ERF Transcription Factor OsBIERF3 Positively Contributes to Immunity against Fungal and Bacterial Diseases but Negatively Regulates Cold Tolerance in Rice
Abstract
:1. Introduction
2. Results
2.1. Responsiveness of OsBIERF3 to a Bacterial Pathogen, Abiotic Stress, and Hormones
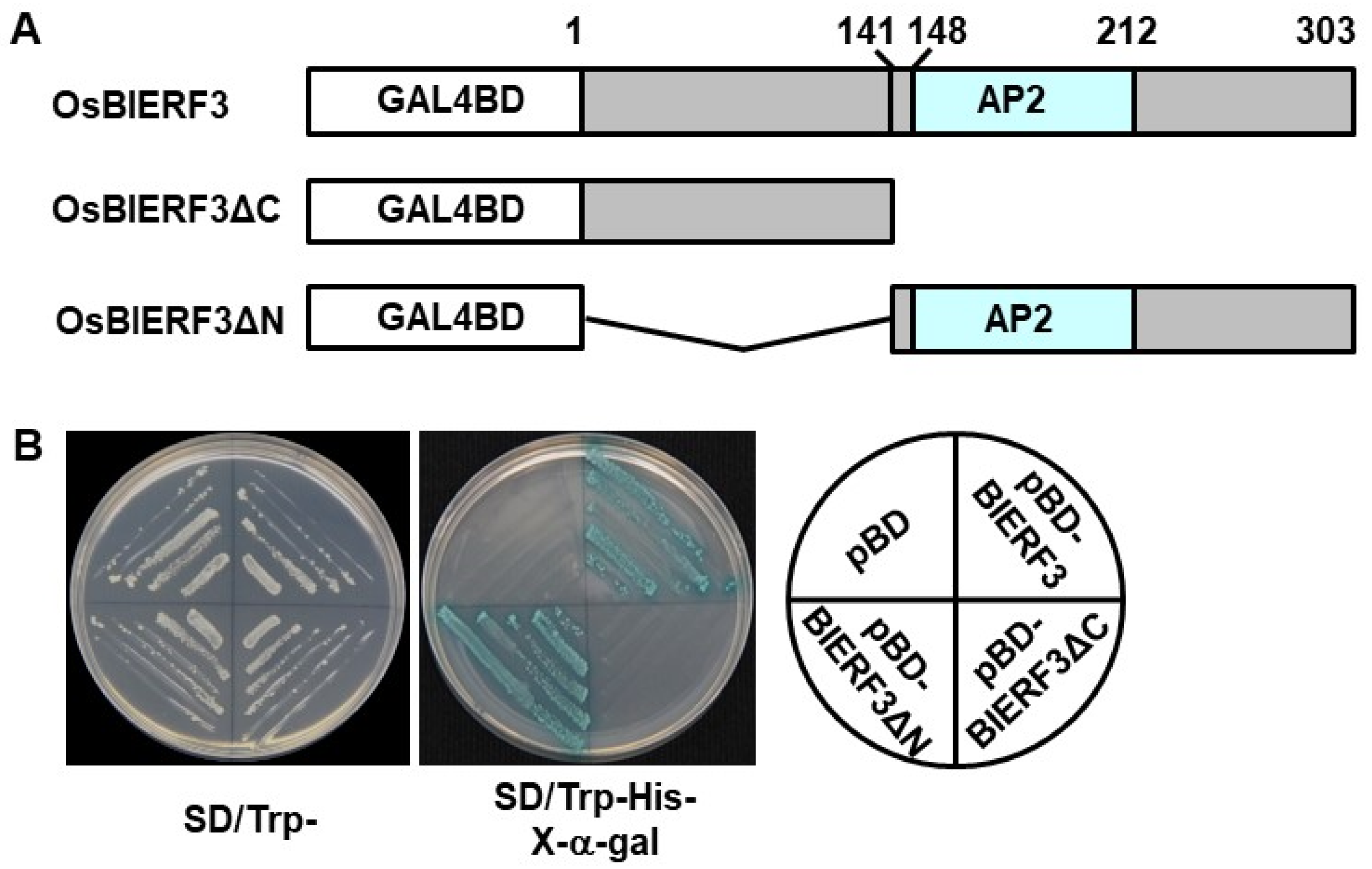
2.2. OsBIERF3 Is a Transcriptional Activator
2.3. Generation of OsBIERF3-OE and OsBIERF3-Ri Transgenic Lines
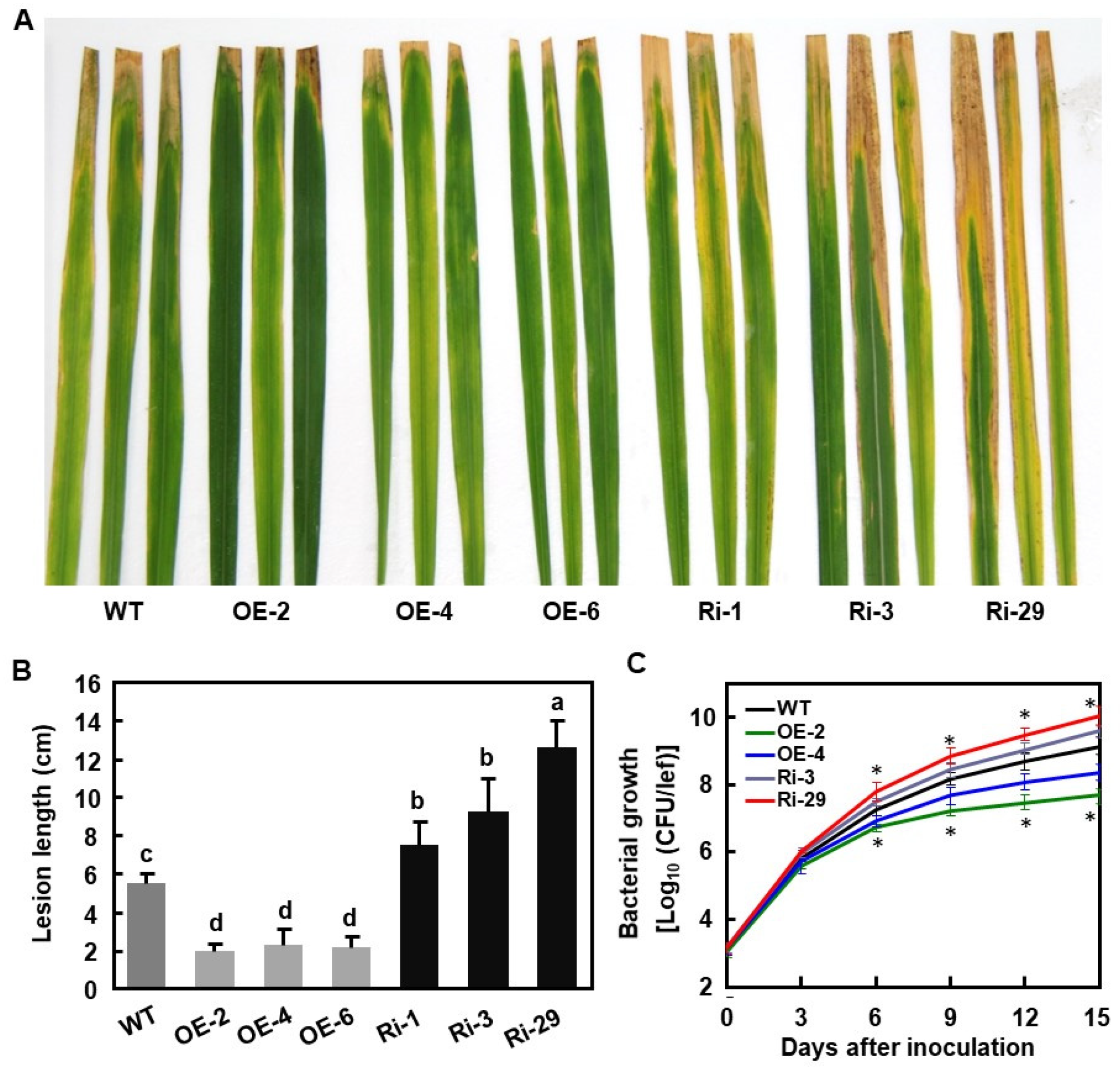
2.4. OsBIERF3 Positively Regulates Resistance to M. oryzae
2.5. OsBIERF3 Positively Regulates Resistance to X. oryzae pv. oryzae
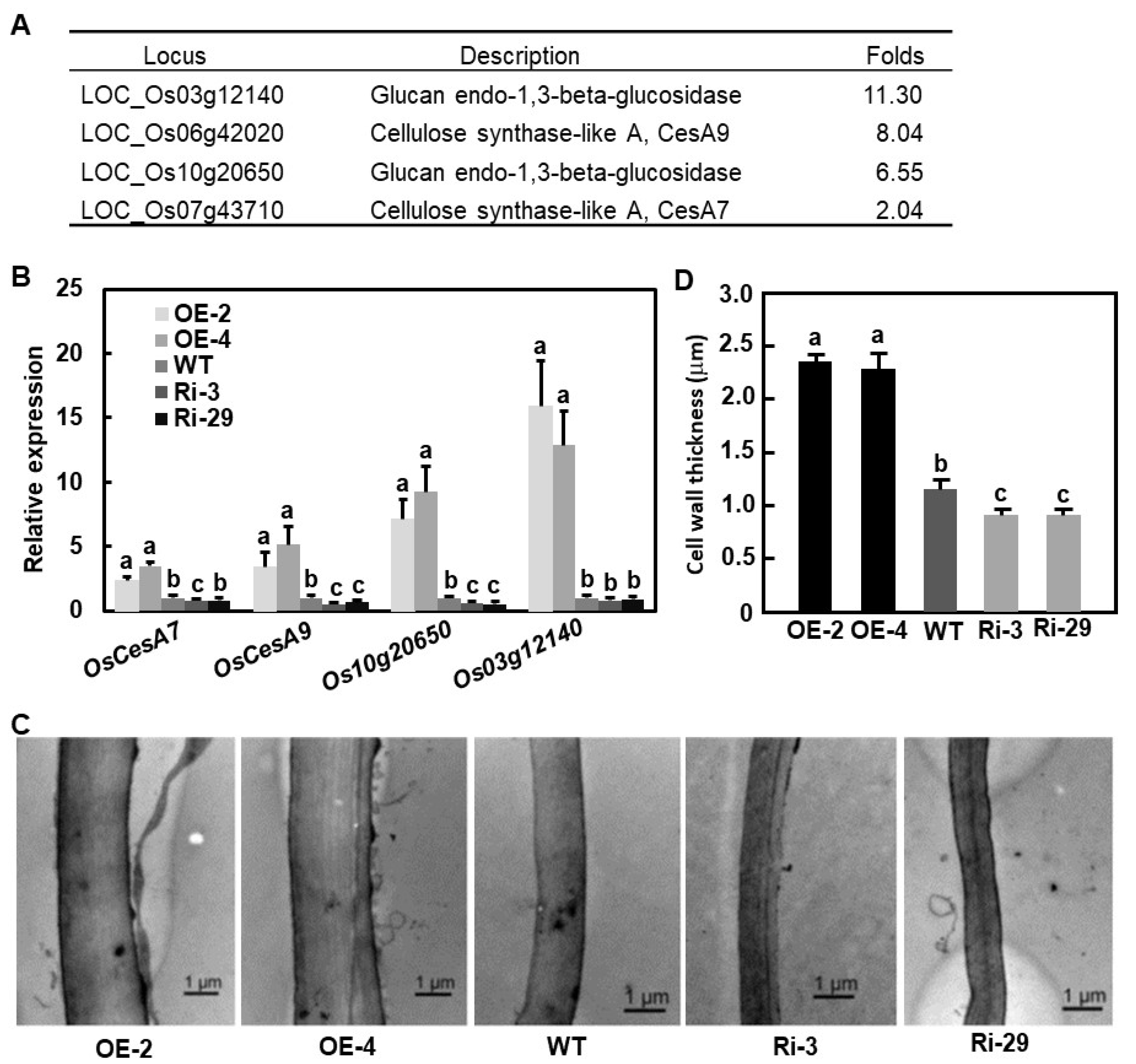
2.6. Identification of Differentially Expressed Genes in OsBIERF3-OE Plants
2.7. Altered Expression of OsBIERF3 Affected Cell Wall Thickness
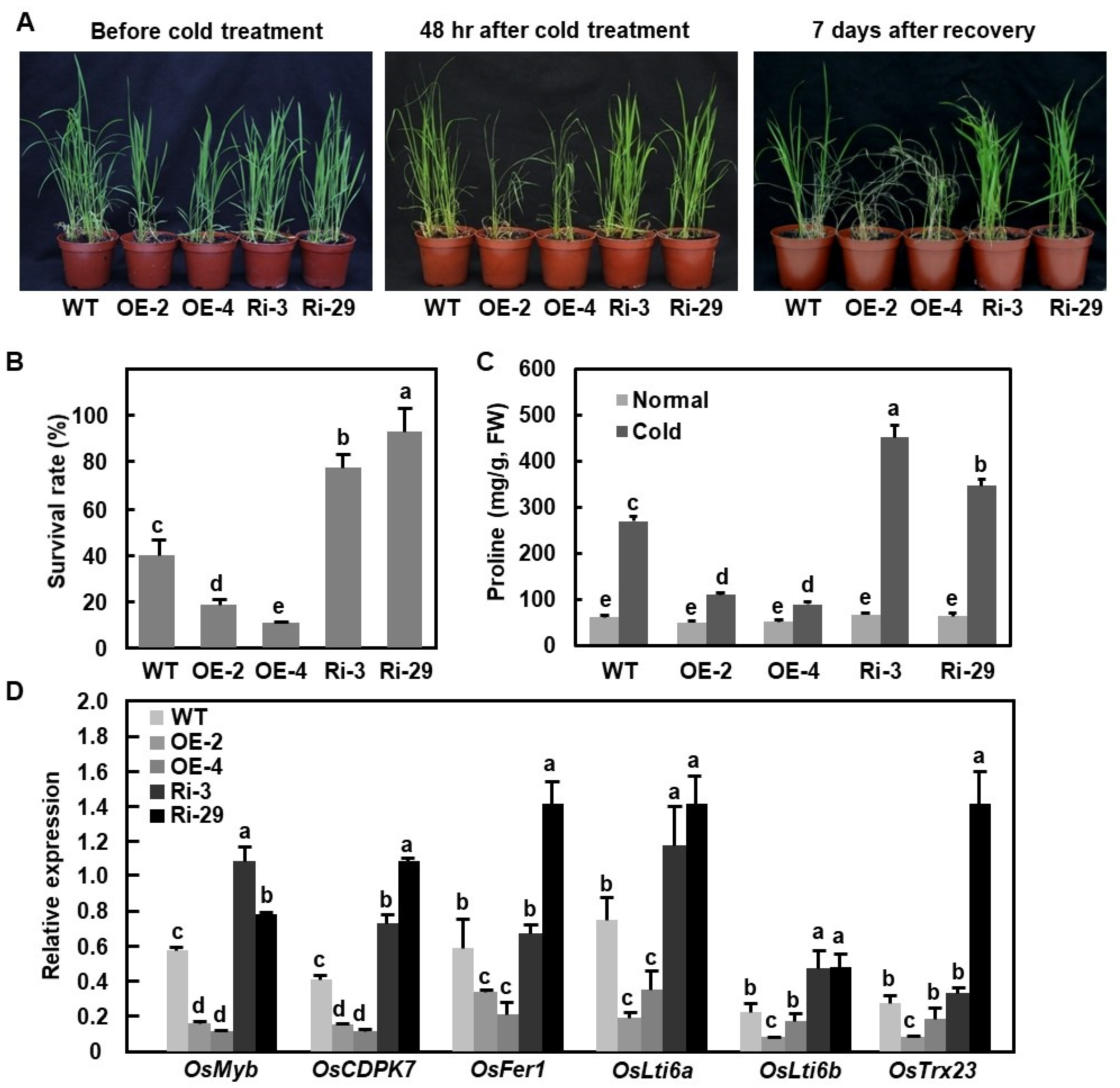
2.8. OsBIERF3 Negatively Regulates Cold Tolerance but Does Not Affect Drought and Salt Tolerance
2.9. Involvement of ET and ABA in OsBIERF3-Mediated Cold Response in Rice
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Treatments
4.2. Generation and Characterization of OsBIERF3-OE and OsBIERF3-Ri Lines
4.3. Transcriptional Activation Assays
4.4. Disease Assays
4.5. Abiotic Stress Tolerance Assays
4.6. Observation and Measurement of Cell Wall Thickness by TEM
4.7. Microarray Analyses of Differentially Expressed Genes in OsBIERF3-OE Plants
4.8. qRT-PCR Analyses of Gene Expression
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought stress responses and resistance in plants: From cellular responses to long-distance intercellular communication. Front. Plant Sci. 2020, 11, 556972. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Lamers, J.; van der Meer, T.; Testerink, C. How plants sense and respond to stressful environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhao, Y.; Zhu, J.K. Thriving under stress: How plants balance growth and the stress response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Zhou, J.M.; Zhang, Y. Plant immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bürger, M.; Chory, J. Stressed out about hormones: How plants orchestrate immunity. Cell Host Microbe 2019, 26, 163–172. [Google Scholar] [CrossRef]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.J. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [Green Version]
- Zandalinas, S.I.; Fritschi, F.B.; Mittler, R. Signal transduction networks during stress combination. J. Exp. Bot. 2020, 71, 1734–1741. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Meng, X.; Shan, L.; He, P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 2016, 19, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H. Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Clinton, M.; Qi, G.; Wang, D.; Liu, F.; Fu, Z.Q. Reprogramming and remodeling: Transcriptional and epigenetic regulation of salicylic acid-mediated plant defense. J. Exp. Bot. 2020, 71, 5256–5268. [Google Scholar] [CrossRef]
- Buscaill, P.; Rivas, S. Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 2014, 20, 35–46. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Liu, S.; Somssich, I.E. Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 2017, 38, 1–9. [Google Scholar] [CrossRef]
