Systematic Down-Selection of Repurposed Drug Candidates for COVID-19
Abstract
:1. Introduction
2. Results
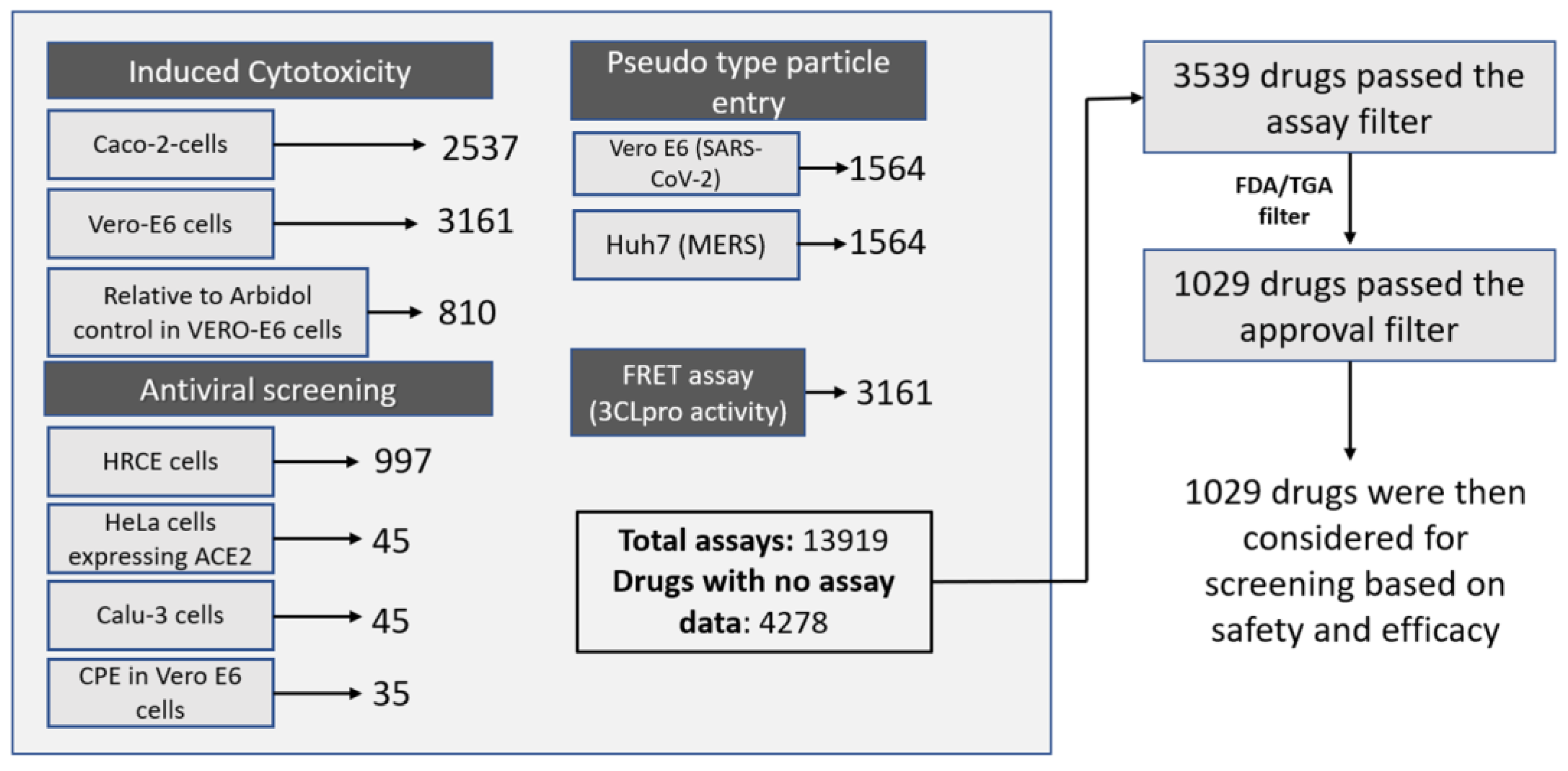
2.1. Assembly of the Data
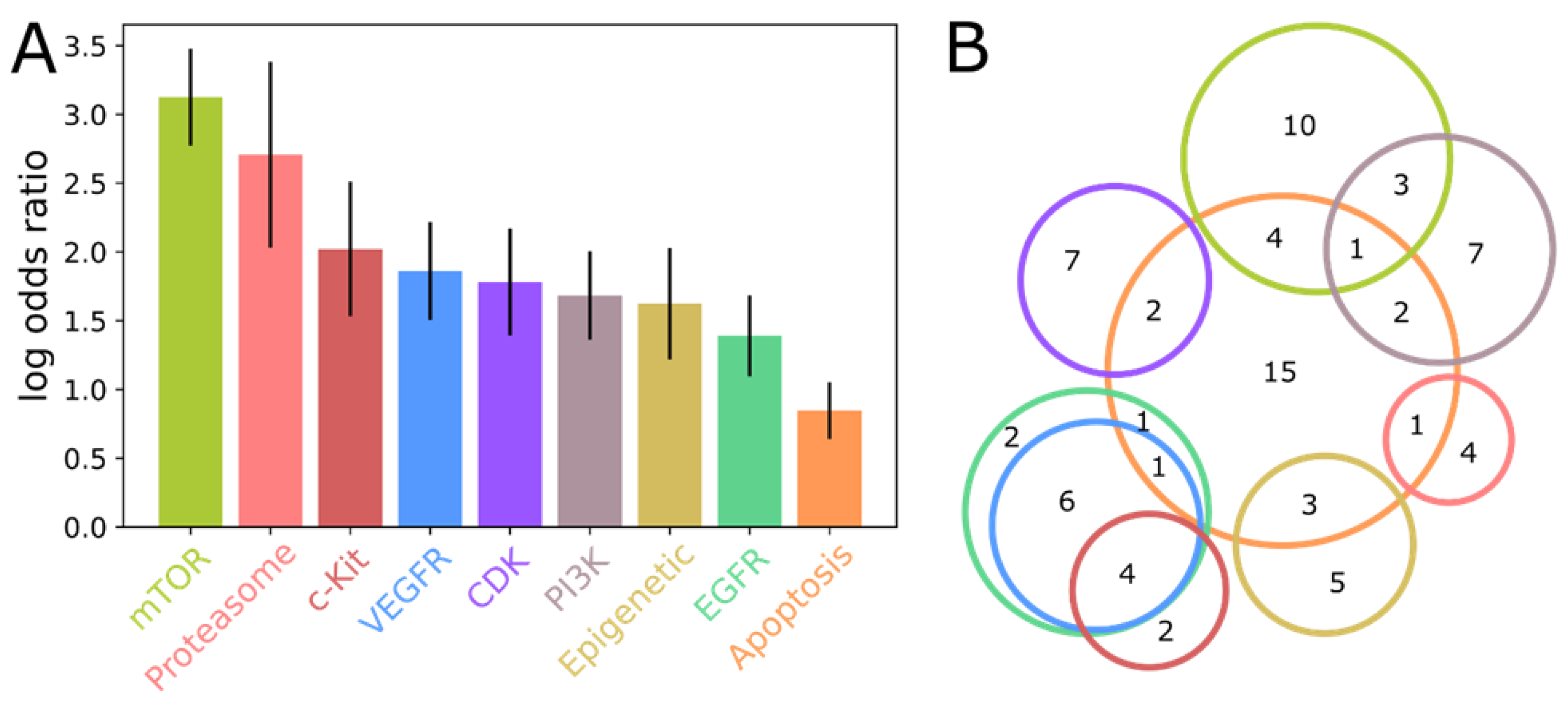
2.2. The CoviRx Database Reveals Host-Cell Pathways
2.3. Down-Selection of Drugs Based on Available Data and Regulatory Approval Status
2.4. Down-Selection of Drugs Based on Indicators of Efficacy and Safety
3. Discussion
4. Materials and Methods
4.1. Database Assembly, Data Collection and Processing
4.2. SARS-CoV-2 Assay Data Collection and Activity Rank Score Calculation
4.3. Filtration Methodology
4.4. Enrichment Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Director-General’s Opening Remarks at the Media Briefing on COVID-19. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 2 February 2022).
- Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 2 February 2022).
- The Pandemic’s True Death Toll. Available online: https://www.economist.com/graphic-detail/coronavirus-excess-deaths-estimates (accessed on 29 September 2022).
- Avila, J.; Long, B.; Holladay, D.; Gottlieb, M. Thrombotic Complications of COVID-19. Am. J. Emerg. Med. 2021, 39, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Talasaz, A.H.; Kakavand, H.; van Tassell, B.; Aghakouchakzadeh, M.; Sadeghipour, P.; Dunn, S.; Geraiely, B. Cardiovascular