HFR1, a bHLH Transcriptional Regulator from Arabidopsis thaliana, Improves Grain Yield, Shade and Osmotic Stress Tolerances in Common Wheat
Abstract
:1. Introduction
2. Results
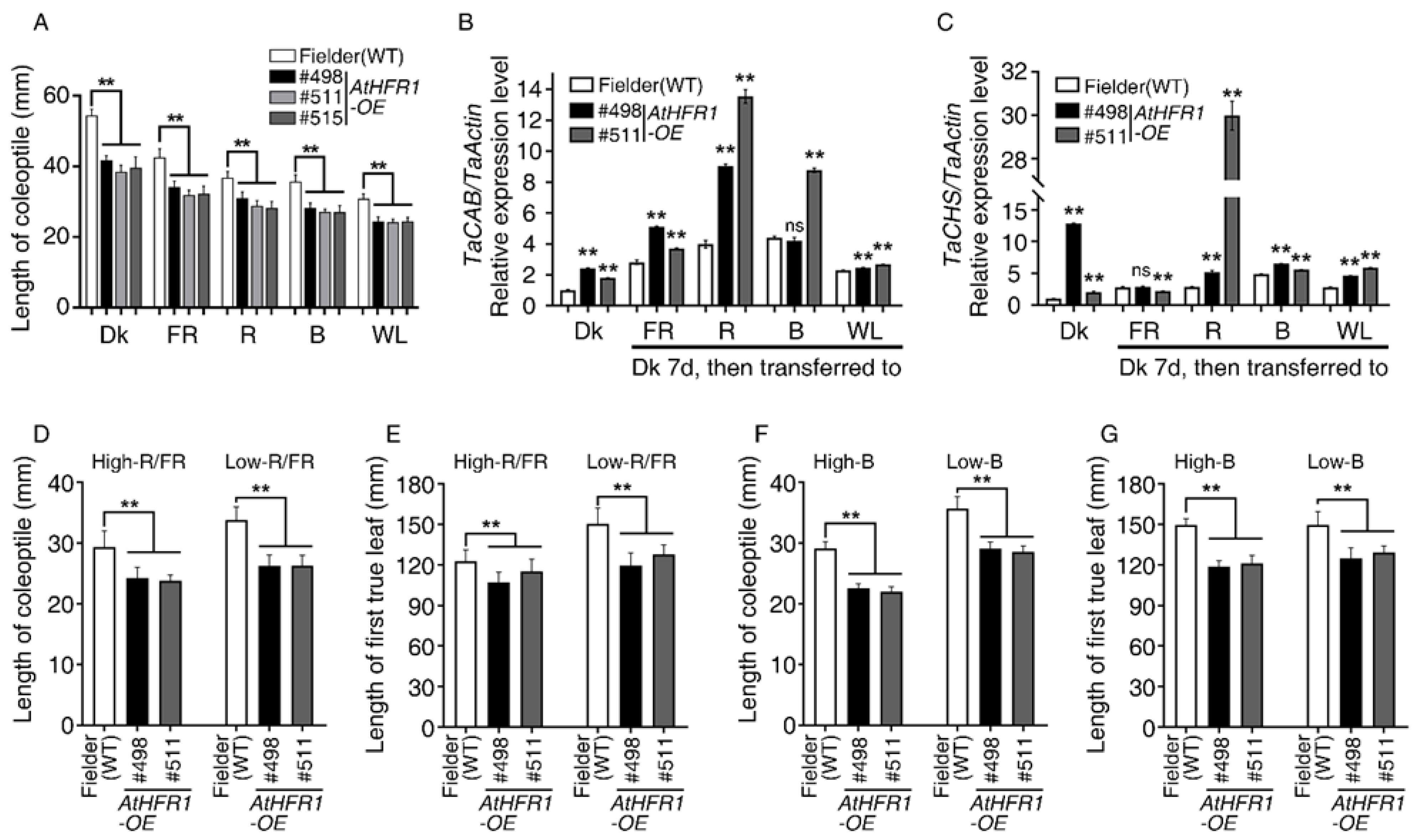
2.1. The AtHFR1 Transgene Represses Seedling Etiolation and Shade Avoidance in Common Wheat
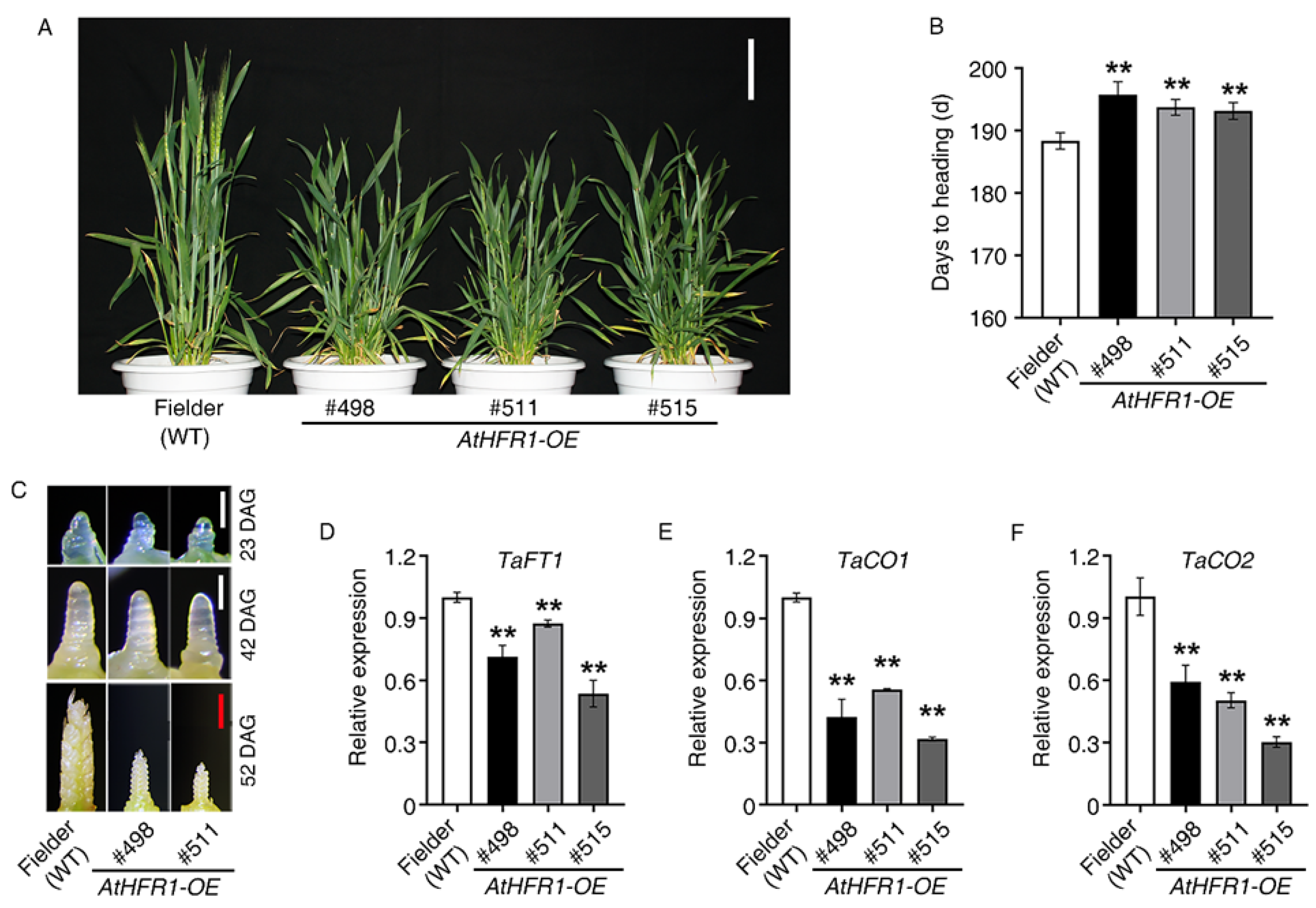
2.2. The AtHFR1 Transgene Delays Heading in Common Wheat
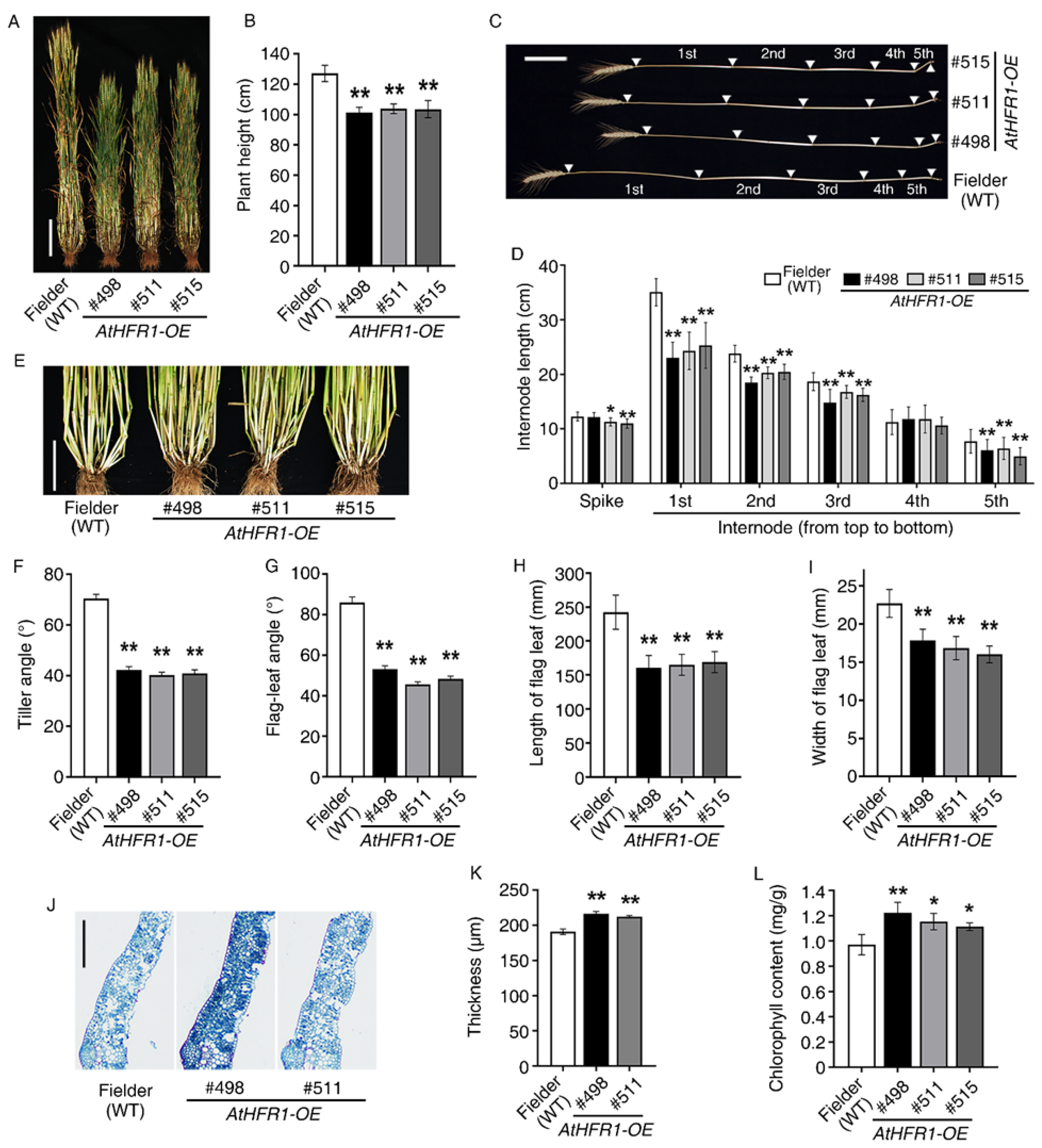
2.3. The AtHFR1 Transgene Leads to Compact Plant Architecture in the Field
2.4. The AtHFR1 Transgene Enhances Grain Yield per Plant in Wheat
2.5. The AtHFR1 Transgene Enhances Wheat Tolerance to Osmotic Stress during Seed Germination
2.6. The AtHFR1 Transgene Inhibits Dark-Induced Senescence in Wheat
3. Discussion
3.1. Overexpression of AtHFR1 Alters Plant Architecture by Activating the Light Signal Transduction Pathway in Wheat
3.2. Overexpression of AtHFR1 Effectively Improves Tolerance to Shade and Osmotic Stress in Wheat
3.3. Genetic Modification of the Light Signaling Pathways Has Positive Effects on Grain Yield through Changes in Resource Allocation
4. Materials and Methods
4.1. Plasmid Construction and Genetic Transformation
4.2. Plant Growth Conditions
4.3. Agronomic Traits
4.4. PCR and Real-Time Reverse Transcription PCR
4.5. Chlorophyll Extraction
4.6. Statistical Analysis
4.7. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hickey, L.T.; Hafeez, N.A.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Coffel, E.D.; Horton, R.M.; de Sherbinin, A. Temperature and humidity based projections of a rapid rise in global heat stress exposure during the 21(st) century. Environ. Res. Lett. 2018, 13, 14001. [Google Scholar] [CrossRef]
- Mora, C.; Dousset, B.; Caldwell, I.R.; Powell, F.E.; Geronimo, R.C.; Bielecki, C.R.; Counsell, C.W.W.; Dietrich, B.S.; Johnston, E.T.; Louis, L.V.; et al. Global risk of deadly heat. Nat. Clim. Chang. 2017, 7, 501–506. [Google Scholar] [CrossRef] [Green Version]
- Meehl, G.A.; Tebaldi, C. More intense, more frequent, and longer lasting heat waves in the 21st century. Science 2004, 305, 994–997. [Google Scholar] [CrossRef] [Green Version]
