The lncRNAs at X Chromosome Inactivation Center: Not Just a Matter of Sex Dosage Compensation
Abstract
:1. Introduction
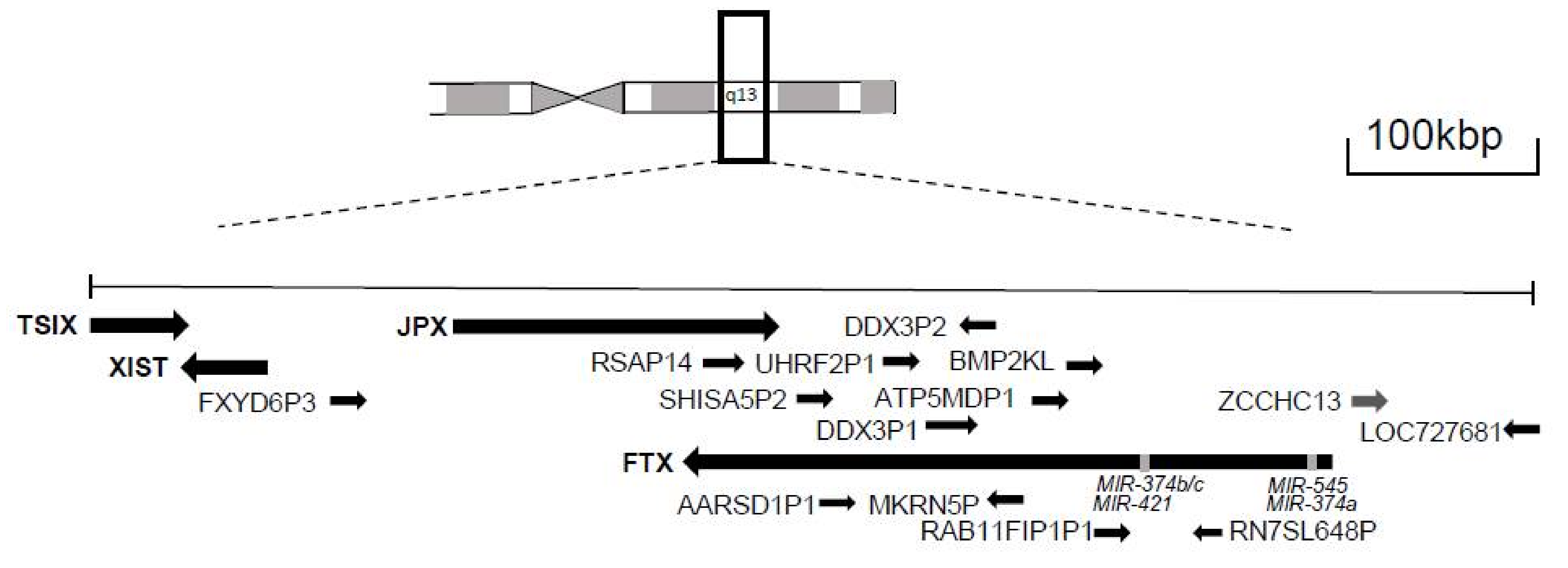
2. Dosage Compensation and X-Inactivation Center
2.1. XIST
2.2. JPX
2.3. FTX
2.4. TSIX
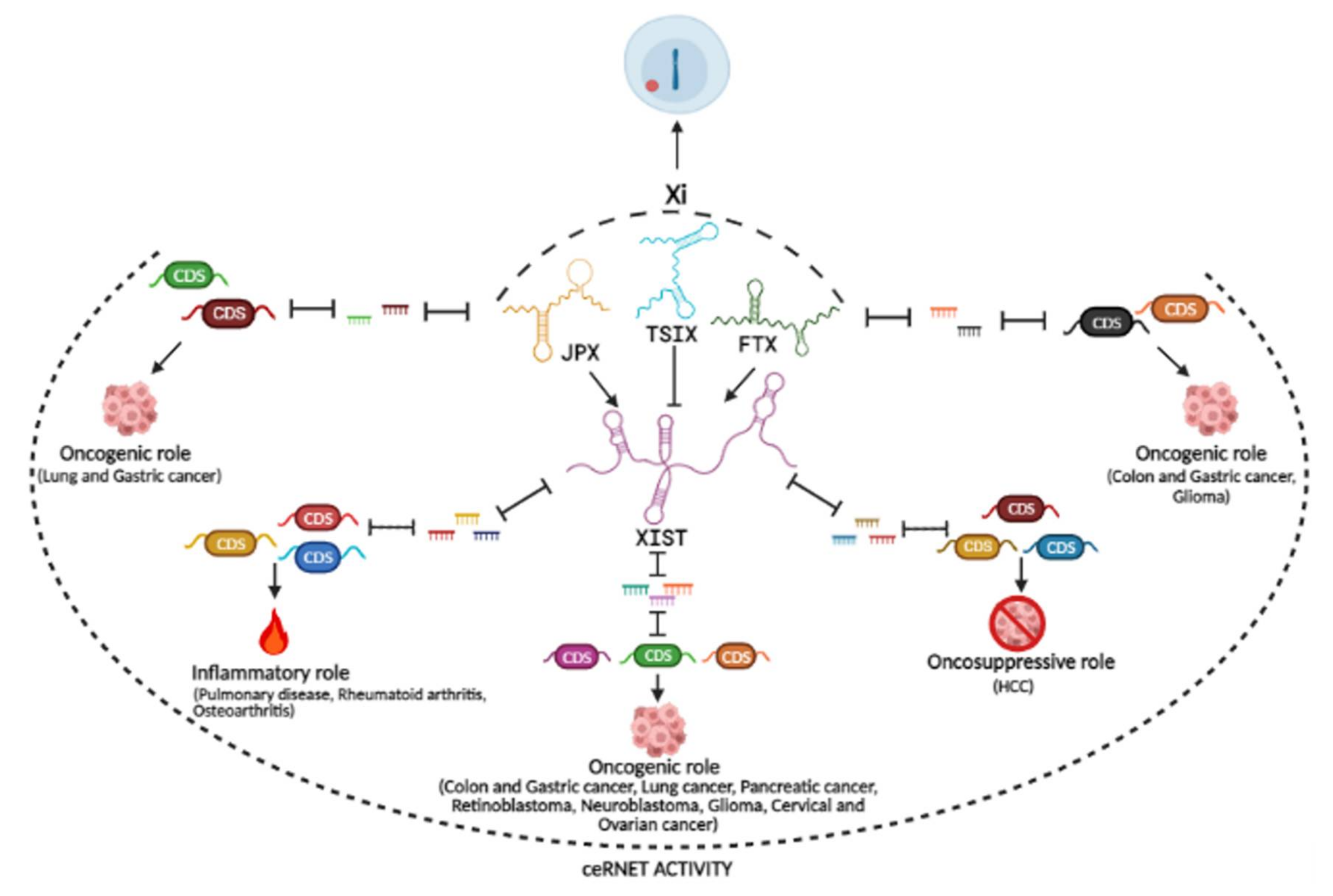
3. ceRNA Activity of the lncRNAs from the XIC
3.1. ceRNETs Involving XIST
| Competing Endogenous RNAs | |||||
|---|---|---|---|---|---|
| lncRNAs | mRNAs | Shared miRNAs | Context | Effect | Ref. |
| XIST | WNT1 | miR-34a | Colon cancer | Oncogenic role | [64] |
| neuropilin-2 | miR-486-5p | Oncogenic role | [65] | ||
| XBP-1 | miR-500a-3p,miR-370-3p, miR-2467-3p, miR-512-3p | Oncogenic role | [67] | ||
| PAX5 | miR-338-3p | Oncogenic role | [105] | ||
| HIF-1A | miR-93-5p | [106] | |||
| SGK1 | miR-124 | Doxorubicin resistance | [66] | ||
| ROR1 | miR-30a-5p | Chemoresistance | [107] | ||
| WEE1 | miR-125b-2-3p | Oncogenic role and chemoresistance | [108] | ||
| EZH2 | miR-101 | Gastric cancer | Oncogenic role | [68] | |
| PXN | miR-132 | Oncogenic role | [69] | ||
| JAK2 | miR-337 | Oncogenic role | [109] | ||
| ZEB2 | miR-367, miR-141 | Oncogenic role | [70] | ||
| Notch-1 | miR-137 | NSCLC | Oncogenic role | [110] | |
| SOD2 | miR-335 | Oncogenic role | [111] | ||
| RING1 | miR-744 | Oncogenic role | [112] | ||
| PAX6 | miR-142-5p | Oncogenic role | [113] | ||
| ATG7 | miR-17 | Cisplatin resistance | [114] | ||
| MDM2 | miR-363-3p | LUAD | Oncogenic role | [115] | |
