Transcriptome Profiling of the Hippocampal Seizure Network Implicates a Role for Wnt Signaling during Epileptogenesis in a Mouse Model of Temporal Lobe Epilepsy
Abstract
1. Introduction
2. Results
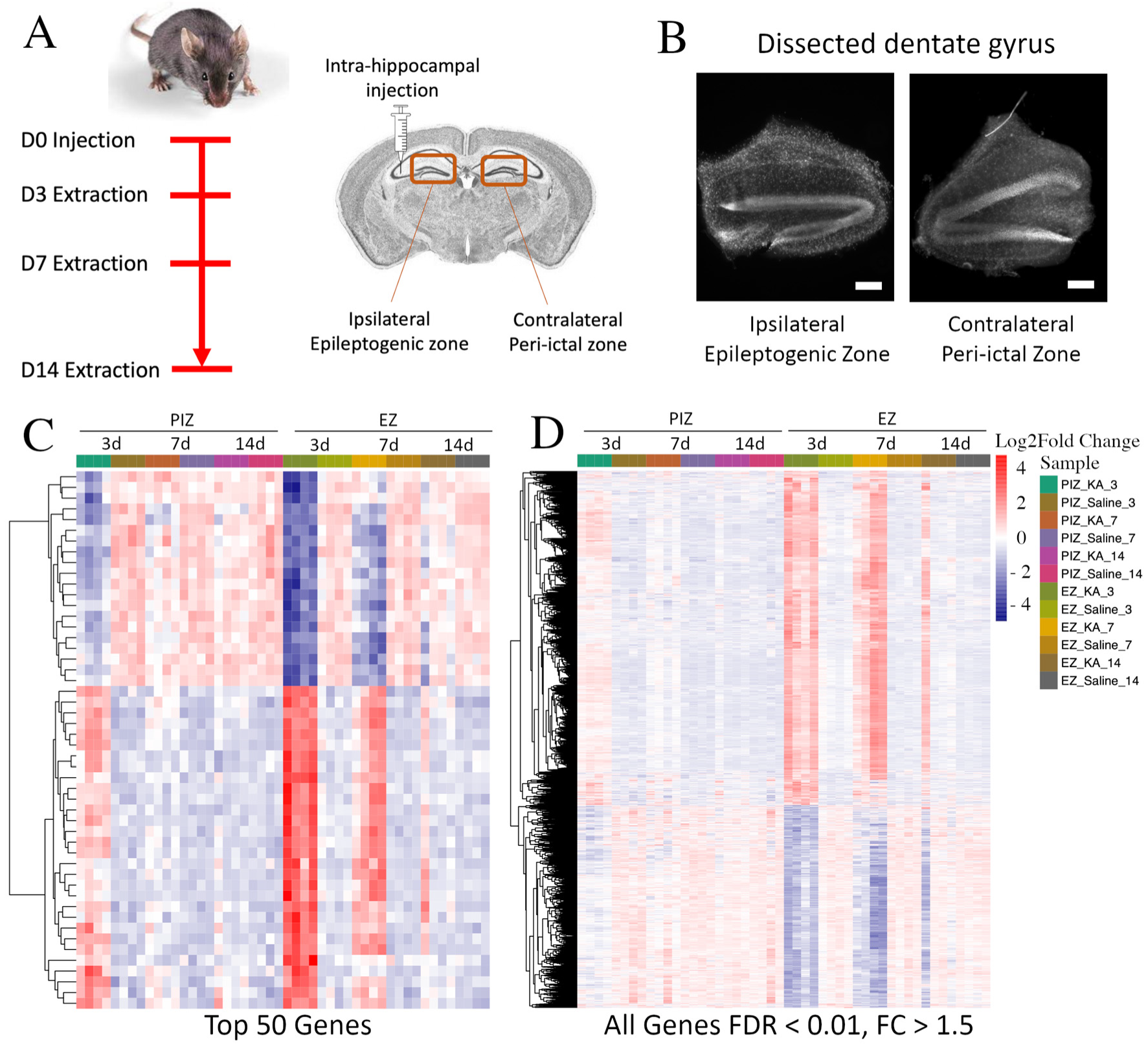
2.1. Mesial Temporal Lobe Epilepsy Model and Experimental Design
2.2. Transcriptomic Dysregulation across the Seizure Network in Early Epileptogenesis
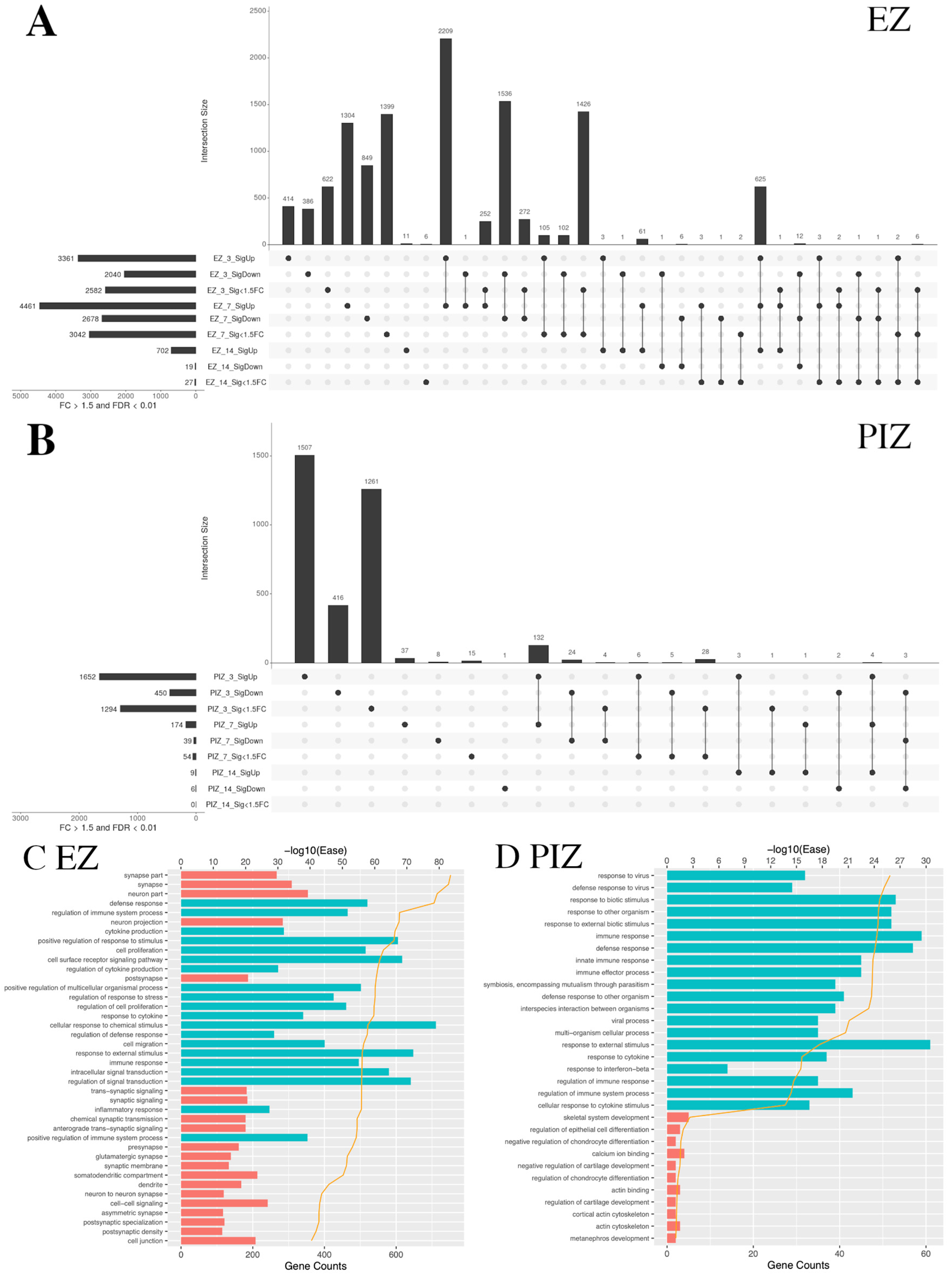
2.3. Shared Patterns of Gene Dysregulation between the Epileptogenic Zone and Peri-Ictal Zone across Early Epileptogenesis
2.4. Enrichment of Functional DAVID Categories in the Epileptogenic Zone and Seizure Network across Early Epileptogenesis
2.5. Wnt Pathway Gene Dysregulation during Early Epileptogenesis
2.6. Activated-Beta-Catenin Was Upregulated Early after SE Induction in the Epileptogenic and Peri-Ictal Zones
3. Discussion
3.1. Transcriptomic Data Implicated a Role for Wnt Pathway Dysregulation in the Seizure Network in Early Epileptogenesis
3.2. Study Limitations
4. Materials and Methods
4.1. Animal Husbandry and Experimental Design
4.2. Intrahippocampal Kainate-Induced Status Epilepticus
4.3. RNA Extraction and Library Preparation
4.4. RNA Sequencing and Bioinformatic Analysis
4.5. Immuno-Histochemistry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kobau, R.; Zahran, H.; Thurman, D.J.; Zack, M.M.; Henry, T.R.; Schachter, S.C.; Price, P.H.; Centers for Disease and Control Prevention. Epilepsy surveillance among adults—19 States, Behavioral Risk Factor Surveillance System, 2005. MMWR Surveill. Summ. 2008, 57, 1–20. [Google Scholar] [PubMed]
- Kwan, P.; Brodie, M.J. Early identification of refractory epilepsy. N. Engl. J. Med. 2000, 342, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Kwan, P.; Sander, J.W. The natural history of epilepsy: An epidemiological view. J. Neurol. Neurosurg. Psychiatry 2004, 75, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
- Picot, M.C.; Baldy-Moulinier, M.; Daures, J.P.; Dujols, P.; Crespel, A. The prevalence of epilepsy and pharmacoresistant epilepsy in adults: A population-based study in a Western European country. Epilepsia 2008, 49, 1230–1238. [Google Scholar] [CrossRef]
- Chen, D.Y.; Chen, C.C.; Crawford, J.R.; Wang, S.G. Tumor-related epilepsy: Epidemiology, pathogenesis and management. J. Neurooncol. 2018, 139, 13–21. [Google Scholar] [CrossRef] [PubMed]
- French, J.A. Febrile seizures: Possible outcomes. Neurology 2012, 79, e80–e82. [Google Scholar] [CrossRef]
- Frey, L.C. Epidemiology of posttraumatic epilepsy: A critical review. Epilepsia 2003, 44, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Myint, P.K.; Staufenberg, E.F.; Sabanathan, K. Post-stroke seizure and post-stroke epilepsy. Postgrad. Med. J. 2006, 82, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Ramantani, G.; Holthausen, H. Epilepsy after cerebral infection: Review of the literature and the potential for surgery. Epileptic Disord. Int. Epilepsy J. Videotape 2017, 19, 117–136. [Google Scholar] [CrossRef]
- Cho, K.-O.O.; Lybrand, Z.R.; Ito, N.; Brulet, R.; Tafacory, F.; Zhang, L.; Good, L.; Ure, K.; Kernie, S.G.; Birnbaum, S.G.; et al. Aberrant hippocampal neurogenesis contributes to epilepsy and associated cognitive decline. Nat. Commun. 2015, 6, 6606. [Google Scholar] [CrossRef] [PubMed]
