Physiological and Transcriptomic Responses of Growth in Neolamarckia cadamba Stimulated by Exogenous Gibberellins
Abstract
1. Introduction
2. Results
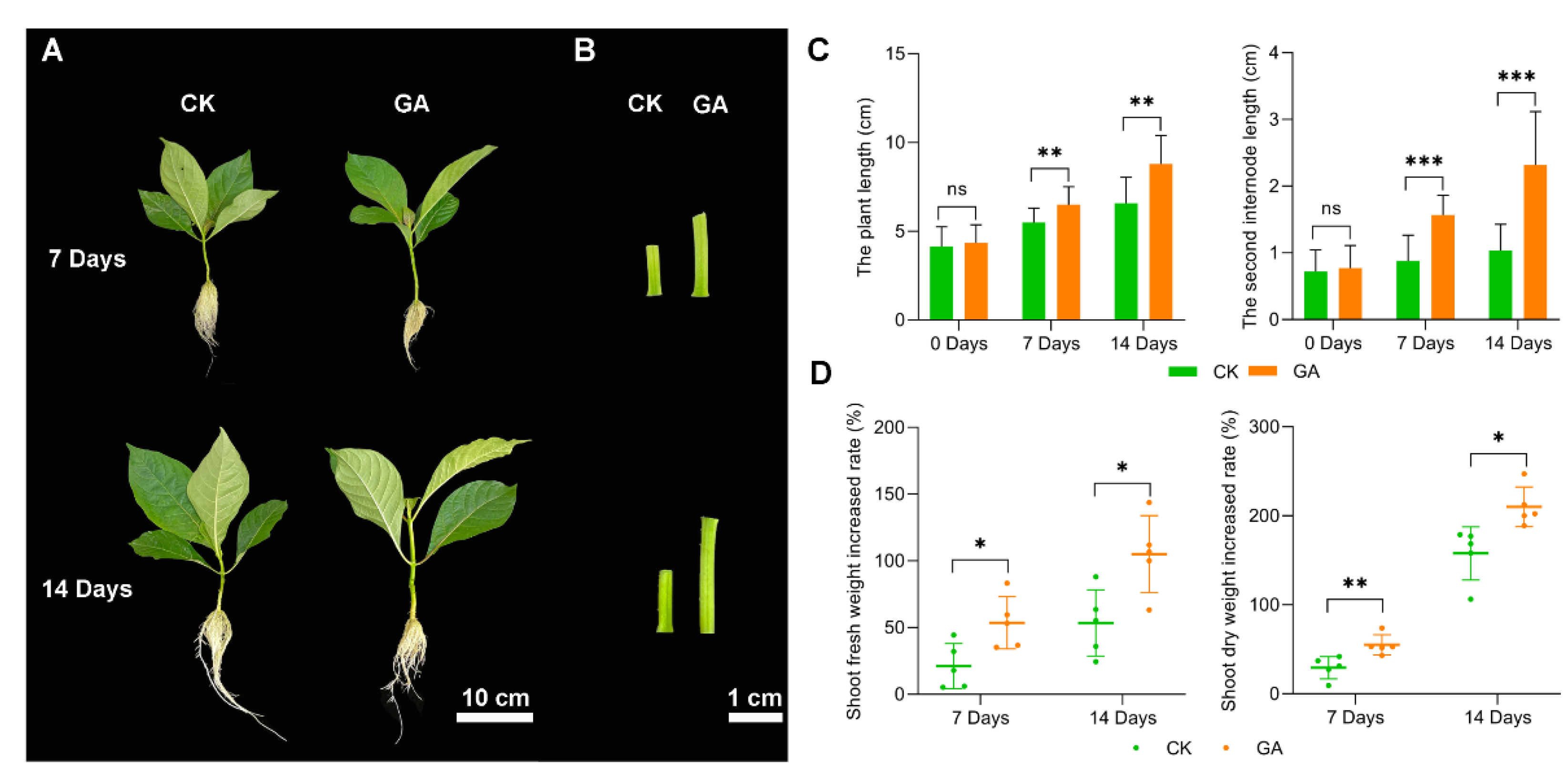
2.1. Effect of Exogenous GA Application on N. cadamba Growth
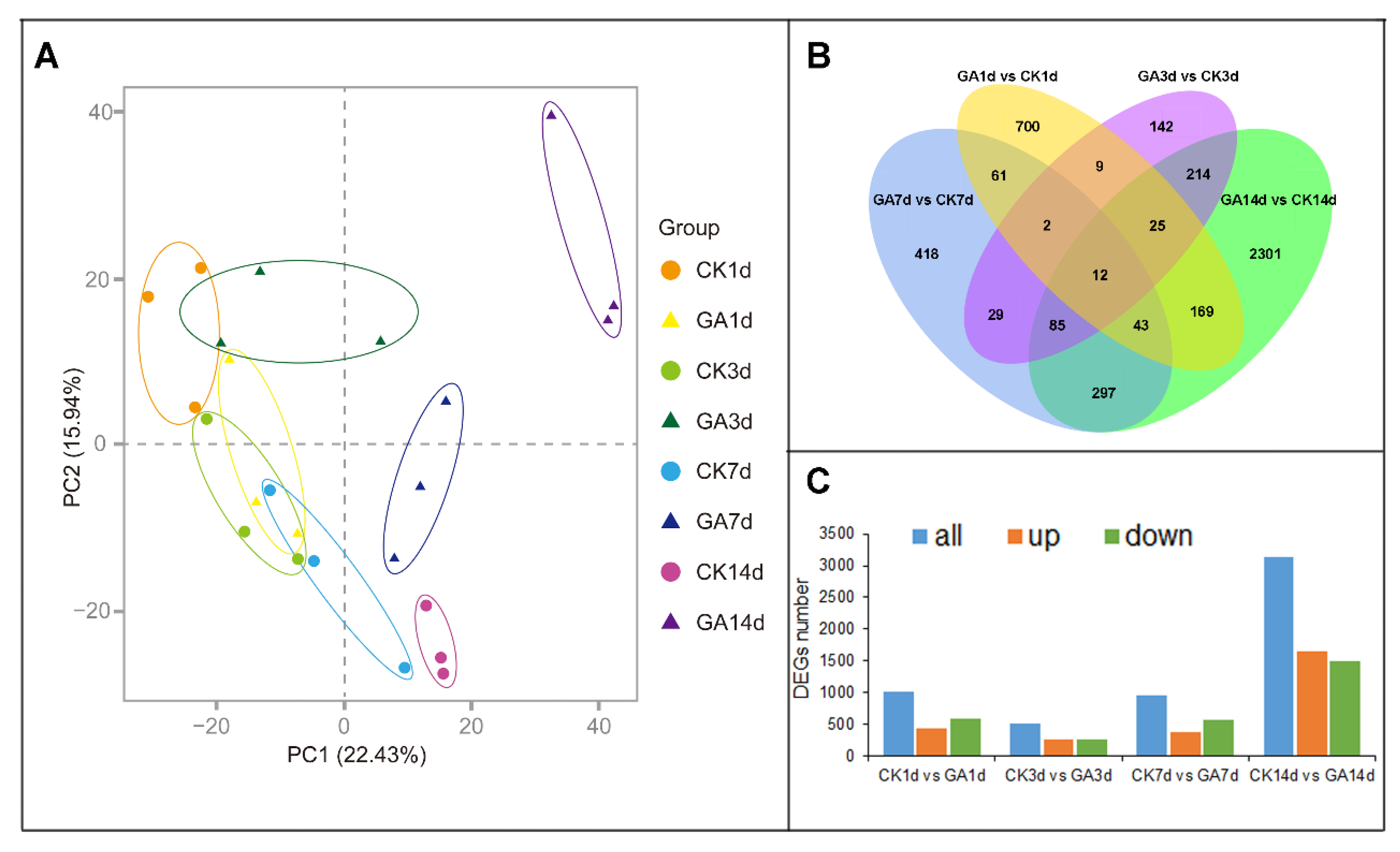
2.2. Identification of DEGs in N. cadamba in GA Treatment
2.3. Expression Profiles of DEGs
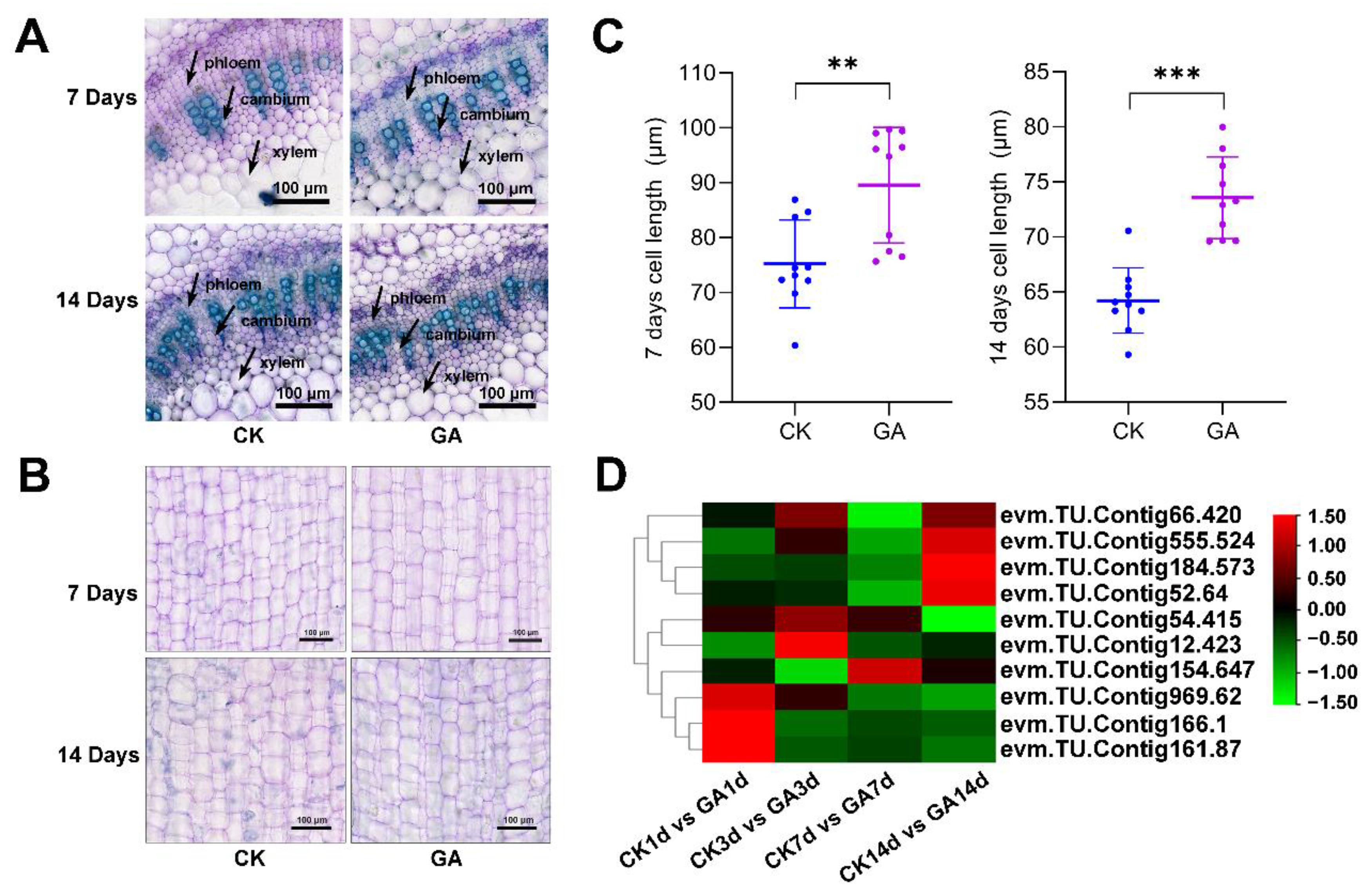
2.4. GA Regulates Cell Elongation in the Second Internode of N. cadamba
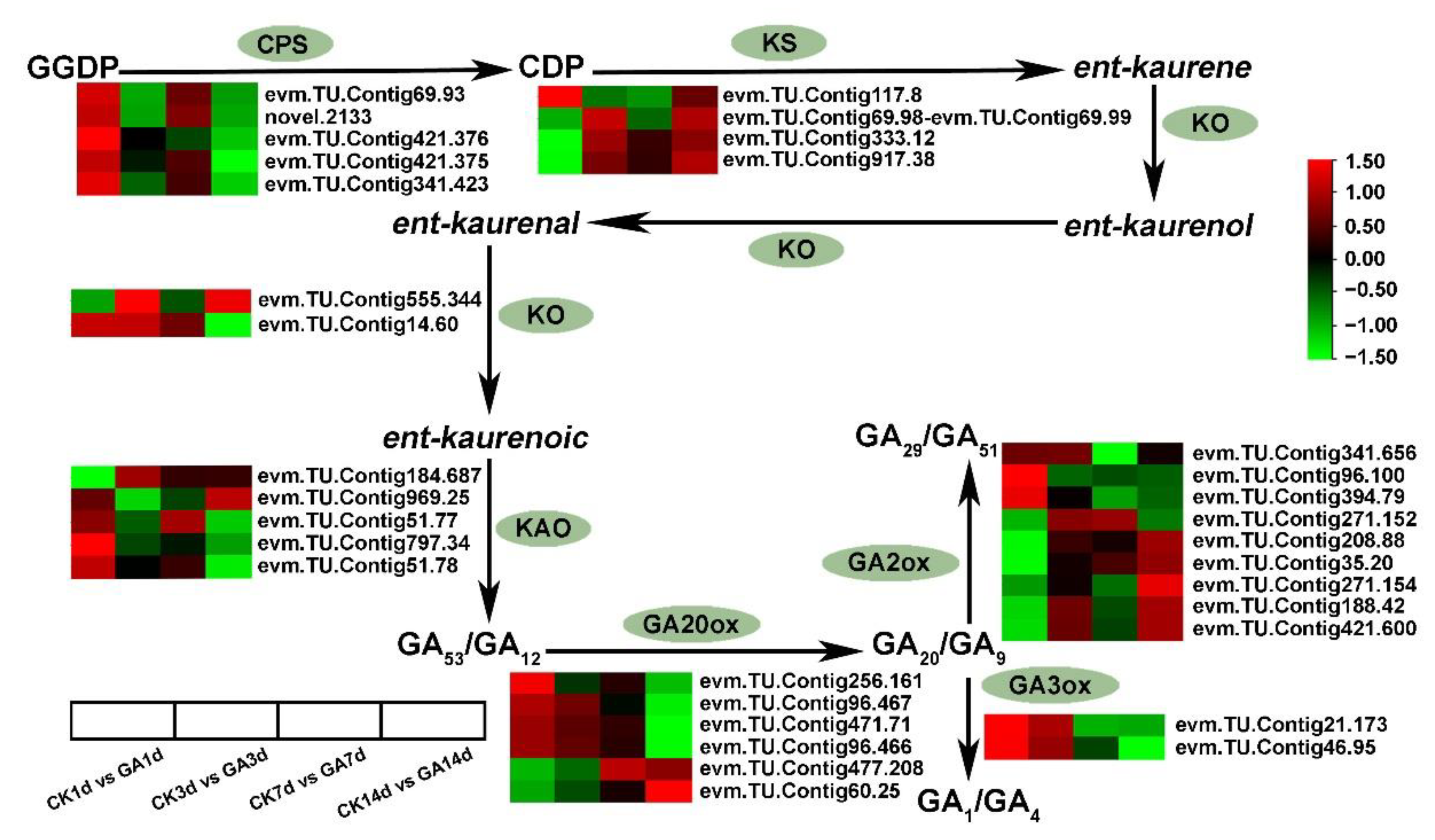
2.5. DEGs Related to GA Biosynthesis and Signaling Transduction
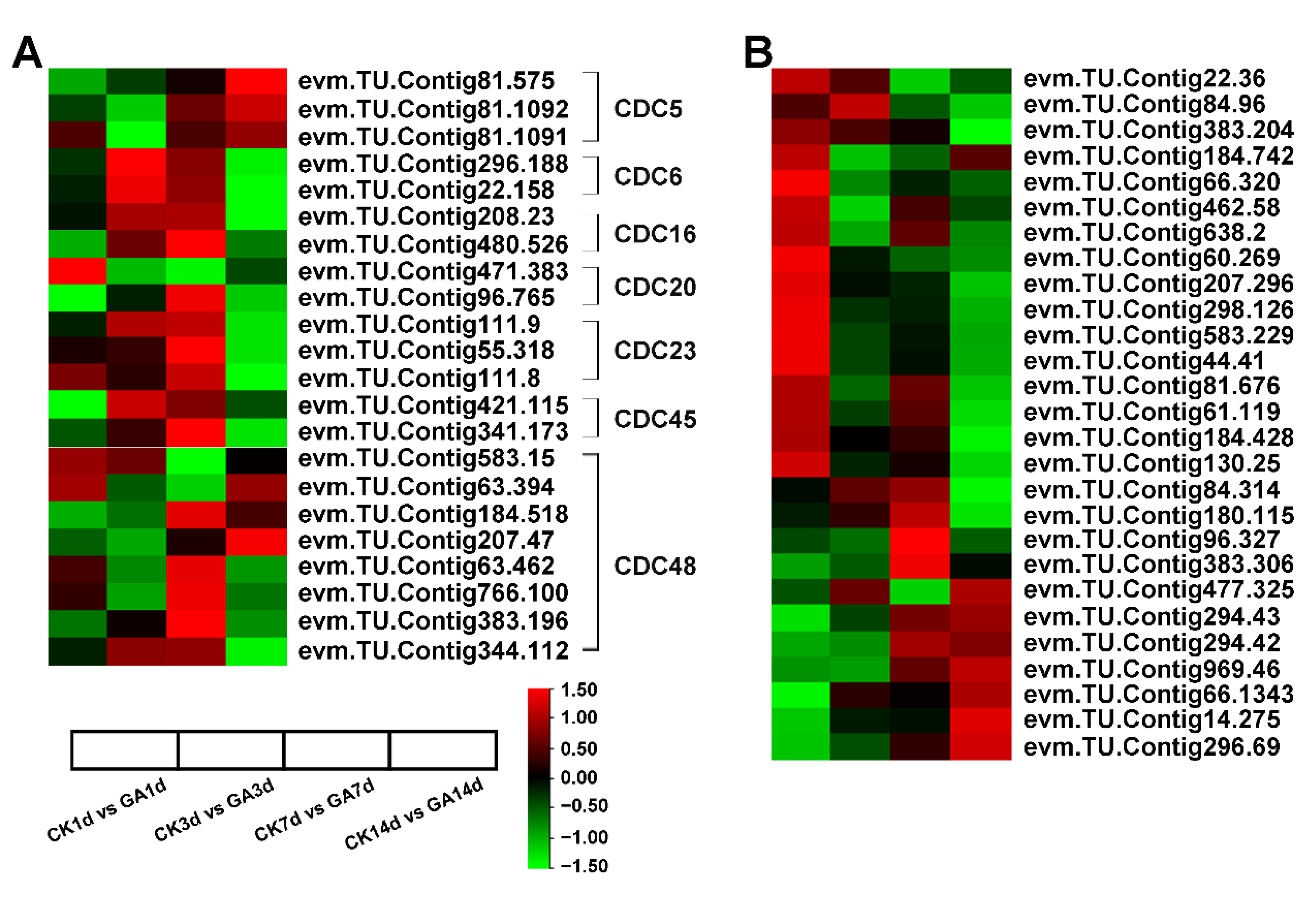
2.6. Exogenous GA Promotes Cell Division and Expansion
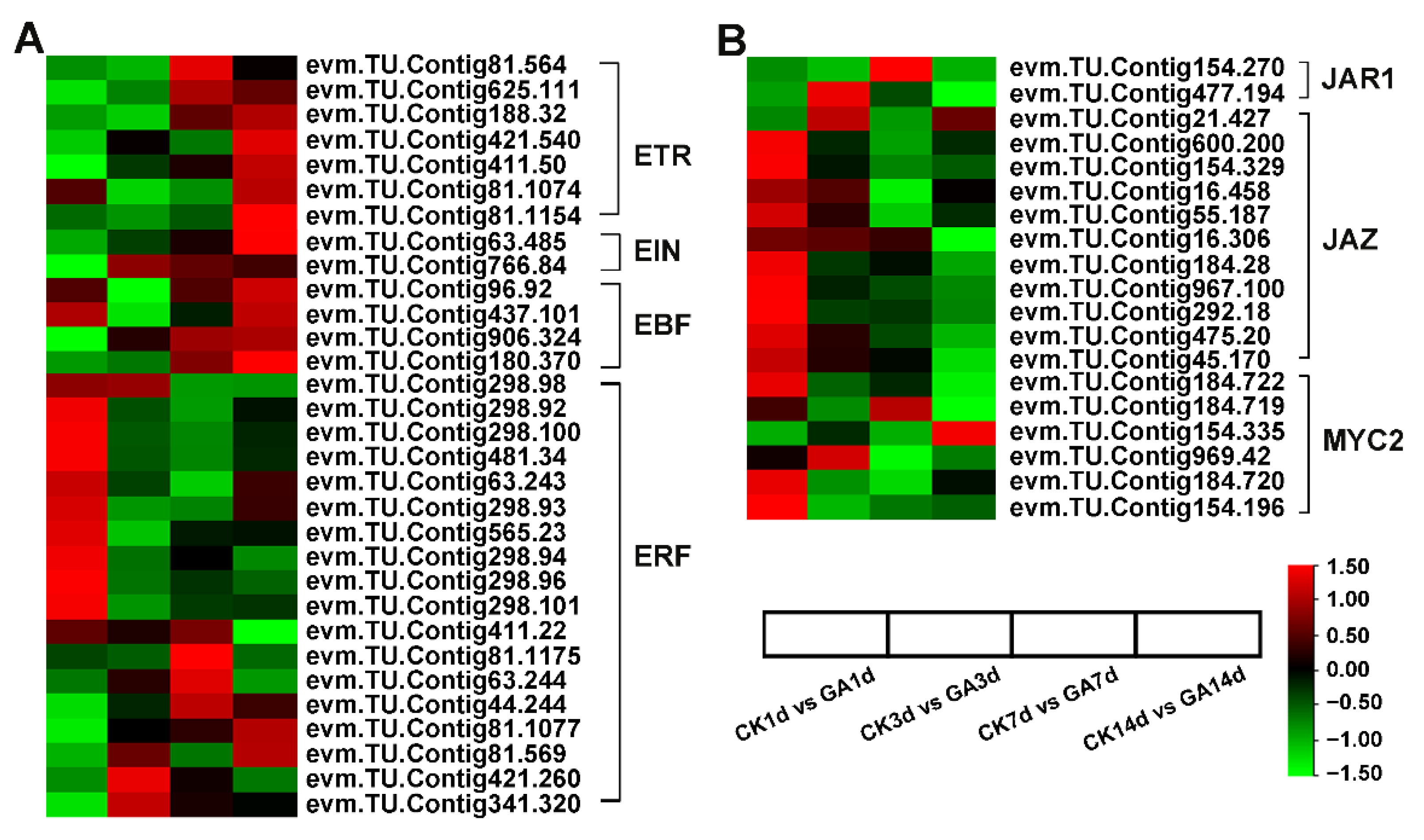
2.7. DEGs with Phytohormone-Related Genes
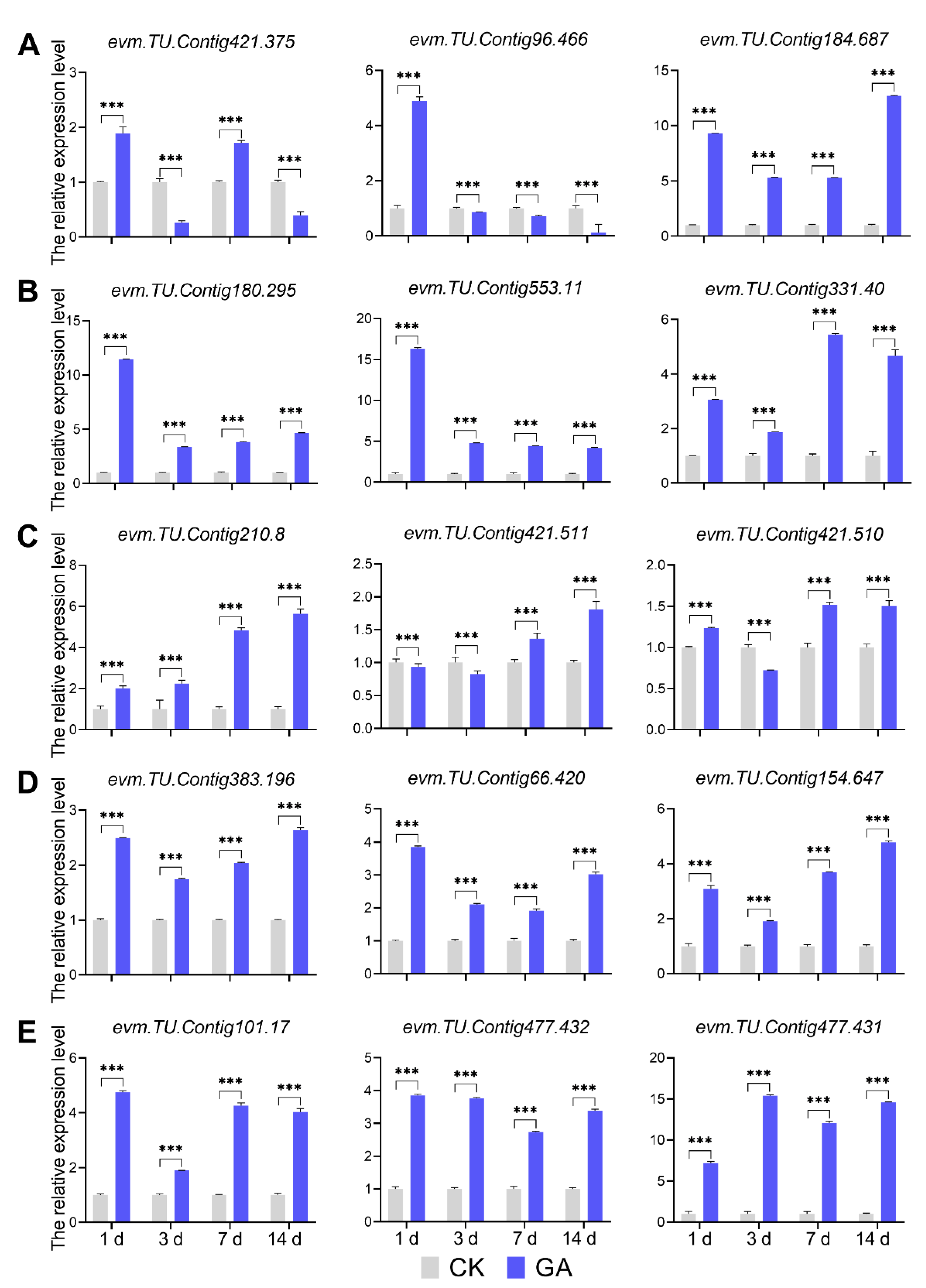
2.8. Assay of DEGs Expression Level by qRT-PCR
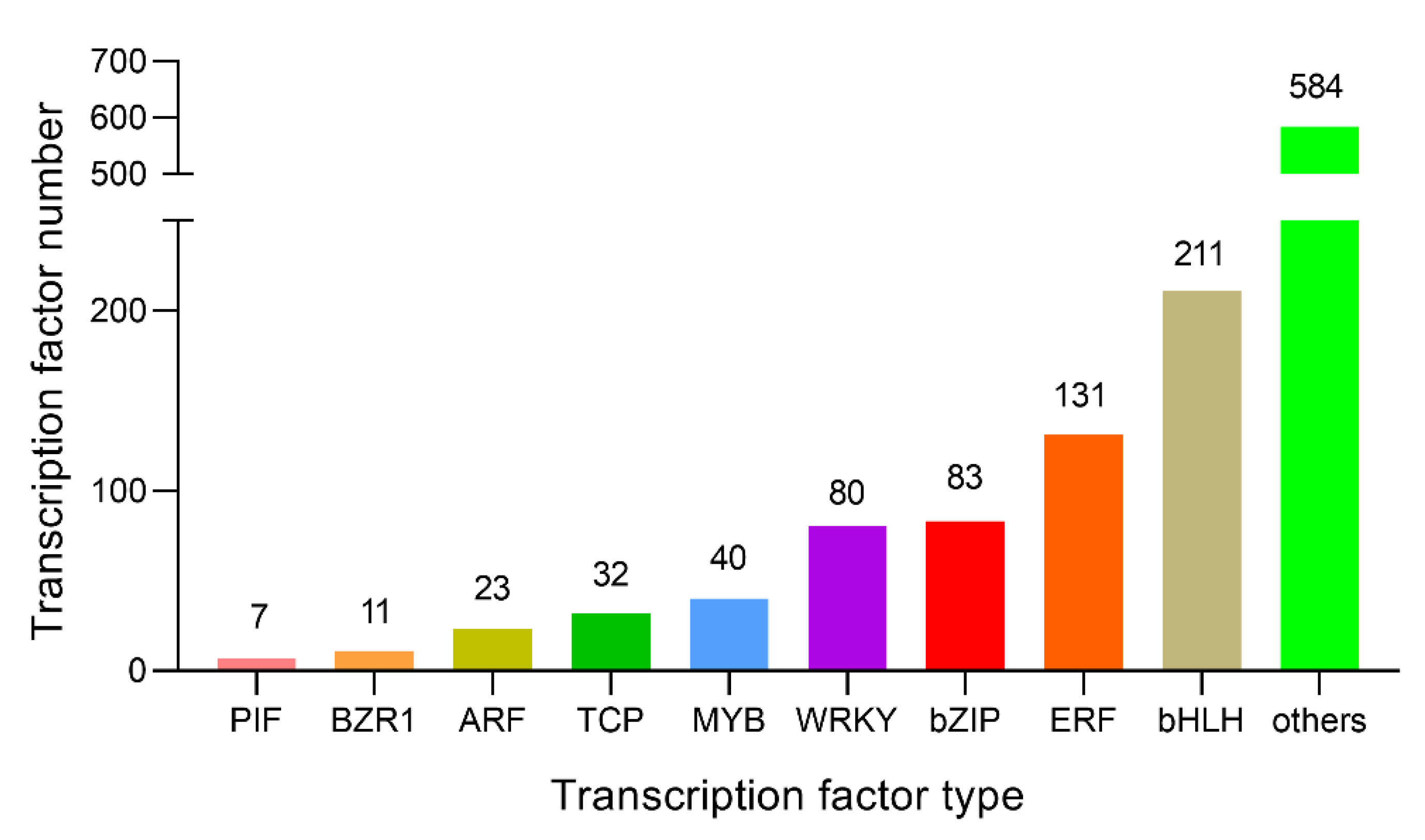
2.9. Changes in Related Transcription Factors and Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. GA Treatment
4.3. Plant Physiological Parameters Measurement
4.4. Microscope Observation
4.5. RNA-Seq Analysis
4.6. Differential Expression Genes Pathway Function Analysis
4.7. Quantitative Real Time PCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pozo, M.J.; Lopez-Raez, J.A.; Azcon-Aguilar, C.; Garcia-Garrido, J.M. Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 2015, 205, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Hauvermale, A.L.; Ariizumi, T.; Steber, C.M. Gibberellin signaling: A theme and variations on DELLA repression. Plant Physiol. 2012, 160, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Kende, H.; Zeevaart, J. The Five “Classical” Plant Hormones. Plant Cell 1997, 9, 1197–1210. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 2000, 18, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Ragni, L.; Nieminen, K.; Pacheco-Villalobos, D.; Sibout, R.; Schwechheimer, C.; Hardtke, C.S. Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. Plant Cell 2011, 23, 1322–1336. [Google Scholar] [CrossRef]
- Immanen, J.; Nieminen, K.; Smolander, O.P.; Kojima, M.; Alonso, S.J.; Koskinen, P.; Zhang, J.; Elo, A.; Mahonen, A.P.; Street, N.; et al. Cytokinin and Auxin Display Distinct but Interconnected Distribution and Signaling Profiles to Stimulate Cambial Activity. Curr. Biol. 2016, 26, 1990–1997. [Google Scholar] [CrossRef]
- Ben-Targem, M.; Ripper, D.; Bayer, M.; Ragni, L. Auxin and gibberellin signaling cross-talk promotes hypocotyl xylem expansion and cambium homeostasis. J. Exp. Bot. 2021, 72, 3647–3660. [Google Scholar] [CrossRef]