Complications of COVID-19: Pharmacotherapy Perspective. Cardiovasc. Drugs Ther. 2021, 35, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.D.; Ji, C.; Connolly, B.A.; Couper, K.; Lall, R.; Baillie, J.K.; Bradley, J.M.; Dark, P.; Dave, C.; de Soyza, A.; et al. Effect of Noninvasive Respiratory Strategies on Intubation or Mortality Among Patients with Acute Hypoxemic Respiratory Failure and COVID-19. JAMA 2022, 327, 546. [Google Scholar] [CrossRef]
- Sudre, C.H.; Murray, B.; Varsavsky, T.; Graham, M.S.; Penfold, R.S.; Bowyer, R.C.; Pujol, J.C.; Klaser, K.; Antonelli, M.; Canas, L.S.; et al. Attributes and Predictors of Long COVID. Nat. Med. 2021, 27, 626–631. [Google Scholar] [CrossRef]
- Pilkington, E.H.; Suys, E.J.A.; Trevaskis, N.L.; Wheatley, A.K.; Zukancic, D.; Algarni, A.; Al-Wassiti, H.; Davis, T.P.; Pouton, C.W.; Kent, S.J.; et al. From Influenza to COVID-19: Lipid Nanoparticle MRNA Vaccines at the Frontiers of Infectious Diseases. Acta Biomater. 2021, 131, 16–40. [Google Scholar] [CrossRef]
- Eroglu, B.; Nuwarda, R.F.; Ramzan, I.; Kayser, V. A Narrative Review of COVID-19 Vaccines. Vaccines 2021, 10, 62. [Google Scholar] [CrossRef]
- WHO. Coronavirus Disease (COVID-19): Vaccines. Available online: https://www.who.int/news-room/questions-and-answers/item/coronavirus-disease-(covid-19)-vaccines (accessed on 30 September 2022).