- Kralic, J.E.; Ledergerber, D.A.; Fritschy, J.-M.M. Disruption of the neurogenic potential of the dentate gyrus in a mouse model of temporal lobe epilepsy with focal seizures. Eur. J. Neurosci. 2005, 22, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Kron, M.M.; Zhang, H.; Parent, J.M. The developmental stage of dentate granule cells dictates their contribution to seizure-induced plasticity. J. Neurosci. 2010, 30, 2051–2059. [Google Scholar] [CrossRef] [PubMed]
- Parent, J.M.; Yu, T.W.; Leibowitz, R.T.; Geschwind, D.H.; Sloviter, R.S.; Lowenstein, D.H. Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus. J. Neurosci. 1997, 17, 3727–3738. [Google Scholar] [CrossRef] [PubMed]
- Twele, F.; Schidlitzki, A.; Tollner, K.; Loscher, W. The intrahippocampal kainate mouse model of mesial temporal lobe epilepsy: Lack of electrographic seizure-like events in sham controls. Epilepsia Open 2017, 2, 180–187. [Google Scholar] [CrossRef]
- Twele, F.; Tollner, K.; Bankstahl, M.; Loscher, W. The effects of carbamazepine in the intrahippocampal kainate model of temporal lobe epilepsy depend on seizure definition and mouse strain. Epilepsia Open 2016, 1, 45–60. [Google Scholar] [CrossRef]
- Twele, F.; Tollner, K.; Brandt, C.; Loscher, W. Significant effects of sex, strain, and anesthesia in the intrahippocampal kainate mouse model of mesial temporal lobe epilepsy. Epilepsy Behav. 2016, 55, 47–56. [Google Scholar] [CrossRef]
- Nadler, J.V.; Spencer, D.D. What is a seizure focus? Adv. Exp. Med. Biol. 2014, 813, 55–62. [Google Scholar] [CrossRef]
- Sheybani, L.; Birot, G.; Contestabile, A.; Seeck, M.; Kiss, J.Z.; Schaller, K.; Michel, C.M.; Quairiaux, C. Electrophysiological Evidence for the Development of a Self-Sustained Large-Scale Epileptic Network in the Kainate Mouse Model of Temporal Lobe Epilepsy. J. Neurosci. 2018, 38, 3776–3791. [Google Scholar] [CrossRef]
- Wendling, F.; Chauvel, P.; Biraben, A.; Bartolomei, F. From intracerebral EEG signals to brain connectivity: Identification of epileptogenic networks in partial epilepsy. Front. Syst. Neurosci. 2010, 4, 154. [Google Scholar] [CrossRef]
- Barba, C.; Rheims, S.; Minotti, L.; Guenot, M.; Hoffmann, D.; Chabardes, S.; Isnard, J.; Kahane, P.; Ryvlin, P. Temporal plus epilepsy is a major determinant of temporal lobe surgery failures. Brain 2016, 139, 444–451. [Google Scholar] [CrossRef]
- McIntosh, A.M.; Kalnins, R.M.; Mitchell, L.A.; Fabinyi, G.C.; Briellmann, R.S.; Berkovic, S.F. Temporal lobectomy: Long-term seizure outcome, late recurrence and risks for seizure recurrence. Brain 2004, 127, 2018–2030. [Google Scholar] [CrossRef] [PubMed]
- Canto, A.M.; Matos, A.H.B.; Godoi, A.B.; Vieira, A.S.; Aoyama, B.B.; Rocha, C.S.; Henning, B.; Carvalho, B.S.; Pascoal, V.D.B.; Veiga, D.F.T.; et al. Multi-omics analysis suggests enhanced epileptogenesis in the Cornu Ammonis 3 of the pilocarpine model of mesial temporal lobe epilepsy. Hippocampus 2021, 31, 122–139. [Google Scholar] [CrossRef]
- Dingledine, R.; Coulter, D.A.; Fritsch, B.; Gorter, J.A.; Lelutiu, N.; McNamara, J.; Nadler, J.V.; Pitkänen, A.; Rogawski, M.A.; Skene, P.; et al. Transcriptional profile of hippocampal dentate granule cells in four rat epilepsy models. Sci. Data 2017, 4, 170061. [Google Scholar] [CrossRef] [PubMed]
- Dixit, A.B.; Banerjee, J.; Srivastava, A.; Tripathi, M.; Sarkar, C.; Kakkar, A.; Jain, M.; Chandra, P.S. RNA-seq analysis of hippocampal tissues reveals novel candidate genes for drug refractory epilepsy in patients with MTLE-HS. Genomics 2016, 107, 178–188. [Google Scholar] [CrossRef]