- Jeon, H.W.; Cho, J.S.; Park, E.J.; Han, K.H.; Choi, Y.I.; Ko, J.H. Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 2016, 14, 1161–1170. [Google Scholar] [CrossRef]
- Iftikhar, A.; Ali, S.; Yasmeen, T.; Arif, M.S.; Zubair, M.; Rizwan, M.; Alhaithloul, H.; Alayafi, A.; Soliman, M.H. Effect of gibberellic acid on growth, photosynthesis and antioxidant defense system of wheat under zinc oxide nanoparticle stress. Environ. Pollut. 2019, 254, 113109. [Google Scholar] [CrossRef]
- Huerta, L.; Forment, J.; Gadea, J.; Fagoaga, C.; Pena, L.; Perez-Amador, M.A.; Garcia-Martinez, J.L. Gene expression analysis in citrus reveals the role of gibberellins on photosynthesis and stress. Plant Cell Environ. 2008, 31, 1620–1633. [Google Scholar] [CrossRef]
- Xie, J.; Tian, J.; Du, Q.; Chen, J.; Li, Y.; Yang, X.; Li, B.; Zhang, D. Association genetics and transcriptome analysis reveal a gibberellin-responsive pathway involved in regulating photosynthesis. J. Exp. Bot. 2016, 67, 3325–3338. [Google Scholar] [CrossRef] [PubMed]
- Piskurewicz, U.; Jikumaru, Y.; Kinoshita, N.; Nambara, E.; Kamiya, Y.; Lopez-Molina, L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 2008, 20, 2729–2745. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, N.; Girin, T.; Sorefan, K.; Fuentes, S.; Wood, T.A.; Lawrenson, T.; Sablowski, R.; Ostergaard, L. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010, 24, 2127–2132. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Ljung, K.; Sorefan, K.; Alvey, E.; Harberd, N.P.; Ostergaard, L. Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell 2012, 24, 3982–3996. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P.; Kamiya, Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 1994, 6, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef]
- Chhun, T.; Aya, K.; Asano, K.; Yamamoto, E.; Morinaka, Y.; Watanabe, M.; Kitano, H.; Ashikari, M.; Matsuoka, M.; Ueguchi-Tanaka, M. Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 2007, 19, 3876–3888. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Nakajima, M.; Motoyuki, A.; Matsuoka, M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 2007, 58, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Richards, D.E.; Ait-Ali, T.; Hynes, L.W.; Ougham, H.; Peng, J.; Harberd, N.P. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 2002, 14, 3191–3200. [Google Scholar] [CrossRef] [PubMed]