- Amorim, L.L.B.; da Fonseca Dos Santos, R.; Neto, J.P.B.; Guida-Santos, M.; Crovella, S.; Benko-Iseppon, A.M. Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 2017, 18, 335–351. [Google Scholar] [CrossRef]
- Ng, D.W.; Abeysinghe, J.K.; Kamali, M. Regulating the regulators: The control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef] [Green Version]
- Ashapkin, V.V.; Kutueva, L.I.; Aleksandrushkina, N.I.; Vanyushin, B.F. Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. Int. J. Mol. Sci. 2020, 21, 7457. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.Y.; Catinot, J.; Zimmerli, L. Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 2016, 67, 1231–1241. [Google Scholar] [CrossRef] [Green Version]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanetti, M.E.; Rípodas, C.; Niebel, A. Plant NF-Y transcription factors: Key players in plant-microbe interactions, root devel-opment and adaptation to stress. Biochim. Biophys. Acta 2017, 1860, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Liu, Z.; Aqeel, M.; Zainab, M.; Khan, M.I.; Hussain, A.; Ashraf, M.F.; Li, X.; Weng, Y.; He, S. Basic leucine zipper domain transcription factors: The vanguards in plant immunity. Biotechnol. Lett. 2017, 39, 1779–1791. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.-Y.; Zhuang, J.; Xiong, A.-S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [Green Version]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharoni, A.M.; Nuruzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar]
- Hao, D.; Ohme-Takagi, M.; Sarai, A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element binding factor (ERF domain) in plant. J. Biol. Chem. 1998, 273, 26857–26861. [Google Scholar] [CrossRef] [Green Version]
- Hao, D.; Yamasaki, K.; Sarai, A.; Ohme-Takagi, M. Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 2002, 41, 4202–4208. [Google Scholar] [CrossRef]
- Brown, R.L.; Kazan, K.; McGrath, K.C.; Maclean, D.J.; Manners, J.M. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003, 132, 1020–1032. [Google Scholar] [CrossRef] [Green Version]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [PubMed] [Green Version]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, S.; Vlot, A.C. Ethylene responsive factors in the orchestration of stress responses in monocotyledonous plants. Front. Plant Sci. 2015, 6, 640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuntoli, B.; Perata, P. Group VII ethylene response factors in Arabidopsis: Regulation and physiological roles. Plant Physiol. 2018, 176, 1143–1155. [Google Scholar] [CrossRef] [Green Version]
- Shoji, T.; Yuan, L. ERF gene clusters: Working together to regulate metabolism. Trends Plant Sci. 2021, 26, 23–32. [Google Scholar] [CrossRef]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Kumar, R. The expanding roles of APETALA2/Ethylene Responsive Factors and their potential applications in crop improvement. Br. Funct. Genom. 2018, 18, 240–254. [Google Scholar] [CrossRef]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C. Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR-Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: A review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef]