- Zheng, C.; Shao, W.; Chen, X.; Zhang, B.; Wang, G.; Zhang, W. Real-World Effectiveness of COVID-19 Vaccines: A Literature Review and Meta-Analysis. Int. J. Infect. Dis. 2022, 114, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, D. Paxlovid: What We Know about Pfizer’s COVID-19 Pill. Available online: https://www.pharmaceutical-technology.com/analysis/paxlovid-pfizer-covid-19-pill/ (accessed on 30 September 2022).
- Merck. Merck and Ridgeback Biotherapeutics Provide Update on Results from MOVe-OUT Study of Molnupiravir, an Investigational Oral Antiviral Medicine, in At Risk Adults with Mild-to-Moderate COVID-19. Available online: https://www.merck.com/news/merck-and-ridgeback-biotherapeutics-provide-update-on-results-from-move-out-study-of-molnupiravir-an-investigational-oral-antiviral-medicine-in-at-risk-adults-with-mild-to-moderate-covid-19/ (accessed on 7 February 2022).
- Zhou, S.; Hill, C.S.; Sarkar, S.; Tse, L.V.; Woodburn, B.M.D.; Schinazi, R.F.; Sheahan, T.P.; Baric, R.S.; Heise, M.T.; Swanstrom, R. β-D-N4-Hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis but is Also Mutagenic to Mammalian Cells. J. Infect. Dis. 2021, 224, 415–419. [Google Scholar] [CrossRef]
- NIH. The COVID-19 Treatment Guidelines Panel’s Statement on Potential Drug-Drug Interactions Between Ritonavir-Boosted Nirmatrelvir (Paxlovid) and Concomitant Medications. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/statement-on-paxlovid-drug-drug-interactions/ (accessed on 7 February 2022).
- NIH. Anti-SARS-CoV-2 Monoclonal Antibodies. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/anti-sars-cov-2-antibody-products/anti-sars-cov-2-monoclonal-antibodies/ (accessed on 7 February 2022).
- Tsirtsakis, A. RACGP—What is the Latest on COVID-19 Treatments Available in Australia? Available online: https://www1.racgp.org.au/newsgp/clinical/what-is-the-latest-on-covid-19-treatments-availabl (accessed on 30 September 2022).
- Robinson, B.W.S.; Tai, A.; Springer, K. Why We Still Need Drugs for COVID-19 and Can’t Just Rely on Vaccines. Respirology 2022, 27, 109–111. [Google Scholar] [CrossRef]
- NIH. COVID-19 Treatment Guidelines: Corticosteroids. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/immunomodulators/corticosteroids/ (accessed on 30 September 2022).
- Jain, H.A.; Agarwal, V.; Bansal, C.; Kumar, A.; Faheem, F.; Mohammed, M.-U.-R.; Murugesan, S.; Simpson, M.M.; Karpe, A.V.; Chandra, R.; et al. CoviRx: A User-Friendly Interface for Systematic Down-Selection of Repurposed Drug Candidates for COVID-19. 2022. Available online: https://www.preprints.org/manuscript/202209.0323/v1 (accessed on 30 September 2022).