- Locascio, A.; Blazquez, M.A.; Alabadi, D. Genomic analysis of DELLA protein activity. Plant Cell Physiol. 2013, 54, 1229–1237. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Matsuoka, M. The perception of gibberellins: Clues from receptor structure. Curr. Opin. Plant Biol. 2010, 13, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Shimada, A.; Ueguchi-Tanaka, M.; Nakatsu, T.; Nakajima, M.; Naoe, Y.; Ohmiya, H.; Kato, H.; Matsuoka, M. Structural basis for gibberellin recognition by its receptor GID1. Nature 2008, 456, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Tanimoto, E.; Hirai, T.; Miyanoiri, Y.; Mitani, R.; Kawamura, M.; Takeda, M.; Takehara, S.; Hirano, K.; Kainosho, M.; et al. Evolution and diversification of the plant gibberellin receptor GID1. Proc. Natl. Acad. Sci. USA 2018, 115, E7844–E7853. [Google Scholar] [CrossRef]
- Daviere, J.M.; Achard, P. A Pivotal Role of DELLAs in Regulating Multiple Hormone Signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef]
- Wang, G.L.; Que, F.; Xu, Z.S.; Wang, F.; Xiong, A.S. Exogenous gibberellin altered morphology, anatomic and transcriptional regulatory networks of hormones in carrot root and shoot. BMC Plant Biol. 2015, 15, 290. [Google Scholar] [CrossRef]
- Ouyang, K.; Li, J.; Zhao, X.; Que, Q.; Li, P.; Huang, H.; Deng, X.; Singh, S.K.; Wu, A.M.; Chen, X. Transcriptomic Analysis of Multipurpose Timber Yielding Tree Neolamarckia cadamba during Xylogenesis Using RNA-Seq. PLoS ONE 2016, 11, e159407. [Google Scholar] [CrossRef]
- Zhao, X.; Tong, T.; Li, H.; Lu, H.; Ren, J.; Zhang, A.; Deng, X.; Chen, X.; Wu, A.M. Characterization of hemicelluloses from Neolamarckia cadamba (Rubiaceae) during xylogenesis. Carbohydr. Polym. 2017, 156, 333–339. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, Q.; Dai, B.; Wang, K.; Lu, L.; Qaseem, M.F.; Wang, J.; Li, H.; Wu, A. The key physiology and molecular responses to potassium deficiency in Neolamarckia cadamba. Ind. Crops Prod. 2021, 162, 113260. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, L.; Zhang, Y.; Yin, Q.; Yi, N.; Qaseem, M.F.; Li, H.; Wu, A.M. Potassium deficiency inhibits leaf growth and promotes leaf necrotic spots in Neolamarckia cadamba (Roxb.) Bosser. Tree Physiol. 2022, 42, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Chen, C.; Liu, Y.; Liu, L.; Qaseem, M.F.; Wang, J.; Li, H.; Wu, A.M. Physiological, Biochemical, and Transcriptomic Responses of Neolamarckia cadamba to Aluminum Stress. Int. J. Mol. Sci. 2020, 21, 9624. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhang, Y.; Li, L.; Yi, N.; Liu, Y.; Qaseem, M.F.; Li, H.; Wu, A.M. Physiological and Transcriptomic Responses to Nitrogen Deficiency in Neolamarckia cadamba. Front. Plant Sci. 2021, 12, 747121. [Google Scholar] [CrossRef]
- Hernandez-Garcia, J.; Briones-Moreno, A.; Blazquez, M.A. Origin and evolution of gibberellin signaling and metabolism in plants. Semin. Cell Dev. Biol. 2021, 109, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Beemster, G.T.; Fiorani, F.; Inze, D. Cell cycle: The key to plant growth control? Trends Plant Sci. 2003, 8, 154–158. [Google Scholar] [CrossRef]