- Cao, Y.; Song, F.; Goodman, R.; Zheng, Z. Molecular characterization of four rice ethylene-responsive element binding protein genes and their expressions in response to biotic and abiotic stresses. J. Plant Physiol. 2006, 163, 1167–1178. [Google Scholar] [CrossRef]
- Lin, R.; Zhao, W.; Meng, X.; Peng, Y. Molecular cloning and characterization of a rice gene encoding AP2/EREBP-type transcription factor and its expression in response to infection with blast fungus and abiotic stresses. Physiol. Mol. Plant Pathol. 2007, 70, 60–68. [Google Scholar] [CrossRef]
- Liu, D.F.; Chen, X.J.; Liu, J.Q.; Ye, J.C.; Guo, Z.J. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 2012, 63, 3899–3912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.Y.; Lee, S.; Park, H.C.; Chang, C.G.; Cheong, Y.H.; Choi, Y.J.; Han, C.; Lee, S.Y.; Lim, C.O.; Cho, M.J. Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol. Plant-Microbe Interact. 2000, 13, 470–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tezuka, D.; Kawamata, A.; Kato, H.; Saburi, W.; Mori, H.; Imai, R. The rice ethylene response factor OsERF83 positively regulates disease resistance to Magnaporthe oryzae. Plant Physiol. Biochem. 2019, 135, 263–271. [Google Scholar] [CrossRef]
- Cao, Y.F.; Wu, Y.; Zheng, Z.; Song, F.M. Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiol. Mol. Plant Pathol. 2006, 67, 202–211. [Google Scholar] [CrossRef]
- Pillai, S.E.; Kumar, C.; Dasgupta, M.; Kumar, B.K.; Vungarala, S.; Patel, H.K.; Sonti, R.V. Ectopic expression of a cell-wall-degrading enzyme-induced OsAP2/ERF152 leads to resistance against bacterial and fungal infection in Arabidopsis. Phytopathology 2020, 110, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.J.; Wang, C.L.; Liu, P.Q.; Lei, C.L.; Hao, W.; Liu, Y.; Zhao, K. Enhanced rice blast resistance by CRISPR/Cas9-tagarted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar]
- Sahu, A.; Das, A.; Saikia, K.; Barah, P. Temperature differentially modulates the transcriptome response in Oryza sativa to Xanthomonas oryzae pv. oryzae infection. Genomics 2020, 112, 4842–4852. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Moon, B.C.; Kim, J.K.; Kim, C.Y.; Kim, M.C.; Kim, I.H.; Park, C.Y.; Park, B.O.; Koo, S.C.; Yoon, H.W.; et al. BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 2003, 132, 1961–1972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, Y.S.; Chern, M.; Bartley, L.E.; Han, M.; Jung, K.H.; Lee, I.; Walia, H.; Richter, T.; Xu, X.; Cao, P.; et al. Towards establishment of a rice stress response interactome. PLoS Genet. 2011, 7, e1002020. [Google Scholar] [CrossRef]
- Jisha, V.; Dampanaboina, L.; Vadassery, J.; Mithofer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 2015, 10, e0127831. [Google Scholar] [CrossRef]
- Tran, T.T.; Pérez-Quintero, A.L.; Wonni, I.; Carpenter, S.C.D.; Yu, Y.; Wang, L.; Leach, J.E.; Verdier, V.; Cunnac, S.; Bogdanove, A.J.; et al. Functional analysis of African Xanthomonas oryzae pv. oryzae TALomes reveals a new susceptibility gene in bacterial leaf blight of rice. PLoS Pathog. 2018, 14, e1007092. [Google Scholar] [CrossRef]