- Simpson, M.; Poulsen, S.-A. An Overview of Australia’s Compound Management Facility: The Queensland Compound Library. ACS Chem. Biol. 2014, 9, 28–33. [Google Scholar] [CrossRef]
- Chen, C.Z.; Xu, M.; Pradhan, M.; Gorshkov, K.; Petersen, J.D.; Straus, M.R.; Zhu, W.; Shinn, P.; Guo, H.; Shen, M.; et al. Identifying SARS-CoV-2 Entry Inhibitors through Drug Repurposing Screens of SARS-S and MERS-S Pseudotyped Particles. ACS Pharmacol. Transl. Sci. 2020, 3, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Pache, L.; Burgstaller-Muehlbacher, S.; de Jesus, P.D.; Teriete, P.; Hull, M.V.; et al. Discovery of SARS-CoV-2 Antiviral Drugs through Large-Scale Compound Repurposing. Nature 2020, 586, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Bakowski, M.A.; Beutler, N.; Wolff, K.C.; Kirkpatrick, M.G.; Chen, E.; Nguyen, T.-T.H.; Riva, L.; Shaabani, N.; Parren, M.; Ricketts, J.; et al. Drug Repurposing Screens Identify Chemical Entities for the Development of COVID-19 Interventions. Nat. Commun. 2021, 12, 3309. [Google Scholar] [CrossRef] [PubMed]
- Touret, F.; Gilles, M.; Barral, K.; Nougairède, A.; van Helden, J.; Decroly, E.; de Lamballerie, X.; Coutard, B. In Vitro Screening of a FDA Approved Chemical Library Reveals Potential Inhibitors of SARS-CoV-2 Replication. Sci. Rep. 2020, 10, 13093. [Google Scholar] [CrossRef]
- Kuzikov, M.; Costanzi, E.; Reinshagen, J.; Esposito, F.; Vangeel, L.; Wolf, M.; Ellinger, B.; Claussen, C.; Geisslinger, G.; Corona, A.; et al. Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen. ACS Pharmacol. Transl. Sci. 2021, 4, 1096–1110. [Google Scholar] [CrossRef]
- Ellinger, B.; Zaliani, A.; Claussen, C.; Reinshagen, J.; Kuzikov, M.; Wolf, M.; Gribbon, P.; Ciesek, S. Identication of Inhibitors of SARS-CoV-2 in-Vitro Cellular Toxicity in Human (Caco-2) Cells Using a Large Scale Drug Repurposing Collection. 2020. Available online: https://www.researchsquare.com/article/rs-23951/v1 (accessed on 30 September 2022).
- Zaliani, A.; Vangeel, L.; Reinshagen, J.; Iaconis, D.; Kuzikov, M.; Keminer, O.; Wolf, M.; Ellinger, B.; Esposito, F.; Corona, A.; et al. Cytopathic SARS-CoV-2 Screening on VERO-E6 Cells in a Large-Scale Repurposing Effort. Sci. Data 2022, 9, 405. [Google Scholar] [CrossRef]
- Heiser, K.; Mclean, P.F.; Davis, C.T.; Fogelson, B.; Gordon, H.B.; Jacobson, P.; Hurst, B.; Miller, B.; Alfa, R.W.; Earnshaw, B.A.; et al. Identification of Potential Treatments for COVID-19 through Artificial Intelligence-Enabled Phenomic Analysis of Human Cells Infected with SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Selleckchem.Com. Available online: https://www.selleckchem.com/ (accessed on 7 February 2022).
- Microsource Discovery Systems. Available online: http://www.msdiscovery.com/spectrum.html (accessed on 7 February 2022).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 7 February 2022).
- DrugBank. Available online: https://go.drugbank.com/ (accessed on 7 February 2022).
- Drug Central 2021—Online Drug Compendium—Database Update. Available online: https://drugcentral.org/ (accessed on 7 February 2022).
- ChEMBL. Available online: https://www.ebi.ac.uk/chembl/ (accessed on 7 February 2022).
- ClinicalTrials.Gov. A Database of Privately and Publicly Funded Clinical Studies Conducted around the World. Available online: https://clinicaltrials.gov/ (accessed on 7 February 2022).
- International Clinical Trials Registery Platform. Available online: https://trialsearch.who.int/Default.aspx (accessed on 7 February 2022).
- Cule Repurposing. Available online: https://clue.io/repurposing-app (accessed on 7 February 2022).
- Binding DB. The Binding Database. Available online: https://www.bindingdb.org/bind/index.jsp (accessed on 7 February 2022).
- Drugs@FDA: FDA—Approved Drugs. Available online: https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm (accessed on 7 February 2022).
- ARTG Search. Available online: https://tga-search.clients.funnelback.com/s/search.html?query=&collection=tga-artg (accessed on 7 February 2022).
- Prescribing Medicines in Pregnancy Database. Available online: https://www.tga.gov.au/prescribing-medicines-pregnancy-database (accessed on 7 February 2022).
- (LactMed)—National Library of Medicine (US). Drugs and Lactation Database. Available online: https://www.ncbi.nlm.nih.gov/books/NBK501922/ (accessed on 7 February 2022).
- Medscape. Available online: https://reference.medscape.com/ (accessed on 7 February 2022).
- KEGG Database. Available online: https://www.kegg.jp/kegg/ (accessed on 7 February 2022).
- ATC Index. Available online: https://www.whocc.no/atc_ddd_index/ (accessed on 7 February 2022).