- Hansen, K.F.; Sakamoto, K.; Pelz, C.; Impey, S.; Obrietan, K. Profiling status epilepticus-induced changes in hippocampal RNA expression using high-throughput RNA sequencing. Sci. Rep. 2014, 4, 6930. [Google Scholar] [CrossRef]
- Li, A.; Choi, Y.S.; Dziema, H.; Cao, R.; Cho, H.Y.; Jung, Y.J.; Obrietan, K. Proteomic profiling of the epileptic dentate gyrus. Brain Pathol. 2010, 20, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Theilhaber, J.; Rakhade, S.N.; Sudhalter, J.; Kothari, N.; Klein, P.; Pollard, J.; Jensen, F.E. Gene expression profiling of a hypoxic seizure model of epilepsy suggests a role for mTOR and Wnt signaling in epileptogenesis. PLoS ONE 2013, 8, e74428. [Google Scholar] [CrossRef]
- Berger, T.C.; Vigeland, M.D.; Hjorthaug, H.S.; Nome, C.G.; Tauboll, E.; Selmer, K.K.; Heuser, K. Differential Glial Activation in Early Epileptogenesis-Insights From Cell-Specific Analysis of DNA Methylation and Gene Expression in the Contralateral Hippocampus. Front. Neurol. 2020, 11, 573575. [Google Scholar] [CrossRef]
- Alqurashi, R.S.; Yee, A.S.; Malone, T.; Alrubiaan, S.; Tam, M.W.; Wang, K.; Nandedwalla, R.R.; Field, W.; Alkhelb, D.; Given, K.S.; et al. A Warburg-like metabolic program coordinates Wnt, AMPK, and mTOR signaling pathways in epileptogenesis. PLoS ONE 2021, 16, e0252282. [Google Scholar] [CrossRef]
- Gupta, K.; Schnell, E. Neuronal network remodeling and Wnt pathway dysregulation in the intra-hippocampal kainate mouse model of temporal lobe epilepsy. PLoS ONE 2019, 14, e0215789. [Google Scholar] [CrossRef]
- Jean, W.H.; Huang, C.T.; Hsu, J.H.; Chiu, K.M.; Lee, M.Y.; Shieh, J.S.; Lin, T.Y.; Wang, S.J. Anticonvulsive and Neuroprotective Effects of Eupafolin in Rats Are Associated with the Inhibition of Glutamate Overexcitation and Upregulation of the Wnt/beta-Catenin Signaling Pathway. ACS Chem. Neurosci. 2022, 13, 1594–1603. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Fu, J.; Qu, Z.; Jia, L.; Li, D.; Zhen, J.; Wang, W. Chronic Intermittent Hypobaric Hypoxia Restores Hippocampus Function and Rescues Cognitive Impairments in Chronic Epileptic Rats via Wnt/beta-catenin Signaling. Front. Mol. Neurosci. 2020, 13, 617143. [Google Scholar] [CrossRef] [PubMed]
- Oliva, C.A.; Vargas, J.Y.; Inestrosa, N.C. Wnts in adult brain: From synaptic plasticity to cognitive deficiencies. Front. Cell. Neurosci. 2013, 7, 224. [Google Scholar] [CrossRef] [PubMed]
- Arredondo, S.B.; Guerrero, F.G.; Herrera-Soto, A.; Jensen-Flores, J.; Bustamante, D.B.; Onate-Ponce, A.; Henny, P.; Varas-Godoy, M.; Inestrosa, N.C.; Varela-Nallar, L. Wnt5a promotes differentiation and development of adult-born neurons in the hippocampus by noncanonical Wnt signaling. Stem Cells 2020, 38, 422–436. [Google Scholar] [CrossRef] [PubMed]
- Choe, Y.; Pleasure, S.J. Wnt signaling regulates intermediate precursor production in the postnatal dentate gyrus by regulating CXCR4 expression. Dev. Neurosci. 2012, 34, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Arlotta, P.; Macklis, J.D.; Chen, J. Conditional knock-out of beta-catenin in postnatal-born dentate gyrus granule neurons results in dendritic malformation. J. Neurosci. 2007, 27, 14317–14325. [Google Scholar] [CrossRef] [PubMed]
- Lie, D.C.; Colamarino, S.A.; Song, H.J.; Desire, L.; Mira, H.; Consiglio, A.; Lein, E.S.; Jessberger, S.; Lansford, H.; Dearie, A.R.; et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature 2005, 437, 1370–1375. [Google Scholar] [CrossRef]