- Keyes, W.J.; Taylor, J.V.; Apkarian, R.P.; Lynn, D.G. Dancing together. Social controls in parasitic plant development. Plant Physiol. 2001, 127, 1508–1512. [Google Scholar] [CrossRef]
- Oh, E.; Zhu, J.Y.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 2014, 3, e03031. [Google Scholar] [CrossRef]
- Asahina, M.; Iwai, H.; Kikuchi, A.; Yamaguchi, S.; Kamiya, Y.; Kamada, H.; Satoh, S. Gibberellin produced in the cotyledon is required for cell division during tissue reunion in the cortex of cut cucumber and tomato hypocotyls. Plant Physiol. 2002, 129, 201–210. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Rehman, H.M.; Imtiaz, M.; Baloch, F.S.; Lee, J.D.; Yang, S.H.; Lee, S.I.; Chung, G. Systems Identification and Characterization of Cell Wall Reassembly and Degradation Related Genes in Glycine max (L.) Merill, a Bioenergy Legume. Sci. Rep. 2017, 7, 10862. [Google Scholar] [CrossRef]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb Perspect. Biol. 2010, 2, a1446. [Google Scholar] [CrossRef] [PubMed]
- Majda, M.; Robert, S. The Role of Auxin in Cell Wall Expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.R.; Wang, X.C.; Li, X.M.; Yang, C.D. Transcription factors in higher plant. Yi Chuan 2004, 26, 403–408. [Google Scholar]
- Daviere, J.M.; Wild, M.; Regnault, T.; Baumberger, N.; Eisler, H.; Genschik, P.; Achard, P. Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 2014, 24, 1923–1928. [Google Scholar] [CrossRef] [PubMed]
- Little, C.H.; MacDonald, J.E. Effects of exogenous gibberellin and auxin on shoot elongation and vegetative bud development in seedlings of Pinus sylvestris and Picea glauca. Tree Physiol. 2003, 23, 73–83. [Google Scholar] [CrossRef]
- MacDonald, J.E.; Little, C.H. Foliar application of GA3 during terminal long-shoot bud development stimulates shoot apical meristem activity in Pinus sylvestris seedlings. Tree Physiol. 2006, 26, 1271–1276. [Google Scholar] [CrossRef][Green Version]
- Liu, Q.Y.; Guo, G.S.; Qiu, Z.F.; Li, X.D.; Zeng, B.S.; Fan, C.J. Exogenous GA3 application altered morphology, anatomic and transcriptional regulatory networks of hormones in Eucalyptus grandis. Protoplasma 2018, 255, 1107–1119. [Google Scholar] [CrossRef]
- Xu, Y.L.; Gage, D.A.; Zeevaart, J.A. Gibberellins and stem growth in Arabidopsis thaliana. Effects of photoperiod on expression of the GA4 and GA5 loci. Plant Physiol. 1997, 114, 1471–1476. [Google Scholar] [CrossRef]