- Hirano, K.; Ordonio, R.L.; Matsuoka, M. Engineering the lodging resistance mechanism of post-Green Revolution rice to meet future demands. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 220–233. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Bohra, A.; Roorkiwal, M.; Barmukh, R.; Cowling, W.A.; Chitikineni, A.; Lam, H.M.; Hickey, L.T.; Croser, J.S.; Bayer, P.E.; et al. Fast-forward breeding for a food-secure world. Trends Genet. 2021, 37, 1124–1136. [Google Scholar] [CrossRef]
- Razzaq, A.; Kaur, P.; Akhter, N.; Wani, S.H.; Saleem, F. Next-Generation Breeding Strategies for Climate-Ready Crops. Front. Plant Sci. 2021, 12, 620420. [Google Scholar] [CrossRef]
- Dwivedi, S.; Perotti, E.; Ortiz, R. Towards molecular breeding of reproductive traits in cereal crops. Plant Biotechnol. J. 2008, 6, 529–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavridou, E.; Schmid-Siegert, E.; Fankhauser, C.; Ulm, R. UVR8-mediated inhibition of shade avoidance involves HFR1 stabilization in Arabidopsis. PLoS Genet. 2020, 16, e1008797. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Huang, X.; Deng, X.W. The Photomorphogenic Central Repressor COP1: Conservation and Functional Diversification during Evolution. Plant Commun. 2020, 1, 100044. [Google Scholar] [CrossRef]
- Alves, F.R.R.; Lira, B.S.; Pikart, F.C.; Monteiro, S.S.; Furlan, C.M.; Purgatto, E.; Pascoal, G.B.; Andrade, S.; Demarco, D.; Rossi, M.; et al. Beyond the limits of photoperception: Constitutively active Phytochrome B2 overexpression as a means of improving fruit nutritional quality in tomato. Plant Biotechnol. J. 2020, 18, 2027–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, R.; Ulm, R. How plants cope with UV-B: From perception to response. Curr. Opin. Plant Biol. 2017, 37, 42–48. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, G.; Wang, H.; Wang Deng, X. Phytochrome signaling mechanisms. Arab. Book 2011, 9, e0148. [Google Scholar] [CrossRef] [Green Version]
- Schafer, E.; Bowle, C. Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Rep. 2002, 3, 1042–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C. Blue light receptors and signal transduction. Plant Cell 2002, 14 (Suppl. 1), S207–S225. [Google Scholar] [CrossRef] [Green Version]