- Mardones, M.D.; Andaur, G.A.; Varas-Godoy, M.; Henriquez, J.F.; Salech, F.; Behrens, M.I.; Couve, A.; Inestrosa, N.C.; Varela-Nallar, L. Frizzled-1 receptor regulates adult hippocampal neurogenesis. Mol. Brain 2016, 9, 29. [Google Scholar] [CrossRef]
- Fasen, K.; Beck, H.; Elger, C.E.; Lie, A.A. Differential regulation of cadherins and catenins during axonal reorganization in the adult rat CNS. J. Neuropathol. Exp. Neurol. 2002, 61, 903–913. [Google Scholar] [CrossRef]
- Qu, Z.; Su, F.; Qi, X.; Sun, J.; Wang, H.; Qiao, Z.; Zhao, H.; Zhu, Y. Wnt/beta-catenin signalling pathway mediated aberrant hippocampal neurogenesis in kainic acid-induced epilepsy. Cell Biochem. Funct. 2017, 35, 472–476. [Google Scholar] [CrossRef]
- Häussler, U.; Bielefeld, L.; Froriep, U.P.; Wolfart, J.; Haas, C.A. Septotemporal position in the hippocampal formation determines epileptic and neurogenic activity in temporal lobe epilepsy. Cereb. Cortex 2012, 22, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Krook-Magnuson, E.; Armstrong, C.; Bui, A.; Lew, S.; Oijala, M.; Soltesz, I. In vivo evaluation of the dentate gate theory in epilepsy. J. Physiol. 2015, 593, 2379–2388. [Google Scholar] [CrossRef] [PubMed]
- Buckmaster, P.S.; Dudek, F.E. Neuron loss, granule cell axon reorganization, and functional changes in the dentate gyrus of epileptic kainate-treated rats. J. Comp. Neurol. 1997, 385, 385–404. [Google Scholar] [CrossRef]
- Liu, C.; Kato, Y.; Zhang, Z.; Do, V.M.; Yankner, B.A.; He, X. beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc. Natl. Acad. Sci. USA 1999, 96, 6273–6278. [Google Scholar] [CrossRef] [PubMed]
- Marikawa, Y.; Elinson, R.P. beta-TrCP is a negative regulator of Wnt/beta-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech. Dev. 1998, 77, 75–80. [Google Scholar] [CrossRef]
- Bouilleret, V.; Ridoux, V.; Depaulis, A.; Marescaux, C.; Nehlig, A.; Le Gal La Salle, G. Recurrent seizures and hippocampal sclerosis following intrahippocampal kainate injection in adult mice: Electroencephalography, histopathology and synaptic reorganization similar to mesial temporal lobe epilepsy. Neuroscience 1999, 89, 717–729. [Google Scholar] [CrossRef]
- Jessberger, S.; Romer, B.; Babu, H.; Kempermann, G. Seizures induce proliferation and dispersion of doublecortin-positive hippocampal progenitor cells. Exp. Neurol. 2005, 196, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, O.K.; Janjoppi, L.; Bonone, F.M.; Pansani, A.P.; da Silva, A.V.; Scorza, F.A.; Cavalheiro, E.A. Whole transcriptome analysis of the hippocampus: Toward a molecular portrait of epileptogenesis. BMC Genom. 2010, 11, 230. [Google Scholar] [CrossRef]
- Choi, J.; Ko, J.; Racz, B.; Burette, A.; Lee, J.R.; Kim, S.; Na, M.; Lee, H.W.; Kim, K.; Weinberg, R.J.; et al. Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. J. Neurosci. 2005, 25, 869–879. [Google Scholar] [CrossRef]
- Bjellvi, J.; Olsson, I.; Malmgren, K.; Wilbe Ramsay, K. Epilepsy duration and seizure outcome in epilepsy surgery: A systematic review and meta-analysis. Neurology 2019, 93, e159–e166. [Google Scholar] [CrossRef]
- Salasova, A.; Yokota, C.; Potesil, D.; Zdrahal, Z.; Bryja, V.; Arenas, E. A proteomic analysis of LRRK2 binding partners reveals interactions with multiple signaling components of the WNT/PCP pathway. Mol. Neurodegener. 2017, 12, 54. [Google Scholar] [CrossRef]