- Hamano, M.; Yamato, Y.; Yamazaki, H.; Miura, H. Endogenous gibberellins and their effects on flowering and stem elongation in cabbage (Brassica oleracea var. capitata). J. Hortic. Sci. Biotech. 2002, 77, 220–225. [Google Scholar] [CrossRef]
- Hedden, P.; Kamiya, Y. GIBBERELLIN BIOSYNTHESIS: Enzymes, Genes and Their Regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 431–460. [Google Scholar] [CrossRef]
- Ross, J.J.; Murfet, I.C.; Reid, J.B. Distribution of Gibberellins in Lathyrus odoratus L. and Their Role in Leaf Growth. Plant Physiol. 1993, 102, 603–608. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Silverstone, A.L.; Chang, C.; Krol, E.; Sun, T.P. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 1997, 12, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Fleet, C.M.; Yamaguchi, S.; Hanada, A.; Kawaide, H.; David, C.J.; Kamiya, Y.; Sun, T.P. Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol. 2003, 132, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tian, X.; Xue, L.; Zhang, X.; Yang, S.; Traw, M.B.; Huang, J. CRISPR-Based Assessment of Gene Specialization in the Gibberellin Metabolic Pathway in Rice. Plant Physiol. 2019, 180, 2091–2105. [Google Scholar] [CrossRef]
- Thomas, S.G.; Phillips, A.L.; Hedden, P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 1999, 96, 4698–4703. [Google Scholar] [CrossRef]
- Hedden, P. The Current Status of Research on Gibberellin Biosynthesis. Plant Cell Physiol. 2020, 61, 1832–1849. [Google Scholar] [CrossRef]
- Li, Q.F.; Wang, C.; Jiang, L.; Li, S.; Sun, S.S.; He, J.X. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 2012, 5, a72. [Google Scholar] [CrossRef]
- Bai, M.Y.; Shang, J.X.; Oh, E.; Fan, M.; Bai, Y.; Zentella, R.; Sun, T.P.; Wang, Z.Y. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012, 14, 810–817. [Google Scholar] [CrossRef]
- De Lucas, M.; Daviere, J.M.; Rodriguez-Falcon, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blazquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484. [Google Scholar] [CrossRef]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef]
- Gallego-Bartolome, J.; Minguet, E.G.; Grau-Enguix, F.; Abbas, M.; Locascio, A.; Thomas, S.G.; Alabadi, D.; Blazquez, M.A. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 13446–13451. [Google Scholar] [CrossRef] [PubMed]
- Bernardo-Garcia, S.; de Lucas, M.; Martinez, C.; Espinosa-Ruiz, A.; Daviere, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Björklund, S.; Antti, H.; Uddestrand, I.; Moritz, T.; Sundberg, B. Cross-talk between gibberellin and auxin in development of Populus wood: Gibberellin stimulates polar auxin transport and has a common transcriptome with auxin: Cross-talk between GA and auxin in wood development. Plant J. 2007, 52, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Israelsson, M.; Sundberg, B.; Moritz, T. Tissue-specific localization of gibberellins and expression of gibberellin-biosynthetic and signaling genes in wood-forming tissues in aspen. Plant J. 2005, 44, 494–504. [Google Scholar] [CrossRef]