- Briggs, W.R.; Christie, J.M. Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 2002, 7, 204–210. [Google Scholar] [CrossRef]
- Deng, X.W.; Quail, P.H. Signaling in light-controlled development. Semin. Cell Dev. Biol. 1999, 10, 121–129. [Google Scholar] [CrossRef]
- Cashmore, A.R.; Jarillo, J.A.; Wu, Y.J.; Liu, D. Cryptochromes: Blue light receptors for plants and animals. Science 1999, 284, 760–765. [Google Scholar] [CrossRef]
- Zhou, P.; Song, M.; Yang, Q.; Su, L.; Hou, P.; Guo, L.; Zheng, X.; Xi, Y.; Meng, F.; Xiao, Y.; et al. Both Phytochrome Rapidly Regulated1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol. 2014, 164, 841–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, I.C.; Henriques, R.; Seo, H.S.; Nagatani, A.; Chua, N.H. Arabidopsis Phytochrome Interacting Factor proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 2010, 22, 2370–2383. [Google Scholar] [CrossRef]
- Zhu, D.; Maier, A.; Lee, J.H.; Laubinger, S.; Saijo, Y.; Wang, H.; Qu, L.J.; Hoecker, U.; Deng, X.W. Biochemical characterization of Arabidopsis complexes containing Constitutively Photomorphogenic1 and Suppressor of PHYA proteins in light control of plant development. Plant Cell 2008, 20, 2307–2323. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Lin, R.; Sullivan, J.; Hoecker, U.; Liu, B.; Xu, L.; Deng, X.W.; Wang, H. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 2005, 17, 804–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Lin, R.; Hoecker, U.; Liu, B.; Xu, L.; Wang, H. Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J. 2005, 43, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Jang, I.C.; Yang, J.Y.; Seo, H.S.; Chua, N.H. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005, 19, 593–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, H.S.; Watanabe, E.; Tokutomi, S.; Nagatani, A.; Chua, N.H. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004, 18, 617–622. [Google Scholar] [CrossRef] [Green Version]
- Duek, P.D.; Elmer, M.V.; van Oosten, V.R.; Fankhauser, C. The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 2004, 14, 2296–2301. [Google Scholar] [CrossRef] [Green Version]
