 | −1.0217 | −1.1084 | −1.2312 |
| Algeria | 7 | 1 | 2 | 0.286 ± 0.03856 | 0.00063 | −1.0062 | −1.0488 | −1.1015 |
| CAR | 2 | 2 | 2 | 1.0 ± 0.25 | 0.00442 | n.d. | n.d. | n.d. |
| DRC | 45 | 2 | 3 | 0.457 ± 0.00562 | 0.00129 | 0.5144 | 0.7583 | 0.7967 |
| Mauritius | 2 | 2 | 2 | 1.0 ± 0.25 | 0.00442 | n.d. | n.d. | n.d. |
| Mayotte | 3 | 2 | 2 | 0.667 ± 0.09877 | 0.00294 | n.d. | n.d. | n.d. |
| Cameroon | 9 | 5 | 6 | 0.917 ± 0.00526 | 0.00343 | −0.6542 | −0.5973 | −0.6796 |
| Morocco | 2 | 0 | 1 | 0.0 ± 0.00 | 0.0 | n.d. | n.d. | n.d. |
| R Congo | 3 | 2 | 3 | 1.0 ± 0.07407 | 0.00294 | n.d. | n.d. | n.d. |
| La Réunion | 14 | 3 | 4 | 0.648 ± 0.1353 | 0.0017 | −0.5651 | 0.0168 | −0.1524 |
| Sudan | 3 | 0 | 1 | 0.0 ± 0.00 | 0.0 | n.d. | n.d. | n.d. |
| Nigeria | 1 | 0 | n.a. | 0.0 ± 0.00 | 0.0 | n.d. | n.d. | n.d. |
| Seychelles | 1 | 0 | n.a. | 0.0 ± 0.00 | 0.0 | n.d. | n.d. | n.d. |
| Benin | 1 | 0 | n.a. | 0.0 ± 0.00 | 0.0 | n.d. | n.d. | n.d. |
| DRC | Madagascar | Mauritius | Mayotte | Morocco | R Congo | Sudan | Cameroon | Reunion | CAR | |
|---|---|---|---|---|---|---|---|---|---|---|
| DRC | - | - | - | - | - | - | - | - | - | - |
| Madagascar | 0.613 | - | - | - | - | - | - | - | - | - |
| Mauritius | 0.538 | 0.058 | - | - | - | - | - | - | - | - |
| Mayotte | 0.657 | 0.004 | 0.167 | - | - | - | - | - | - | - |
| Morocco | 0.837 | 0.127 | 0.000 | 0.333 | - | - | - | - | - | - |
| R Congo | 0.028 | 0.442 | 0.375 | 0.500 | 0.600 | - | - | - | - | - |
| Sudan | 0.787 | 0.603 | 0.500 | 0.667 | 1.000 | 0.667 | - | - | - | - |
| Cameroon | 0.051 | 0.062 | 0.021 | 0.178 | 0.842 | 0.054 | 0.662 | - | - | - |
| Reunion | 0.648 | 0.047 | 0.020 | 0.151 | 0.103 | 0.462 | 0.615 | 0.0721 | - | - |
| CAR | 0.456 | 0.390 | 0.333 | 0.444 | 0.500 | 0.444 | 0.000 | 0.493 | 0.308 | - |
| Algeria | 0.776 | 0.098 | 0.000 | 0.292 | 0.000 | 0.553 | 0.875 | 0.943 | 0.077 | 0.467 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, A.; Abubakr, M.; Sami, H.; Mahdi, I.; Mohamed, N.S.; Zinsstag, J. The First Molecular Detection of Aedes albopictus in Sudan Associates with Increased Outbreaks of Chikungunya and Dengue. Int. J. Mol. Sci. 2022, 23, 11802. https://doi.org/10.3390/ijms231911802
Ahmed A, Abubakr M, Sami H, Mahdi I, Mohamed NS, Zinsstag J. The First Molecular Detection of Aedes albopictus in Sudan Associates with Increased Outbreaks of Chikungunya and Dengue. International Journal of Molecular Sciences. 2022; 23(19):11802. https://doi.org/10.3390/ijms231911802
Chicago/Turabian StyleAhmed, Ayman, Mustafa Abubakr, Hamza Sami, Isam Mahdi, Nouh S. Mohamed, and Jakob Zinsstag. 2022. "The First Molecular Detection of Aedes albopictus in Sudan Associates with Increased Outbreaks of Chikungunya and Dengue" International Journal of Molecular Sciences 23, no. 19: 11802. https://doi.org/10.3390/ijms231911802
APA StyleAhmed, A., Abubakr, M., Sami, H., Mahdi, I., Mohamed, N. S., & Zinsstag, J. (2022). The First Molecular Detection of Aedes albopictus in Sudan Associates with Increased Outbreaks of Chikungunya and Dengue. International Journal of Molecular Sciences, 23(19), 11802. https://doi.org/10.3390/ijms231911802