- Berwick, D.C.; Harvey, K. LRRK2 functions as a Wnt signaling scaffold, bridging cytosolic proteins and membrane-localized LRP6. Hum. Mol. Genet. 2012, 21, 4966–4979. [Google Scholar] [CrossRef]
- Kim, S.Y.; Senatorov, V.V., Jr.; Morrissey, C.S.; Lippmann, K.; Vazquez, O.; Milikovsky, D.Z.; Gu, F.; Parada, I.; Prince, D.A.; Becker, A.J.; et al. TGFbeta signaling is associated with changes in inflammatory gene expression and perineuronal net degradation around inhibitory neurons following various neurological insults. Sci. Rep. 2017, 7, 7711. [Google Scholar] [CrossRef]
- Bar-Klein, G.; Cacheaux, L.P.; Kamintsky, L.; Prager, O.; Weissberg, I.; Schoknecht, K.; Cheng, P.; Kim, S.Y.; Wood, L.; Heinemann, U.; et al. Losartan prevents acquired epilepsy via TGF-beta signaling suppression. Ann. Neurol. 2014, 75, 864–875. [Google Scholar] [CrossRef]
- Cacheaux, L.P.; Ivens, S.; David, Y.; Lakhter, A.J.; Bar-Klein, G.; Shapira, M.; Heinemann, U.; Friedman, A.; Kaufer, D. Transcriptome profiling reveals TGF-beta signaling involvement in epileptogenesis. J. Neurosci. 2009, 29, 8927–8935. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Lubin, F.D.; Ren, Y.; Xu, X.; Anderson, A.E. Nuclear factor-kappa B regulates seizure threshold and gene transcription following convulsant stimulation. J. Neurochem. 2007, 103, 1381–1395. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, S.D.; Lin, T.K.; Chang, W.N.; Liou, C.W.; Chang, A.Y.; Chan, S.H.; Chuang, Y.C. Heat shock protein 70 protects against seizure-induced neuronal cell death in the hippocampus following experimental status epilepticus via inhibition of nuclear factor-kappaB activation-induced nitric oxide synthase II expression. Neurobiol. Dis. 2014, 62, 241–249. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Chen, S.D.; Lin, T.K.; Chang, W.N.; Lu, C.H.; Liou, C.W.; Chan, S.H.; Chang, A.Y. Transcriptional upregulation of nitric oxide synthase II by nuclear factor-kappaB promotes apoptotic neuronal cell death in the hippocampus following experimental status epilepticus. J. Neurosci. Res. 2010, 88, 1898–1907. [Google Scholar] [CrossRef]
- Park, C.Y.; Kim, H.S.; Jang, J.; Lee, H.; Lee, J.S.; Yoo, J.E.; Lee, D.R.; Kim, D.W. ABCD2 is a direct target of beta-catenin and TCF-4: Implications for X-linked adrenoleukodystrophy therapy. PLoS ONE 2013, 8, e56242. [Google Scholar] [CrossRef]
- Naito, A.T.; Sumida, T.; Nomura, S.; Liu, M.L.; Higo, T.; Nakagawa, A.; Okada, K.; Sakai, T.; Hashimoto, A.; Hara, Y.; et al. Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell 2012, 149, 1298–1313. [Google Scholar] [CrossRef]
- Xia, L.; Wu, L.; Bao, J.; Li, Q.; Chen, X.; Xia, H.; Xia, R. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/beta-catenin pathway. Biochem. Biophys. Res. Commun. 2018, 503, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Ponferrada, V.G.; Fan, J.; Vallance, J.E.; Hu, S.; Mamedova, A.; Rankin, S.A.; Kofron, M.; Zorn, A.M.; Hegde, R.S.; Lang, R.A. CRIM1 complexes with ss-catenin and cadherins, stabilizes cell-cell junctions and is critical for neural morphogenesis. PLoS ONE 2012, 7, e32635. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; Cornelis, F.M.F.; Lories, R.J.; Monteagudo, S. Exostosin-1 enhances canonical Wnt signaling activity during chondrogenic differentiation. Osteoarthr. Cartil. 2019, 27, 1702–1710. [Google Scholar] [CrossRef]