- Saijo, Y.; Sullivan, J.A.; Wang, H.; Yang, J.; Shen, Y.; Rubio, V.; Ma, L.; Hoecker, U.; Deng, X.W. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003, 17, 2642–2647. [Google Scholar] [CrossRef] [Green Version]
- Paulišić, S.; Qin, W.; Arora Verasztó, H.; Then, C.; Alary, B.; Nogue, F.; Tsiantis, M.; Hothorn, M.; Martínez-García, J.F. Adjustment of the PIF7-HFR1 transcriptional module activity controls plant shade adaptation. EMBO J. 2021, 40, e104273. [Google Scholar] [CrossRef]
- Oh, J.; Park, E.; Song, K.; Bae, G.; Choi, G. Phytochrome Interacting Factor8 Inhibits Phytochrome A-Mediated Far-Red Light Responses in Arabidopsis. Plant Cell 2020, 32, 186–205. [Google Scholar] [CrossRef]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and Phytochrome Interacting Factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, I.C.; Yang, S.W.; Yang, J.Y.; Chua, N.H. Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev. 2007, 21, 2100–2111. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.M.; Woo, J.C.; Song, P.S.; Soh, M.S. HFR1, a phytochrome A-signalling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J. 2002, 30, 711–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fairchild, C.D.; Schumaker, M.A.; Quail, P.H. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000, 14, 2377–2391. [Google Scholar] [PubMed]
- Zhang, X.N.; Wu, Y.; Tobias, J.W.; Brunk, B.P.; Deitzer, G.F.; Liu, D. HFR1 is crucial for transcriptome regulation in the cryptochrome 1-mediated early response to blue light in Arabidopsis thaliana. PLoS ONE 2008, 3, e3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duek, P.D.; Fankhauser, C. HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 2003, 34, 827–836. [Google Scholar] [CrossRef]
- Luo, Q.; Lian, H.L.; He, S.B.; Li, L.; Jia, K.P.; Yang, H.Q. COP1 and phyB Physically Interact with PIL1 to Regulate Its Stability and Photomorphogenic Development in Arabidopsis. Plant Cell 2014, 26, 2441–2456. [Google Scholar] [CrossRef] [Green Version]
- Hayes, S.; Sharma, A.; Fraser, D.P.; Trevisan, M.; Cragg-Barber, C.K.; Tavridou, E.; Fankhauser, C.; Jenkins, G.I.; Franklin, K.A. UV-B Perceived by the UVR8 Photoreceptor Inhibits Plant Thermomorphogenesis. Curr. Biol. 2017, 27, 120–127. [Google Scholar] [CrossRef] [Green Version]