- Lu, J.; Ju, H.; Zhou, G.; Zhu, C.; Erb, M.; Wang, X.; Wang, P.; Lou, Y. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 2011, 68, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [Green Version]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Schmitz, A.J.; Folsom, J.J.; Jikamaru, Y.; Ronald, P.; Walia, H. SUB1A-mediated submergence tolerance response in rice involves differential regulation of the brassinosteroid pathway. New Phytol. 2013, 198, 1060–1070. [Google Scholar] [CrossRef] [Green Version]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Chen, J.Q.; Meng, X.P.; Zhang, Y.; Xia, M.; Wang, X.P. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 2008, 30, 2191–2198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guan, Y.; Wu, Y.; Chen, H.; Chen, F.; Chu, C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 589–602. [Google Scholar] [CrossRef]
- Cui, M.; Zhang, W.; Zhang, Q.; Xu, Z.; Zhu, Z.; Duan, F.; Wu, R. Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 2011, 49, 1384–1391. [Google Scholar] [CrossRef]
- Mallikarjuna, G.; Mallikarjuna, K.; Reddy, M.K.; Kaul, T. Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol. Lett. 2011, 33, 1689–1697. [Google Scholar] [CrossRef]
- Joo, J.; Choi, H.J.; Lee, Y.H.; Kim, Y.K.; Song, S.I. A transcriptional repressor of the ERF family confers drought tolerance to rice and regulates genes preferentially located on chromosome 11. Planta 2013, 238, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Quan, R.; Pan, X.; Wan, L.; Huang, R. EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 2013, 237, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Chung, P.J.; Park, S.-H.; Redillas, M.C.F.R.; Kim, Y.S.; Suh, J.-W.; Kim, J.-K. Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 2017, 15, 1295–1308. [Google Scholar] [CrossRef] [Green Version]
- Xiong, H.; Yu, J.; Miao, J.; Li, J.; Zhang, H.; Wang, X.; Liu, P.; Zhao, Y.; Jiang, C.; Yin, Z.; et al. Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol. 2018, 178, 451–467. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Pan, W.; Zheng, X.; Cheng, X.; Liu, M.; Ma, H.; Ge, X. OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues. Plant Mol. Biol. 2018, 98, 51–65. [Google Scholar] [CrossRef]
- Lee, D.-K.; Jung, H.; Jang, G.; Jeong, J.S.; Kim, Y.S.; Ha, S.-H.; Choi, Y.D.; Kim, J.-K. Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol. 2016, 172, 575–588. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Guo, X.; Zhang, M.; Wang, X.; Zhao, Y.; Yin, Z.; Zhang, Z.; Wang, Y.; Xiong, H.; Zhang, H.; et al. OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci. 2018, 270, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.; Jung, I.; Shin, S.J.; Park, J.; Rhee, S.; Kim, J.K.; Jung, W.; Kwon, H.-B.; Kim, S. Transcriptional network analysis reveals drought resistance mechanisms of AP2/ERF transgenic rice. Front. Plant Sci. 2017, 8, 1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, S.E.; Bang, S.W.; Kim, S.H.; Seo, J.S.; Yoon, H.B.; Kim, Y.S.; Kim, J. Overexpression of OsERF83, a vascular tissue-specific tran-scription factor gene, confers drought tolerance in rice. Int. J. Mol. Sci. 2021, 22, 7656. [Google Scholar] [CrossRef] [PubMed]