- Conrad, W.; Major, M.B.; Cleary, M.A.; Ferrer, M.; Roberts, B.; Marine, S.; Chung, N.; Arthur, W.T.; Moon, R.T.; Berndt, J.D.; et al. FAM129B is a novel regulator of Wnt/beta-catenin signal transduction in melanoma cells. F1000Research 2013, 2, 134. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.G.; Kaufman, A.M.; Gong, Y.; Ramaswami, D.; Walsh, L.A.; Turcan, S.; Eng, S.; Kannan, K.; Zou, Y.; Peng, L.; et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat. Genet. 2013, 45, 253–261. [Google Scholar] [CrossRef]
- Katoh, Y.; Katoh, M. Comparative integromics on BMP/GDF family. Int. J. Mol. Med. 2006, 17, 951–955. [Google Scholar] [CrossRef]
- Gao, W.; Ho, M. The role of glypican-3 in regulating Wnt in hepatocellular carcinomas. Cancer Rep. 2011, 1, 14–19. [Google Scholar] [PubMed]
- Ukai, S.; Sakamoto, N.; Taniyama, D.; Harada, K.; Honma, R.; Maruyama, R.; Naka, K.; Hinoi, T.; Takakura, Y.; Shimizu, W.; et al. KHDRBS3 promotes multi-drug resistance and anchorage-independent growth in colorectal cancer. Cancer Sci. 2021, 112, 1196–1208. [Google Scholar] [CrossRef]
- Noack, C.; Zafiriou, M.P.; Schaeffer, H.J.; Renger, A.; Pavlova, E.; Dietz, R.; Zimmermann, W.H.; Bergmann, M.W.; Zelarayan, L.C. Krueppel-like factor 15 regulates Wnt/beta-catenin transcription and controls cardiac progenitor cell fate in the postnatal heart. EMBO Mol. Med. 2012, 4, 992–1007. [Google Scholar] [CrossRef]
- Che, C.; Li, C.; Lin, J.; Zhang, J.; Jiang, N.; Yuan, K.; Zhao, G. Wnt5a contributes to dectin-1 and LOX-1 induced host inflammatory response signature in Aspergillus fumigatus keratitis. Cell. Signal. 2018, 52, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Ingraham, C.A.; Park, G.C.; Makarenkova, H.P.; Crossin, K.L. Matrix metalloproteinase (MMP)-9 induced by Wnt signaling increases the proliferation and migration of embryonic neural stem cells at low O2 levels. J. Biol. Chem. 2011, 286, 17649–17657. [Google Scholar] [CrossRef] [PubMed]
- Patapoutian, A.; Backus, C.; Kispert, A.; Reichardt, L.F. Regulation of neurotrophin-3 expression by epithelial-mesenchymal interactions: The role of Wnt factors. Science 1999, 283, 1180–1183. [Google Scholar] [CrossRef][Green Version]
- Cironi, L.; Petricevic, T.; Fernandes Vieira, V.; Provero, P.; Fusco, C.; Cornaz, S.; Fregni, G.; Letovanec, I.; Aguet, M.; Stamenkovic, I. The fusion protein SS18-SSX1 employs core Wnt pathway transcription factors to induce a partial Wnt signature in synovial sarcoma. Sci. Rep. 2016, 6, 22113. [Google Scholar] [CrossRef] [PubMed]
- van Tienen, L.M.; Mieszczanek, J.; Fiedler, M.; Rutherford, T.J.; Bienz, M. Constitutive scaffolding of multiple Wnt enhanceosome components by Legless/BCL9. Elife 2017, 6, e20882. [Google Scholar] [CrossRef]
- Skah, S.; Uchuya-Castillo, J.; Sirakov, M.; Plateroti, M. The thyroid hormone nuclear receptors and the Wnt/beta-catenin pathway: An intriguing liaison. Dev. Biol 2017, 422, 71–82. [Google Scholar] [CrossRef]
- Das, S.; Yu, S.; Sakamori, R.; Stypulkowski, E.; Gao, N. Wntless in Wnt secretion: Molecular, cellular and genetic aspects. Front. Biol. 2012, 7, 587–593. [Google Scholar] [CrossRef]
- Lomeli, H. ZMIZ proteins: Partners in transcriptional regulation and risk factors for human disease. J. Mol. Med. 2022, 100, 973–983. [Google Scholar] [CrossRef]
- Overstreet-Wadiche, L.S.; Bromberg, D.A.; Bensen, A.L.; Westbrook, G.L. Seizures accelerate functional integration of adult-generated granule cells. J. Neurosci. 2006, 26, 4095–4103. [Google Scholar] [CrossRef] [PubMed]