- WHO Solidarity Trial Consortium. Repurposed Antiviral Drugs for COVID-19—Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021, 384, 497–511. [Google Scholar] [CrossRef]
- Martinez, M.A. Lack of Effectiveness of Repurposed Drugs for COVID-19 Treatment. Front. Immunol. 2021, 12, 653. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Baker, E.H.; Gnjidic, D.; Kirkpatrick, C.M.J.; Pirmohamed, M.; Wright, D.F.B.; Zecharia, A.Y. A Call for the Appropriate Application of Clinical Pharmacological Principles in the Search for Safe and Efficacious COVID-19 (SARS-CoV-2) Treatments. Br. J. Clin. Pharmacol. 2021, 87, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Tummino, T.A.; Rezelj, V.V.; Fischer, B.; Fischer, A.; O’Meara, M.J.; Monel, B.; Vallet, T.; White, K.M.; Zhang, Z.; Alon, A.; et al. Drug-Induced Phospholipidosis Confounds Drug Repurposing for SARS-CoV-2. Science 2021, 373, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, J.L.; Auld, D.S.; Rothenaigner, I.; Haney, S.; Sexton, J.Z.; Nissink, J.W.M.; Walsh, J.; Lee, J.A.; Strelow, J.M.; Willard, F.S.; et al. Nuisance Compounds in Cellular Assays. Cell Chem. Biol. 2021, 28, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [Green Version]
- FDA. Why You Should Not Use Ivermectin to Treat or Prevent COVID-19. Available online: https://www.fda.gov/consumers/consumer-updates/why-you-should-not-use-ivermectin-treat-or-prevent-covid-19 (accessed on 7 February 2022).
- Matt Woodley TGA Issues Fresh Warning over Ivermectin as COVID Treatment. Available online: https://www1.racgp.org.au/newsgp/clinical/tga-issues-warning-over-ivermectin-as-covid-treatm (accessed on 7 February 2022).
- Bray, M.; Rayner, C.; Noël, F.; Jans, D.; Wagstaff, K. Ivermectin and COVID-19: A Report in Antiviral Research, Widespread Interest, an FDA Warning, Two Letters to the Editor and the Authors’ Responses. Antivir. Res. 2020, 178, 104805. [Google Scholar] [CrossRef]
- Popp, M.; Stegemann, M.; Metzendorf, M.-I.; Gould, S.; Kranke, P.; Meybohm, P.; Skoetz, N.; Weibel, S. Ivermectin for Preventing and Treating COVID-19. Cochrane Database Syst. Rev. 2021, 2021, CD015017. [Google Scholar] [CrossRef]
- Pulakurthi, Y.S.; Pederson, J.M.; Saravu, K.; Gupta, N.; Balasubramanian, P.; Kamrowski, S.; Schmidt, M.; Vegivinti, C.T.R.; Dibas, M.; Reierson, N.L.; et al. Corticosteroid Therapy for COVID-19. Medicine 2021, 100, e25719. [Google Scholar] [CrossRef]
- NIH. Table: Systemic Corticosteroids Clinical Data|COVID-19 Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/tables/systemic-corticosteroids-data/ (accessed on 30 September 2022).
- Dayer, M.R. Old Drugs for Newly Emerging Viral Disease, COVID-19: Bioinformatic Prospective. arXiv 2020, arXiv:2003.04524. [Google Scholar]
- Alonzi, T.; Aiello, A.; Petrone, L.; Najafi Fard, S.; D’Eletto, M.; Falasca, L.; Nardacci, R.; Rossin, F.; Delogu, G.; Castilletti, C.; et al. Cysteamine with In Vitro Antiviral Activity and Immunomodulatory Effects Has the Potential to Be a Repurposing Drug Candidate for COVID-19 Therapy. Cells 2021, 11, 52. [Google Scholar] [CrossRef]
- Thoene, J.; Gavin, R.F.; Towne, A.; Wattay, L.; Ferrari, M.G.; Pal, R. In Vitro Activity of Cysteamine Against SARS-CoV-2 Variants. bioRxiv 2022. [Google Scholar] [CrossRef]
- McAuley, A.J.; van Vuren, P.J.; Mohammed, M.-U.-R.; Faheem, F.; Goldie, S.; Riddell, S.; Gödde, N.J.; Styles, I.K.; Bruce, M.P.; Chahal, S.; et al. Use of Human Lung Tissue Models for Screening of Drugs Against SARS-CoV-2 Infection. 2022. Available online: https://www.preprints.org/manuscript/202209.0288/v1 (accessed on 30 September 2022).