- Park, S.I.; Kwon, H.J.; Cho, M.H.; Song, J.S.; Kim, B.G.; Baek, J.; Kim, S.L.; Ji, H.S.; Kwon, T.; Kim, K.; et al. The OsERF115/AP2EREBP110 transcription factor is involved in the multiple stress tolerance to heat and drought in rice plants. Int. J. Mol. Sci. 2021, 22, 7181. [Google Scholar] [CrossRef]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.-M.; Rothstein, S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Zhang, J.; Zhang, H.; Zhang, Z.; Quan, R.; Zhou, S.-R.; Huang, R. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 2011, 6, e25216. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Yang, D.; Zhou, S.; Gu, J.; Wang, F.; Dong, J.; Huang, R. The ethylene response factor OsERF109 negatively affects ethylene biosynthesis and drought tolerance in rice. Protoplasma 2017, 254, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Chen, S.; Ma, X.; Wei, H.; Chen, C.; Gao, N.; Zou, Y.; Kong, D.; Li, T.; et al. An APETALA2/ethylene responsive factor, OsEBP89 knockout enhances adaptation to direct-seeding on wet land and tolerance to drought stress in rice. Mol. Genet. Genom. 2020, 295, 941–956. [Google Scholar] [CrossRef]
- Huang, L.; Wang, Y.; Wang, W.; Zhao, X.; Qin, Q.; Sun, F.; Hu, F.; Zhao, Y.; Li, Z.; Fu, B.; et al. Characterization of transcription factor gene OsDRAP1 conferring drought tolerance in rice. Front. Plant Sci. 2018, 9, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, J.; Zhao, X.; Yang, S.; Huang, L.; Du, F.; Li, Z.; Zhao, X.; Fu, B.; Wang, W. Overexpression of the transcription factor gene OsSTAP1 increases salt tolerance in iice. Rice 2020, 13, 50. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, L.; Du, F.; Wang, J.; Zhao, X.; Li, Z.; Wang, W.; Xu, J.; Fu, B. Comparative transcriptome and metabolome profiling reveal molecular mechanisms underlying OsDRAP1-mediated salt tolerance in rice. Sci. Rep. 2021, 11, 5166. [Google Scholar] [CrossRef]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of rice transcription factor OsERF19 in response to ABA and salt stress responses. Plant Physiol. Biochem. 2021, 167, 22–30. [Google Scholar] [CrossRef]
- Chen, H.C.; Chien, T.C.; Chen, T.Y.; Chiang, M.H.; Lai, M.H.; Chang, M.C. Overexpression of a novel ERF-X-type transcription factor, OsERF106MZ, reduces shoot growth and tolerance to salinity stress in rice. Rice 2021, 14, 82. [Google Scholar] [CrossRef]
- Serra, T.S.; Figueiredo, D.D.; Cordeiro, A.M.; Almeida, D.M.; Lourenço, T.; Abreu, I.A.; Sebastián, A.; Fernandes, L.; Contreras-Moreira, B.; Oliveira, M.M.; et al. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol. Biol. 2013, 82, 439–455. [Google Scholar] [CrossRef] [Green Version]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.Y.; Zhang, Z.; Peng, R.H.; Xiong, A.S.; Liu, J.G.; Wu, L.J.; Gao, F.; Zhu, H.; Guo, Z.; Yao, Q. Isolation and characterization of a novel cDNA encoding ERF/AP2-type transcription factor OsAP25 from Oryza sativa L. Biotechnol. Lett. 2007, 29, 1293–1299. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Qiang, Y.; Zhong, H.; Zhao, G.; Li, Y. Integrated RNA-seq analysis and meta-QTLs mapping provide insights into cold stress response in rice seedling roots. Int. J. Mol. Sci. 2020, 21, 4615. [Google Scholar] [CrossRef]