- Madsen, T.M.; Newton, S.S.; Eaton, M.E.; Russell, D.S.; Duman, R.S. Chronic electroconvulsive seizure up-regulates beta-catenin expression in rat hippocampus: Role in adult neurogenesis. Biol. Psychiatry 2003, 54, 1006–1014. [Google Scholar] [CrossRef]
- Koh, S.; Tibayan, F.D.; Simpson, J.N.; Jensen, F.E. NBQX or topiramate treatment after perinatal hypoxia-induced seizures prevents later increases in seizure-induced neuronal injury. Epilepsia 2004, 45, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, R.M.; Koh, S.; Rio, C.; Wang, C.; Lamperti, E.D.; Sharma, D.; Corfas, G.; Jensen, F.E. Decreased glutamate receptor 2 expression and enhanced epileptogenesis in immature rat hippocampus after perinatal hypoxia-induced seizures. J. Neurosci. 2001, 21, 8154–8163. [Google Scholar] [CrossRef] [PubMed]
- Sampson, E.M.; Haque, Z.K.; Ku, M.C.; Tevosian, S.G.; Albanese, C.; Pestell, R.G.; Paulson, K.E.; Yee, A.S. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001, 20, 4500–4511. [Google Scholar] [CrossRef]
- Arredondo, S.B.; Valenzuela-Bezanilla, D.; Mardones, M.D.; Varela-Nallar, L. Role of Wnt Signaling in Adult Hippocampal Neurogenesis in Health and Disease. Front. Cell Dev. Biol. 2020, 8, 860. [Google Scholar] [CrossRef]
- Wiebe, S.; Blume, W.T.; Girvin, J.P.; Eliasziw, M.; Effectiveness and Efficiency of Surgery for Temporal Lobe Epilepsy Study Group. A randomized, controlled trial of surgery for temporal-lobe epilepsy. N. Engl. J. Med. 2001, 345, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Blumcke, I.; Pauli, E.; Clusmann, H.; Schramm, J.; Becker, A.; Elger, C.; Merschhemke, M.; Meencke, H.J.; Lehmann, T.; von Deimling, A.; et al. A new clinico-pathological classification system for mesial temporal sclerosis. Acta Neuropathol. 2007, 113, 235–244. [Google Scholar] [CrossRef]
- Blumcke, I.; Thom, M.; Aronica, E.; Armstrong, D.D.; Bartolomei, F.; Bernasconi, A.; Bernasconi, N.; Bien, C.G.; Cendes, F.; Coras, R.; et al. International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: A Task Force report from the ILAE Commission on Diagnostic Methods. Epilepsia 2013, 54, 1315–1329. [Google Scholar] [CrossRef]
- Crespel, A.; Rigau, V.; Coubes, P.; Rousset, M.C.; de Bock, F.; Okano, H.; Baldy-Moulinier, M.; Bockaert, J.; Lerner-Natoli, M. Increased number of neural progenitors in human temporal lobe epilepsy. Neurobiol. Dis. 2005, 19, 436–450. [Google Scholar] [CrossRef]
- Houser, C.R. Granule cell dispersion in the dentate gyrus of humans with temporal lobe epilepsy. Brain Res. 1990, 535, 195–204. [Google Scholar] [CrossRef]
- Mathern, G.W.; Leiphart, J.L.; De Vera, A.; Adelson, P.D.; Seki, T.; Neder, L.; Leite, J.P. Seizures decrease postnatal neurogenesis and granule cell development in the human fascia dentata. Epilepsia 2002, 43, 68–73. [Google Scholar] [CrossRef]
- Vangoor, V.R.; Reschke, C.R.; Senthilkumar, K.; van de Haar, L.L.; de Wit, M.; Giuliani, G.; Broekhoven, M.H.; Morris, G.; Engel, T.; Brennan, G.P.; et al. Antagonizing Increased miR-135a Levels at the Chronic Stage of Experimental TLE Reduces Spontaneous Recurrent Seizures. J. Neurosci. 2019, 39, 5064–5079. [Google Scholar] [CrossRef] [PubMed]
- Bosco, D.B.; Zheng, J.; Xu, Z.; Peng, J.; Eyo, U.B.; Tang, K.; Yan, C.; Huang, J.; Feng, L.; Wu, G.; et al. RNAseq analysis of hippocampal microglia after kainic acid-induced seizures. Mol. Brain 2018, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Racine, R.J. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr. Clin. Neurophysiol. 1972, 32, 281–294. [Google Scholar] [CrossRef]