- Sessa, G.; Carabelli, M.; Sassi, M.; Ciolfi, A.; Possenti, M.; Mittempergher, F.; Becker, J.; Morelli, G.; Ruberti, I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005, 19, 2811–2815. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Jia, Y.; Yu, L.; Yang, W.; Chen, Z.; Chen, H.; Hu, X. Nitric oxide promotes light-initiated seed germination by repressing PIF1 expression and stabilizing HFR1. Plant Physiol. Biochem. 2018, 123, 204–212. [Google Scholar] [CrossRef]
- Shi, H.; Zhong, S.; Mo, X.; Liu, N.; Nezames, C.D.; Deng, X.W. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 2013, 25, 3770–3784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Yang, C.; Jiang, Y.; Li, L. A PIF7-CONSTANS-Centered Molecular Regulatory Network Underlying Shade-Accelerated Flowering. Mol. Plant 2019, 12, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Duren, Z.; Wang, Y.; Wang, J.; Zhao, X.M.; Lv, L.; Li, X.; Liu, J.; Zhu, X.G.; Chen, L.; Wang, Y. Hierarchical graphical model reveals HFR1 bridging circadian rhythm and flower development in Arabidopsis thaliana. NPJ Syst. Biol. Appl. 2019, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- Ueda, H.; Ito, T.; Inoue, R.; Masuda, Y.; Nagashima, Y.; Kozuka, T.; Kusaba, M. Genetic Interaction Among Phytochrome, Ethylene and Abscisic Acid Signaling During Dark-Induced Senescence in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Mitsuda, N.; Ishizuka, T.; Satoh, M.; Ohme-Takagi, M. The CIB1 transcription factor regulates light- and heat-inducible cell elongation via a two-step HLH/bHLH system. J. Exp. Bot. 2021, 72, 1795–1808. [Google Scholar] [CrossRef] [PubMed]
- Foreman, J.; Johansson, H.; Hornitschek, P.; Josse, E.M.; Fankhauser, C.; Halliday, K.J. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 2011, 65, 441–452. [Google Scholar] [CrossRef]
- Casal, J.J. Shade avoidance. Arab. Book 2012, 10, e0157. [Google Scholar] [CrossRef] [Green Version]
- Franklin, K.A. Shade avoidance. New Phytol. 2008, 179, 930–944. [Google Scholar] [CrossRef]