- Moon, S.-J.; Min, M.K.; Kim, J.-A.; Kim, D.Y.; Yoon, I.S.; Kwon, T.R.; Byun, M.O.; Kim, B.-G. Ectopic expression of OsDREB1G, a member of the OsDREB1 subfamily, confers cold stress tolerance in rice. Front. Plant Sci. 2019, 10, 297. [Google Scholar] [CrossRef]
- Ogawa, T.; Yamamoto, T.; Khush, G.S.; Mew, T.W.; Kaku, H. Near isogenic lines as international differentials for resistance to bacterial blight of rice. Rice Genet. Newsl. 1988, 5, 106–107. [Google Scholar]
- Young, S.A.; Guo, A.; Guikema, J.A.; White, F.F.; Leach, J.E. Rice cationic peroxidase accumulates in xylem vessels during incompatible interactions with Xanthomonas oryzae pv. oryzae. Plant Physiol. 1995, 107, 1333–1341. [Google Scholar] [CrossRef]
- Wang, H.; Hao, J.; Chen, X.; Hao, Z.; Wang, X.; Lou, Y.; Peng, Y.; Guo, Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 2007, 65, 799–815. [Google Scholar] [CrossRef]
- Kishi-Kaboshi, M.; Okada, K.; Kurimoto, L.; Murakami, S.; Umezawa, T.; Shibuya, N.; Yamane, H.; Miyao, A.; Takatsuji, H.; Takahashi, A.; et al. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010, 63, 599–612. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Yuan, B.; Liu, H.; Li, X.; Xu, C.; Wang, S. Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J. 2010, 64, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, J.; Hu, L.; Zhang, T.; Zhang, G.; Lou, Y. OsMPK3 positively regulates the JA signaling pathway and plant resistance to a chewing herbivore in rice. Plant Cell Rep. 2013, 32, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Jalmi, S.K.; Sinha, A.K. Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRK30 in mediating resistance against Xanthomonas oryzae in rice. Sci. Rep. 2016, 6, 37974. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Chen, M.; Zhang, Y.; Gao, Q.; Noman, A.; Wang, Q.; Li, H.; Lu, J.; Lou, Y. OsMKK3, a stress-responsive protein kinase, positively regulates rice resistance to Nilaparvata lugens via phytohormone dynamics. Int. J. Mol. Sci. 2019, 20, 3023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuruzzaman, M.; Gupta, M.; Zhang, C.; Wang, L.; Xie, W.; Xiong, L.; Zhang, Q.; Lian, X. Sequence and expression analysis of the thioredoxin protein gene family in rice. Mol. Genet. Genom. 2008, 280, 139–151. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, J.Y.; Kim, S.J.; An, K.S.; An, G.; Kim, S.R. Isolation of cold stress-responsive genes in the reproductive organs, and characterization of the OsLti6b gene from rice (Oryza sativa L.). Plant Cell Rep. 2007, 26, 1097–1110. [Google Scholar] [CrossRef]
- Morsy, M.R.; Almutairi, A.M.; Gibbons, J.; Yun, S.J.; de Los Reyes, B.G. The OsLti6 genes encoding low-molecular weight membrane proteins are differentially expressed in rice cultivars with contrasting sensitivity to low temperature. Gene 2005, 344, 171–180. [Google Scholar] [CrossRef]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Overexpression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef]