- Hu, J.; Su, H.; Cao, H.; Wei, H.; Fu, X.; Jiang, X.; Song, Q.; He, X.; Xu, C.; Luo, K. AUXIN RESPONSE FACTOR7 integrates gibberellin and auxin signaling via interactions between DELLA and AUX/IAA proteins to regulate cambial activity in poplar. Plant Cell 2022, 34, 2688–2707. [Google Scholar] [CrossRef]
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927. [Google Scholar] [CrossRef]
- Marin-de, L.R.N.; Sotillo, B.; Miskolczi, P.; Gibbs, D.J.; Vicente, J.; Carbonero, P.; Onate-Sanchez, L.; Holdsworth, M.J.; Bhalerao, R.; Alabadi, D.; et al. Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 2014, 166, 1022–1032. [Google Scholar] [CrossRef]
- Lim, S.; Park, J.; Lee, N.; Jeong, J.; Toh, S.; Watanabe, A.; Kim, J.; Kang, H.; Kim, D.H.; Kawakami, N.; et al. ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 2013, 25, 4863–4878. [Google Scholar] [CrossRef]
- Ogas, J. Dissecting the gibberellin response pathway. Curr. Biol. CB 1998, 8, R165–R167. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Yang, R.; Wang, F.; Fu, J.; Yang, W.; Bai, T.; Wang, S.; Yin, H. Effects of gibberellin priming on seedling emergence and transcripts involved in mesocotyl elongation in rice under deep direct-seeding conditions. J. Zhejiang Univ.-Sc. B 2021, 22, 1002–1021. [Google Scholar] [CrossRef]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 2012, 24, 4483–4497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Tanabe, K.; Tamura, F.; Matsumoto, K.; Yoshida, A. 13C-photosynthate accumulation in Japanese pear fruit during the period of rapid fruit growth is limited by the sink strength of fruit rather than by the transport capacity of the pedicel. J. Exp. Bot. 2005, 56, 2713–2719. [Google Scholar] [CrossRef] [PubMed]
- Ji-Eun, P.; YongHee, K.; ByulHaNa, L.; YoSup, P.; Myung, H.J.; Jin-Ho, C.; Hee-Seung, P. Anatomical Structure and Fruit Quality According to the Fruit Developmental Stage as Affected by Gibberellins Treatments in Pyrus pyrifolia Nakai cv. Hanareum. Korean J. Hortic. Sci. 2014, 32, 33–40. [Google Scholar]
- Yosup, P.; Hee-Seung, P. Microstructural changes in the fruit and pedicel of ‘Wonhwang’ oriental pear induced by exogenous gibberellins. Sci. Hortic.-Amst. 2017, 222, 1–6. [Google Scholar]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Mesejo, C.; Yuste, R.; Reig, C.; Martinez-Fuentes, A.; Iglesias, D.J.; Munoz-Fambuena, N.; Bermejo, A.; Germana, M.A.; Primo-Millo, E.; Agusti, M. Gibberellin reactivates and maintains ovary-wall cell division causing fruit set in parthenocarpic Citrus species. Plant Sci. 2016, 247, 13–24. [Google Scholar] [CrossRef]