- Smith, H.; Whitelam, G. The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 1997, 20, 840–844. [Google Scholar] [CrossRef]
- Garg, A.K.; Sawers, R.J.; Wang, H.; Kim, J.K.; Walker, J.M.; Brutnell, T.P.; Parthasarathy, M.V.; Vierstra, R.D.; Wu, R.J. Light-regulated overexpression of an Arabidopsis phytochrome A gene in rice alters plant architecture and increases grain yield. Planta 2006, 223, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Boccalandro, H.E.; PLoSchuk, E.L.; Yanovsky, M.J.; Sanchez, R.A.; Gatz, C.; Casal, J.J. Increased phytochrome B alleviates density effects on tuber yield of field potato crops. Plant Physiol. 2003, 133, 1539–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiele, A.; Herold, M.; Lenk, I.; Quail, P.H.; Gatz, C. Heterologous expression of Arabidopsis phytochrome B in transgenic potato influences photosynthetic performance and tuber development. Plant Physiol. 1999, 120, 73–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Childs, K.L.; Miller, F.R.; Cordonnier-Pratt, M.M.; Pratt, L.H.; Morgan, P.W.; Mullet, J.E. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 1997, 113, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Hanumappa, M.; Pratt, L.H.; Cordonnier-Pratt, M.M.; Deitzer, G.F. A photoperiod-insensitive barley line contains a light-labile phytochrome B. Plant Physiol. 1999, 119, 1033–1040. [Google Scholar] [CrossRef] [Green Version]
- Izawa, T.; Oikawa, T.; Tokutomi, S.; Okuno, K.; Shimamoto, K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 2000, 22, 391–399. [Google Scholar] [CrossRef]
- Sawers, R.J.; Linley, P.J.; Farmer, P.R.; Hanley, N.P.; Costich, D.E.; Terry, M.J.; Brutnell, T.P. Elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiol. 2002, 130, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Li, C.; Hu, W.; Lau, M.Y.; Lin, H.; Rockwell, N.C.; Martin, S.S.; Jernstedt, J.A.; Lagarias, J.C.; Dubcovsky, J. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc. Natl. Acad. Sci. USA 2014, 111, 10037–10044. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wu, M.; Zhang, M.; Jiang, W.; Ren, X.; Liang, E.; Zhang, D.; Zhang, C.; Xiao, N.; Li, Y.; et al. A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol. J. 2018, 16, 1375–1387. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.D.; Yim, W.C.; Liu, D.; Hu, R.; Yang, X.; Cushman, J.C. A Vitis vinifera basic helix-loop-helix transcription factor enhances plant cell size, vegetative biomass and reproductive yield. Plant Biotechnol. J. 2018, 16, 1595–1615. [Google Scholar] [CrossRef]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef]
- Ji, X.; Du, Y.; Li, F.; Sun, H.; Zhang, J.; Li, J.; Peng, T.; Xin, Z.; Zhao, Q. The basic helix-loop-helix transcription factor, OsPIL15, regulates grain size via directly targeting a purine permease gene OsPUP7 in rice. Plant Biotechnol. J. 2019, 17, 1527–1537. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Z.; Chen, Y.; He, J.X.; Bi, Y. Phytochrome-interacting factor 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci. 2015, 237, 57–68. [Google Scholar] [CrossRef]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [Green Version]
- Keller, J.M.; Shanklin, J.; Vierstra, R.D.; Hershey, H.P. Expression of a functional monocotyledonous phytochrome in transgenic tobacco. EMBO J. 1989, 8, 1005–1012. [Google Scholar] [CrossRef]
- Song, M.F.; Zhang, S.; Hou, P.; Shang, H.Z.; Gu, H.K.; Li, J.J.; Xiao, Y.; Guo, L.; Su, L.; Gao, J.W.; et al. Ectopic expression of a phytochrome B gene from Chinese cabbage (Brassica rapa L. ssp. pekinensis) in Arabidopsis thaliana promotes seedling de-etiolation, dwarfing in mature plants, and delayed flowering. Plant Mol. Biol. 2015, 87, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Green, B.R.; Pichersky, E.; Kloppstech, K. Chlorophyll a/b-binding proteins: An extended family. Trends Biochem. Sci. 1991, 16, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef]
- Lyu, X.; Cheng, Q.; Qin, C.; Li, Y.; Xu, X.; Ji, R.; Mu, R.; Li, H.; Zhao, T.; Liu, J.; et al. GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 2021, 14, 298–314. [Google Scholar] [CrossRef]
- Fang, Z.; Liu, W.Z.; Yu, X.F.; Deng, L.C.; Yang, J.S.; Yin, P.B. Discussion on related issues of super wheat breeding. Shandong Agric. Sci. 2004, 3, 27–29. (In Chinese) [Google Scholar]