- Bischof, E.; Siow, R.C.; Zhavoronkov, A.; Kaeberlein, M. The Potential of Rapalogs to Enhance Resilience against SARS-CoV-2 Infection and Reduce the Severity of COVID-19. Lancet Healthy Longev. 2021, 2, e105–e111. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-Based Drug Repurposing for Novel Coronavirus 2019-NCoV/SARS-CoV-2. Cell Discov. 2020, 6, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colmenero, J.; Rodríguez-Perálvarez, M.; Salcedo, M.; Arias-Milla, A.; Muñoz-Serrano, A.; Graus, J.; Nuño, J.; Gastaca, M.; Bustamante-Schneider, J.; Cachero, A.; et al. Epidemiological Pattern, Incidence, and Outcomes of COVID-19 in Liver Transplant Patients. J. Hepatol. 2021, 74, 148–155. [Google Scholar] [CrossRef]
- Terrazzano, G.; Rubino, V.; Palatucci, A.T.; Giovazzino, A.; Carriero, F.; Ruggiero, G. An Open Question: Is It Rational to Inhibit the MTor-Dependent Pathway as COVID-19 Therapy? Front. Pharmacol. 2020, 11, 856. [Google Scholar] [CrossRef] [PubMed]
- Heron, V.C.; Bach, C.-A.T.; Holmes, N.E.; Whitlam, J.B. Complete Recovery from COVID-19 of a Kidney-Pancreas Transplant Recipient: Potential Benefit from Everolimus? BMJ Case Rep. 2021, 14, e238413. [Google Scholar] [CrossRef] [PubMed]
- Mehboob, R.; Lavezzi, A.M. Neuropathological Explanation of Minimal COVID-19 Infection Rate in Newborns, Infants and Children—A Mystery so Far. New Insight into the Role of Substance P. J. Neurol. Sci. 2021, 420, 117276. [Google Scholar] [CrossRef]
- Mehboob, R. Neurokinin-1 Receptor as a Potential Drug Target for COVID-19 Treatment. Biomed. Pharmacother. 2021, 143, 112159. [Google Scholar] [CrossRef] [PubMed]
- Mehboob, R.; Kurdi, M.; Bamaga, A.; Aldardeir, N.; Nasief, H.; Moshref, L.H.; Alsinani, T.; Rayes, A.O.; Jabbad, R.H. Substance P/Neurokinin-1 Receptor, Trigeminal Ganglion, Latency, and Coronavirus Infection-Is There Any Link? Front. Med. 2021, 8, 727593. [Google Scholar] [CrossRef]
- Reinoso-Arija, R.; López-Ramírez, C.; Jimenez-Ruiz, J.A.; López-Campos, J.L. Effectiveness of Aprepitant in Post-acute COVID-19 Syndrome. Clin. Case Rep. 2021, 9, e04646. [Google Scholar] [CrossRef]
- Dodds, M.; Xiong, Y.; Mouksassi, S.; Kirkpatrick, C.M.; Hui, K.; Doyle, E.; Patel, K.; Cox, E.; Wesche, D.; Brown, F.; et al. Model-informed Drug Repurposing: A Pharmacometric Approach to Novel Pathogen Preparedness, Response and Retrospection. Br. J. Clin. Pharmacol. 2021, 87, 3388–3397. [Google Scholar] [CrossRef]
- COVID-19 Pharmacology Resource Centre. In Silico Workbench. Available online: https://www.covidpharmacology.com/in-silico-workbench/ (accessed on 7 February 2022).
- RDKit. Open-Source Cheminformatics. Available online: https://www.rdkit.org/ (accessed on 7 February 2022).
- Jasial, S.; Hu, Y.; Bajorath, J. How Frequently Are Pan-Assay Interference Compounds Active? Large-Scale Analysis of Screening Data Reveals Diverse Activity Profiles, Low Global Hit Frequency, and Many Consistently Inactive Compounds. J. Med. Chem. 2017, 60, 3879–3886. [Google Scholar] [CrossRef] [PubMed]
- Hunter, F.M.I.; Bento, A.P.; Bosc, N.; Gaulton, A.; Hersey, A.; Leach, A.R. Drug Safety Data Curation and Modeling in ChEMBL: Boxed Warnings and Withdrawn Drugs. Chem. Res. Toxicol. 2021, 34, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Mangalaganesh, S.; Wilson, L.O.W.; Kuiper, M.J.; Drew, T.W.; Vasan, S.S. Tracking Co-Occurrence of N501Y, P681R, and Other Key Mutations in SARS-CoV-2 Spike for Surveillance. Zoonotic Dis. 2022, 2, 147–162. [Google Scholar] [CrossRef]


| Database | Purpose |
|---|---|
| Selleckchem epigenetics, Selleck Kinase inhibitors [30] and, MicroSource spectrum libraries [31] | Extraction of drugs for curation |
| PubChem [32] Drug bank [33], Drug central [34], ChEMBL [35] | Drug identifiers; PK-PD parameters |
| Clinicaltrials.gov (accessed on 7 February 2022) [36]; WHO clinical trials [37] | Clinical trial status |
| Drug repurposing hub [38] | Target and mechanism of action |
| Binding DB [39] | IC50 values for original indication |
| Drugs@FDA [40] | FDA approval status |
| Australian Register of Therapeutic goods, Prescribing Medicines in Pregnancy Database [41,42] | TGA approval status; pregnancy category |