- Heinrich, C.; Nitta, N.; Flubacher, A.; Muller, M.; Fahrner, A.; Kirsch, M.; Freiman, T.; Suzuki, F.; Depaulis, A.; Frotscher, M.; et al. Reelin deficiency and displacement of mature neurons, but not neurogenesis, underlie the formation of granule cell dispersion in the epileptic hippocampus. J. Neurosci. 2006, 26, 4701–4713. [Google Scholar] [CrossRef]
- Kiasalari, Z.; Roghani, M.; Khalili, M.; Rahmati, B.; Baluchnejadmojarad, T. Antiepileptogenic effect of curcumin on kainate-induced model of temporal lobe epilepsy. Pharm. Biol. 2013, 51, 1572–1578. [Google Scholar] [CrossRef]
- Lee, J.M.; Hong, J.; Moon, G.J.; Jung, U.J.; Won, S.Y.; Kim, S.R. Morin Prevents Granule Cell Dispersion and Neurotoxicity via Suppression of mTORC1 in a Kainic Acid-induced Seizure Model. Exp. Neurobiol. 2018, 27, 226–237. [Google Scholar] [CrossRef]
- Bradley, J.D.; Brandt, K.D.; Katz, B.P.; Kalasinski, L.A.; Ryan, S.I. Comparison of an antiinflammatory dose of ibuprofen, an analgesic dose of ibuprofen, and acetaminophen in the treatment of patients with osteoarthritis of the knee. N. Engl. J. Med. 1991, 325, 87–91. [Google Scholar] [CrossRef]
- Msolli, M.A.; Sekma, A.; Toumia, M.; Bel Haj Ali, K.; Khalil, M.H.; Grissa, M.H.; Bouida, W.; Beltaief, K.; Zorgati, A.; Methamem, M.; et al. Acetaminophen, Nonsteroidal Anti-inflammatory Drugs, or Combination of Both Analgesics in Acute Posttrauma Pain: A Randomized Controlled Trial. Acad. Emerg. Med. 2021, 28, 155–163. [Google Scholar] [CrossRef]
- Bouilleret, V.; Schwaller, B.; Schurmans, S.; Celio, M.R.; Fritschy, J.M. Neurodegenerative and morphogenic changes in a mouse model of temporal lobe epilepsy do not depend on the expression of the calcium-binding proteins parvalbumin, calbindin, or calretinin. Neuroscience 2000, 97, 47–58. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. GENCODE 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- da Huang, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, D. BioCarta. Biotech Softw. Internet Rep. 2001, 2, 117–120. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mardones, M.D.; Gupta, K. Transcriptome Profiling of the Hippocampal Seizure Network Implicates a Role for Wnt Signaling during Epileptogenesis in a Mouse Model of Temporal Lobe Epilepsy. Int. J. Mol. Sci. 2022, 23, 12030. https://doi.org/10.3390/ijms231912030
Mardones MD, Gupta K. Transcriptome Profiling of the Hippocampal Seizure Network Implicates a Role for Wnt Signaling during Epileptogenesis in a Mouse Model of Temporal Lobe Epilepsy. International Journal of Molecular Sciences. 2022; 23(19):12030. https://doi.org/10.3390/ijms231912030
Chicago/Turabian StyleMardones, Muriel D., and Kunal Gupta. 2022. "Transcriptome Profiling of the Hippocampal Seizure Network Implicates a Role for Wnt Signaling during Epileptogenesis in a Mouse Model of Temporal Lobe Epilepsy" International Journal of Molecular Sciences 23, no. 19: 12030. https://doi.org/10.3390/ijms231912030
APA StyleMardones, M. D., & Gupta, K. (2022). Transcriptome Profiling of the Hippocampal Seizure Network Implicates a Role for Wnt Signaling during Epileptogenesis in a Mouse Model of Temporal Lobe Epilepsy. International Journal of Molecular Sciences, 23(19), 12030. https://doi.org/10.3390/ijms231912030