- Gonzalez, C.V.; Ibarra, S.E.; Piccoli, P.N.; Botto, J.F.; Boccalandro, H.E. Phytochrome B increases drought tolerance by enhancing ABA sensitivity in Arabidopsis thaliana. Plant Cell Environ. 2012, 35, 1958–1968. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Li, H.; Wang, Q.; Liu, B.; Lin, C. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 2013, 25, 4405–4420. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Yoshida, H.; Matsuoka, M. Making the ‘Green Revolution’Truly Green: Improving Crop Nitrogen Use Efficiency. Plant Cell Physiol. 2021, 62, 942–947. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F. ‘Green revolution’genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Wang, S.; Song, W.; Zhang, J.; Wang, Y.; Liu, Q.; Yu, J.; Ye, Y.; Li, S.; Chen, J.; et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 2020, 367, 6478. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, H.; Du, L.; Ye, X. Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 2017, 15, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, C.; Casal, J.J. Phenotypic characterization of a photomorphogenic mutant. Plant J. 2004, 39, 747–760. [Google Scholar] [CrossRef]
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, G.; Yang, L.; Zhan, W.; Chen, S.; Song, M.; Wang, L.; Jiang, L.; Guo, L.; Wang, K.; Ye, X.; et al. HFR1, a bHLH Transcriptional Regulator from Arabidopsis thaliana, Improves Grain Yield, Shade and Osmotic Stress Tolerances in Common Wheat. Int. J. Mol. Sci. 2022, 23, 12057. https://doi.org/10.3390/ijms231912057
Sun G, Yang L, Zhan W, Chen S, Song M, Wang L, Jiang L, Guo L, Wang K, Ye X, et al. HFR1, a bHLH Transcriptional Regulator from Arabidopsis thaliana, Improves Grain Yield, Shade and Osmotic Stress Tolerances in Common Wheat. International Journal of Molecular Sciences. 2022; 23(19):12057. https://doi.org/10.3390/ijms231912057
Chicago/Turabian StyleSun, Guanghua, Luhao Yang, Weimin Zhan, Shizhan Chen, Meifang Song, Lijian Wang, Liangliang Jiang, Lin Guo, Ke Wang, Xingguo Ye, and et al. 2022. "HFR1, a bHLH Transcriptional Regulator from Arabidopsis thaliana, Improves Grain Yield, Shade and Osmotic Stress Tolerances in Common Wheat" International Journal of Molecular Sciences 23, no. 19: 12057. https://doi.org/10.3390/ijms231912057