| Drugs and lactation database, Australian Register of Therapeutic Goods, Medscape [43,44] | Breastfeeding data |
| KEGG database [45] and ATC index [46] | Target, category, and indication |
| Name | Rank Score | Indication | Mechanism of Action | Target |
|---|---|---|---|---|
| L-cycloserine | 0.0098765 | Anti-bacterial (tuberculostatic) | - | - |
| Tolterodine (tartrate) | 0.0271605 | Urinary anti-spasmodics, Overactive bladder agent | Acetylcholine receptor antagonist (anticholinergic) | CHRM1, CHRM2, CHRM3, CHRM4, CHRM5 |
| Moxidectin | 0.0641926 | Anti-parasitic | Chloride channel antagonist | |
| Pyrimethamine | 0.0989685 | Anti-malarial | Dihydrofolate reductase inhibitor | DHFR, SLC47A1 |
| Meclizine hydrochloride | 0.1115153 | Anti-emetic | Constitutive androstane receptor (CAR) agonist, Histamine receptor antagonist (antihistamine) | NR1I3 |
| Cysteamine hydrochloride | 0.1283852 | Anti-urolithic | Tissue transglutaminase inhibitor | NPY2R, SST |
| Deflazacort | 0.153934 | Anti-inflammatory | Glucocorticoid receptor agonist | NR3C1 |
| Nifurtimox | 0.1808927 | Antiprotozoal | DNA inhibitor | HSPD1 |
| Cefaclor | 0.1863393 | Antibacterial | Bacterial cell wall synthesis inhibitor | |
| Mianserin hydrochloride | 0.1925777 | Antidepressant | Serotonin receptor antagonist | ADRA1A, ADRA1B, ADRA1D, HRH1, HRH2, HTR2A, HTR2B, HTR2C, HTR6, HTR7 |
| Procyclidine hydrochloride | 0.2006018 | Antiparkinsonian, Skeletal muscle relaxant | Acetylcholine receptor antagonist (anticholinergic) | |
| Palonosetron (hydrochloride) | 0.2313175 | Anti-emetic | Serotonin receptor antagonist | HTR3A |
| Gefitinib (ZD1839) | 0.2314783 | Anti-cancer | Epidermal Growth Factor Receptor (EGFR) inhibitor | EGFR |
| Dapoxetine | 0.2358875 | Antidepressant | Selective serotonin reuptake inhibitor (SSRI) | HTR1A, HTR1B, HTR2C, SLC6A4 |
| Rilpivirine | 0.2394370 | Antiviral | Non-nucleoside reverse transcriptase inhibitor (NNRTI) | NR1I2, SCN10A |
| Name | Initial Filter Failed | Rank Score |
|---|---|---|
| Quinidine Hydrochloride monohydrate | CAD, Toxicity | 0.0111111 |
| Everolimus | Toxicity, Same class of drug is in clinical trials | 0.0140421 |
| Trihexyphenidyl Hydrochloride | COVID IC50 > 10(*) original IC50 | 0.0170512 |
| Sorafenib tosylate | CC50 < 10, SI < 10, pregnancy | 0.0180246 |
| Rolapitant | SI < 10 | 0.018665 |
| Idarubicin (Hydrochloride) | CC50 < IC50, Pregnancy, PAINS, Side effects | 0.0342517 |
| Regorafenib (BAY 73-4506) | CC50 < 10, SI < 10, pregnancy, Side effects | 0.0371304 |
| Itraconazole Hydrochloride | CC50 < 10; Same class of drug is in clinical trials, PAINS | 0.0411262 |
| Prasterone | Same class of drug in clinical trials | 0.0703704 |
| Gemcitabine Hydrochloride | ROA, Pregnancy, Toxicity | 0.0712136 |
| Pimecrolimus | ROA, Same class of drug is in clinical trials, Toxicity | 0.0742227 |
| Doxepin (Hydrochloride) | Side effects | 0.0790123 |
| Abiraterone acetate | COVID IC50 > 10(*) original IC50, Pregnancy | 0.0868782 |
| Cabozantinib (XL184_ BMS-907351) | COVID IC50 > 10(*) original IC50, Pregnancy, Toxicity | 0.0929004 |
| Raloxifene Hydrochloride | CAD, Pregnancy, Toxicity | 0.0973509 |
| Avobenzone | Indication | 0.1024691 |
| Vinblastine (sulfate) | Pregnancy | 0.1109676 |
| Cobimetinib (racemate) | Pregnancy | 0.1164189 |
| Zotarolimus | Same class of drug is in clinical trials | 0.1578381 |
| Pexidartinib | SI < 10, Side effects | 0.1586386 |
| Digoxin | CC50 < 10 | 0.1666667 |
| Thioguanine | CC50 < 10, SI < 10 | 0.1722083 |
| Mebrofenin | Indication | 0.1798481 |
| Lenvatinib (E7080) | Pregnancy | 0.1801718 |
| Piperonyl butoxide | ROA, Indication | 0.181471 |
| Nortriptyline Hydrochloride | PAINS, Side effects | 0.1830491 |
| Letermovir | COVID IC50 > 10(*) original IC50 | 0.1851185 |
| Thiothixene | PAINS, Side effects | 0.1858974 |
| Drug Name | Cmax (µM) | Protein Binding (%) | Adjusted Cmax Based on Protein Binding (µM) |
|---|---|---|---|
| L-cycloserine | 830 | “No protein binding” | 830 |
| Pyrimethamine | 0.94 | 87 | 0.1222 |
| Ondansetron | 0.43–0.66 | 73 | 0.12–0.18 |
| Cyclizine | 0.26 | 60–76 in rats | 0.0624 |
| Everolimus | 0.186 | 74 | 0.04836 |
| Lapatinib | 4.18 | >99 | 0.0418 |