- Dugardeyn, J.; Vandenbussche, F.; Van Der Straeten, D. To grow or not to grow: What can we learn on ethylene-gibberellin cross-talk by in silico gene expression analysis? J. Exp. Bot. 2008, 59, 1–16. [Google Scholar] [CrossRef]
- Weiss, D.; Ori, N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007, 144, 1240–1246. [Google Scholar] [CrossRef]
- O’Neill, D.P.; Davidson, S.E.; Clarke, V.C.; Yamauchi, Y.; Yamaguchi, S.; Kamiya, Y.; Reid, J.B.; Ross, J.J. Regulation of the gibberellin pathway by auxin and DELLA proteins. Planta 2010, 232, 1141–1149. [Google Scholar] [CrossRef]
- Li, W.; Wang, B.; Sun, H.; Zhang, Z. Transcriptome profiling of runner formation induced by exogenous gibberellin in Fragaria vesca. Sci. Hortic.-Amst. 2021, 281, 109966. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]


| Gene ID | GA1d vs. CK1d | GA3d vs. CK3d | GA7d vs. CK7d | GA14d vs. CK14d | Seq Description | TF Family |
|---|---|---|---|---|---|---|
| evm.TU.Contig63.469 | 2.46 | 0.64 | −1.78 | −3.27 | TCP family transcription factor | TCP |
| evm.TU.Contig84.232 | −2.16 | −2.26 | −3.17 | −3.74 | bHLH transcription factor bHLH051 | bHLH |
| evm.TU.Contig28.410 | −2.57 | 3.21 | 2.49 | 3.69 | bHLH transcription factor bHLH025 | bHLH |
| evm.TU.Contig96.268 | −2.47 | −2.40 | −2.24 | −3.87 | bHLH transcription factor bHLH051 | bHLH |
| evm.TU.Contig28.419 | −0.32 | 3.45 | 0.50 | −0.39 | bHLH transcription factor bHLH025 | bHLH |
| evm.TU.Contig143.21 | −1.97 | 0.44 | 1.97 | 6.37 | Transcription factor MYB44 | MYB |
| evm.TU.Contig294.168 | 2.52 | 2.44 | −1.83 | 4.61 | Transcription factor MYB86 | MYB |
| evm.TU.Contig171.196 | 2.48 | 1.85 | −3.43 | 4.71 | Transcription factor MYB44 | MYB |
| evm.TU.Contig3.39 | 0.20 | 0.90 | −4.36 | −0.93 | MYB-related protein MYB4 | MYB |
| evm.TU.Contig279.118 | −1.63 | 1.18 | −3.66 | 4.37 | Transcription factor MYB44 | MYB |
| evm.TU.Contig12.430 | 2.02 | 2.71 | 2.30 | 4.21 | bZIP transcription factor 53 | bZIP |
| evm.TU.Contig44.177 | 0.02 | −2.77 | −3.79 | −2.52 | bZIP transcription factor 70 | bZIP |
| evm.TU.Contig906.67 | 0.45 | 1.74 | 1.17 | 3.32 | bZIP transcription factor 39 | bZIP |
| evm.TU.Contig66.454 | −0.99 | −1.96 | −1.80 | −6.13 | bZIP transcription factor 43 | bZIP |
| evm.TU.Contig81.569 | −3.72 | −0.45 | −1.27 | −4.38 | Ethylene-responsive transcription factor 1 B | ERF |
| evm.TU.Contig66.1116 | 1.02 | −0.50 | −4.03 | 0.14 | Ethylene-responsive transcription factor 109 | ERF |