- Su, C.F.; Wang, Y.C.; Hsieh, T.H.; Lu, C.A.; Tseng, T.H.; Yu, S.M. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010, 153, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yin, C.C.; Ma, B.; Chen, S.Y.; Zhang, J.S. Ethylene signaling in rice and Arabidopsis: New regulators and mechanisms. J. Integr. Plant Biol. 2021, 63, 102–125. [Google Scholar] [CrossRef] [PubMed]
- Beyer, E.M. A potent inhibitor of ethylene action in plants. Plant Physiol. 1976, 58, 268–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Huang, L.; Dai, Y.; Liu, S.; Hong, Y.; Tian, L.; Huang, L.; Cao, Z.; Li, D.; Song, F. Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Front. Plant Sci. 2015, 6, 686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catinot, J.; Huang, J.B.; Huang, P.Y.; Tseng, M.Y.; Chen, Y.L.; Gu, S.Y.; Lo, W.; Wang, L.; Chen, Y.; Zimmerli, L. ETHYLENE RE-SPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate- and ethylene-responsive defence genes. Plant Cell Environ. 2015, 38, 2721–2734. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Yoshida, Y.; Nakajima, K.; Tominaga, M.; Gyohda, A.; Suzuki, H.; Okamoto, T.; Nishimura, T.; Yokotani, N.; Minami, E.; et al. Expression of RSOsPR10 in rice roots is antagonistically regulated by jasmonate/ethylene and salicylic acid via the activator OsERF87 and the repressor OsWRKY76, respectively. Plant Direct 2018, 2, e00049. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.L.; Yang, Y.; He, Z. Roles of plant hormones and their interplay in rice immunity. Mol. Plant 2013, 6, 675–685. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasir, F.; Tian, L.; Chang, C.; Li, X.; Gao, Y.; Tran, L.S.P.; Tian, C. Current understanding of pattern-triggered immunity and hormone-mediated defense in rice (Oryza sativa) in response to Magnaporthe oryzae infection. Semin. Cell Dev. Biol. 2018, 83, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Underwood, W. The plant cell wall: A dynamic barrier against pathogen invasion. Front. Plant Sci. 2012, 3, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhong, S.; Li, G.; Li, Q.; Mao, B.; Deng, Y.; Zhang, H.; Zeng, L.; Song, F.; He, Z. Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res. 2011, 21, 835–848. [Google Scholar] [CrossRef]
- Hu, K.; Cao, J.; Zhang, J.; Xia, F.; Ke, Y.; Zhang, H.; Xie, W.; Liu, H.; Cui, Y.; Cao, Y.; et al. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 2017, 3, 17009. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, Y.; Chen, Y.; Yu, N.; Liaqat, S.; Wu, W.; Chen, D.; Cheng, S.; Wei, X.; Cao, L.; et al. OsPG1 encodes a polygalacturonase that determines cell wall architecture and affects resistance to bacterial blight pathogen in rice. Rice 2021, 14, 36. [Google Scholar] [CrossRef]
- Wang, D.; Yuan, S.; Yin, L.; Zhao, J.; Guo, B.; Lan, J.; Li, X. A missense mutation in the transmembrane domain of CESA9 affects cell wall biosynthesis and plant growth in rice. Plant Sci. 2012, 196, 117–124. [Google Scholar] [CrossRef]