| Cetirizine | 0.8 | 93–96 | 0.032 |
| Rolapitant | 1.9 | 99.8 | 0.0038 |
| Gefitinib | 0.19 | 90–97 | 0.0095 |
| Mianserin | 0.15 | 95 | 0.0075 |
| Palonosetron | 0.019 | 62 | 0.00722 |
| Meclizine | 0.02 | 99 | 0.0002 |
| Filter Type | Description | Objective |
|---|---|---|
| Clinical trials (A) | Clinicaltrials.gov database was used to search the drugs under clinical evaluation against SARS-CoV-2 infection. Simultaneously, the Tanimoto index was used to look for the analogues of drugs under clinical trials against SARS-CoV-2 and these were also excluded from our study as a similar scaffold produces similar action. | To prevent duplication of existing work. |
| CC50 < 10 µM Or SI < 10 (B) | Compounds with CC50 value < 10µM were considered toxic, while those with >10 µM were deemed non-toxic. Hence, the drugs with CC50 values below 10µM were filtered out. In addition, the selectivity index (SI) was also determined, and SI < 10 was considered the minimum acceptable efficacy. Selectivity index (SI) = CC50/IC50 | To filter out cytotoxic drugs. |
| COVID-19 IC50 > 10(*) Original Indication (C) | The drug that has ten times more than IC50 of original indication are usually toxic, as high doses are needed to show an inhibitory effect. | To filter out drugs that have poor IC50 values. |
| CAD/PAINS (D) | We removed cationic amphiphilic drugs (CAD) that exhibit antiviral activity by inducing phospholipidosis rather than interacting with a specific target. We also removed compound classes that cause pan-assay interference (PAINS) [76]. | To screen out false-positive results. |
| Route of administration (E) | Drugs that are deliverable by oral or inhalation routes were considered in our study, as other routes of administration would limit applicability for the treatment of SARS-CoV-2 infection. Hence, oral and inhalation drugs were retained, and the rest were filtered out. | To filter out drugs that are not orally bioavailable. |
| Pregnancy (F) | Pregnant women with SARS-CoV-2 infection have been a subject of concern as the present drugs approved for COVID-19 cannot be used to treat them. Hence, drug pregnancy categories were found from the ARTG database and category D and X drugs were removed. | To remove drugs unsafe for use in pregnancy. |
| Black box warning (G) | Black box warning refers to serious side effects [77] | Filter out drugs with black box warnings to obtain drugs that are safe to use. |
| Indication (H) | Compounds that have no pharmacological action are also in the database. Hence, all the pharmaceutical aids, diagnostic agents, and supplements were filtered out. | To retain pharmacologically active drugs. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
MacRaild, C.A.; Mohammed, M.-U.-R.; Faheem; Murugesan, S.; Styles, I.K.; Peterson, A.L.; Kirkpatrick, C.M.J.; Cooper, M.A.; Palombo, E.A.; Simpson, M.M.; et al. Systematic Down-Selection of Repurposed Drug Candidates for COVID-19. Int. J. Mol. Sci. 2022, 23, 11851. https://doi.org/10.3390/ijms231911851
MacRaild CA, Mohammed M-U-R, Faheem, Murugesan S, Styles IK, Peterson AL, Kirkpatrick CMJ, Cooper MA, Palombo EA, Simpson MM, et al. Systematic Down-Selection of Repurposed Drug Candidates for COVID-19. International Journal of Molecular Sciences. 2022; 23(19):11851. https://doi.org/10.3390/ijms231911851
Chicago/Turabian StyleMacRaild, Christopher A., Muzaffar-Ur-Rehman Mohammed, Faheem, Sankaranarayanan Murugesan, Ian K. Styles, Amanda L. Peterson, Carl M. J. Kirkpatrick, Matthew A. Cooper, Enzo A. Palombo, Moana M. Simpson, and et al. 2022. "Systematic Down-Selection of Repurposed Drug Candidates for COVID-19" International Journal of Molecular Sciences 23, no. 19: 11851. https://doi.org/10.3390/ijms231911851
APA StyleMacRaild, C. A., Mohammed, M.-U.-R., Faheem, Murugesan, S., Styles, I. K., Peterson, A. L., Kirkpatrick, C. M. J., Cooper, M. A., Palombo, E. A., Simpson, M. M., Jain, H. A., Agarwal, V., McAuley, A. J., Kumar, A., Creek, D. J., Trevaskis, N. L., & Vasan, S. S. (2022). Systematic Down-Selection of Repurposed Drug Candidates for COVID-19. International Journal of Molecular Sciences, 23(19), 11851. https://doi.org/10.3390/ijms231911851