| evm.TU.Contig298.98 | 0.66 | 0.72 | 4.19 | 4.08 | Ethylene-responsive transcription factor 1 B | ERF |
| evm.TU.Contig81.1174 | 0.02 | 2.95 | −2.84 | −4.41 | Ethylene-responsive transcription factor 098 | ERF |
| evm.TU.Contig383.343 | −0.45 | −0.70 | −3.56 | −3.63 | Ethylene-responsive transcription factor 1 A | ERF |
| evm.TU.Contig298.96 | 0.72 | 0.25 | 0.10 | 3.52 | Ethylene-responsive transcription factor 1 B | ERF |
| evm.TU.Contig600.195 | −1.61 | 0.11 | −2.50 | −3.49 | Ethylene-responsive transcription factor 017 | ERF |
| evm.TU.Contig45.300 | −0.73 | 0.45 | −0.71 | −3.38 | Ethylene-responsive transcription factor003 | ERF |
| evm.TU.Contig298.93 | −3.10 | 0.29 | −3.44 | 3.10 | Ethylene-responsive transcription factor 1 B | ERF |
| evm.TU.Contig44.244 | −0.11 | −1.08 | −1.80 | −3.17 | Ethylene-responsive transcription factor 1 B | ERF |
| evm.TU.Contig63.244 | 2.36 | −3.51 | −0.50 | −3.01 | Ethylene-responsive transcription factor 1 B | ERF |
| evm.TU.Contig298.94 | 1.16 | −0.21 | 0.03 | 3.60 | Ethylene-responsive transcription factor 1 B | ERF |
| evm.TU.Contig63.37 | 0.02 | −0.40 | 1.03 | −3.20 | Ethylene-responsive transcription factor 018 | ERF |
| evm.TU.Contig294.72 | 2.46 | 4.76 | −1.83 | 3.97 | Ethylene-responsive transcription factor 2 | ERF |
| evm.TU.Contig435.24 | −0.68 | 0.15 | −4.24 | −3.64 | WRKY transcription factor 72 | WRKY |
| evm.TU.Contig279.48 | 0.57 | 2.46 | −3.75 | 3.77 | WRKY transcription factor 69 | WRKY |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Wang, J.; Chen, J.; Wang, Z.; Qaseem, M.F.; Li, H.; Wu, A. Physiological and Transcriptomic Responses of Growth in Neolamarckia cadamba Stimulated by Exogenous Gibberellins. Int. J. Mol. Sci. 2022, 23, 11842. https://doi.org/10.3390/ijms231911842
Li L, Wang J, Chen J, Wang Z, Qaseem MF, Li H, Wu A. Physiological and Transcriptomic Responses of Growth in Neolamarckia cadamba Stimulated by Exogenous Gibberellins. International Journal of Molecular Sciences. 2022; 23(19):11842. https://doi.org/10.3390/ijms231911842
Chicago/Turabian StyleLi, Lu, Jiaqi Wang, Jiajun Chen, Zhihua Wang, Mirza Faisal Qaseem, Huiling Li, and Aimin Wu. 2022. "Physiological and Transcriptomic Responses of Growth in Neolamarckia cadamba Stimulated by Exogenous Gibberellins" International Journal of Molecular Sciences 23, no. 19: 11842. https://doi.org/10.3390/ijms231911842
APA StyleLi, L., Wang, J., Chen, J., Wang, Z., Qaseem, M. F., Li, H., & Wu, A. (2022). Physiological and Transcriptomic Responses of Growth in Neolamarckia cadamba Stimulated by Exogenous Gibberellins. International Journal of Molecular Sciences, 23(19), 11842. https://doi.org/10.3390/ijms231911842