- Song, X.-Q.; Liu, L.-F.; Jiang, Y.-J.; Zhang, B.-C.; Gao, Y.-P.; Liu, X.-L.; Lin, Q.-S.; Ling, H.-Q.; Zhou, Y.-H. Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants. Mol. Plant 2013, 6, 768–780. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287, 110188. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Sun, X.L.; Cao, Y.L.; Yang, Z.F.; Xu, C.G.; Li, X.H.; Wang, S.P.; Zhang, Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004, 37, 517–527. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Peng, Y.L.; Guo, Z.J. Consititutive expression of pathogen-inducible OsWRKY31 enhance disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008, 18, 508–521. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Yang, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of MoSM1, encoding for an immunity-inducing protein from Magnaporthe oryzae, in rice confers broad-spectrum resistance against fungal and bacterial diseases. Sci. Rep. 2017, 7, 41037. [Google Scholar] [CrossRef]
- Peng, Y.L.; Shishiyama, J. Temporal sequence of cytological events in rice leaves affected with Pyricularia oryzae. Can. J. Bot. 1988, 66, 730–735. [Google Scholar] [CrossRef]
- Qi, M.; Yang, Y.N. Quantification of Magnaporthe grisea during infection of rice plants using real-time polymerase chain re-action and northern blot/phosphoimaging analyses. Phytopathology 2002, 92, 870–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, D.Y.; Xiao, J.; Ding, X.H.; Xiong, M.; Cai, M.; Cao, Y.L.; Li, X.; Xu, C.; Wang, S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate-and jasmonate-dependent signaling. Mol. Plant-Microbe Interact. 2007, 20, 492–499. [Google Scholar] [CrossRef] [Green Version]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Kim, S.G.; Kim, K.W.; Park, E.W.; Choi, D. Silicon-induced cell wall fortification of rice leaves: A possible cellular mechanism of enhanced host resistance to blast. Phytopathology 2002, 92, 1095–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Y.; Wang, H.; Gao, Y.; Bi, Y.; Xiong, X.; Yan, Y.; Wang, J.; Li, D.; Song, F. ERF Transcription Factor OsBIERF3 Positively Contributes to Immunity against Fungal and Bacterial Diseases but Negatively Regulates Cold Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 606. https://doi.org/10.3390/ijms23020606
Hong Y, Wang H, Gao Y, Bi Y, Xiong X, Yan Y, Wang J, Li D, Song F. ERF Transcription Factor OsBIERF3 Positively Contributes to Immunity against Fungal and Bacterial Diseases but Negatively Regulates Cold Tolerance in Rice. International Journal of Molecular Sciences. 2022; 23(2):606. https://doi.org/10.3390/ijms23020606
Chicago/Turabian StyleHong, Yongbo, Hui Wang, Yizhou Gao, Yan Bi, Xiaohui Xiong, Yuqing Yan, Jiajing Wang, Dayong Li, and Fengming Song. 2022. "ERF Transcription Factor OsBIERF3 Positively Contributes to Immunity against Fungal and Bacterial Diseases but Negatively Regulates Cold Tolerance in Rice" International Journal of Molecular Sciences 23, no. 2: 606. https://doi.org/10.3390/ijms23020606
APA StyleHong, Y., Wang, H., Gao, Y., Bi, Y., Xiong, X., Yan, Y., Wang, J., Li, D., & Song, F. (2022). ERF Transcription Factor OsBIERF3 Positively Contributes to Immunity against Fungal and Bacterial Diseases but Negatively Regulates Cold Tolerance in Rice. International Journal of Molecular Sciences, 23(2), 606. https://doi.org/10.3390/ijms23020606