| ZEB1 | miR-429 | Pancreatic cancer | Oncogenic role | [71] | |
| TGF-β2 | miR-141-3p | Oncogenic role | [72] | ||
| Notch1 | miR-137 | Oncogenic role | [116] | ||
| ZEB1, ZEB2 | miR-101 | Retinoblastoma | Oncogenic role | [73] | |
| STAT3 | miR-124 | Oncogenic role | [117] | ||
| SOX4 | miR-140-5p | Oncogenic role | [118] | ||
| BDNF | miR-191-5p | Oncogenic role | [119] | ||
| STX17 | miR-361-3p | Oncogenic role | [120] | ||
| L1CAM | miR-375 | Neuroblastoma | Oncogenic role | [74] | |
| bFGF (FGF2) | miR-424-5p | Pituitary neuroendocrine tumor | Oncogenic role | [121] | |
| IRS1 | miR-126 | Glioma | Oncogenic role | [75] | |
| SOX4 | miR-133a | Oncogenic role | [79] | ||
| CREB1 | miR-329-3p | Oncogenic role | [78] | ||
| ROCK1 | miR-448 | Oncogenic role | [122] | ||
| SP1, MGMT | miR-29c | Temozolomide chemoresistance | [77] | ||
| MET | miR-34a | Thyroid cancer | Oncogenic role | [80] | |
| CLDN1 | miR-101-3p | Oncogenic role | [123] | ||
| E2F3 | miR-34a-5p | Nasopharyngeal carcinoma | Oncogenic role | [81] | |
| ADAM17 | miR-148a-3p | Oncogenic role | [124] | ||
| NEK5 | miR-381-3p | Oncogenic role | [125] | ||
| RECK | miR-30b | Oncogenic role | [126] | ||
| EZH2 | miR-124 | Laryngeal squamous cell carcinoma | Oncogenic role | [127] | |
| IRS1 | miR-144 | Oncogenic role | [128] | ||
| TRIB2 | miR-125b-5p | Oncogenic role | [129] | ||
| CDK6 | miR-494 | Esophageal cancer | Oncogenic role | [130] | |
| mTOR | miR-375-3p | Osteosarcoma | Oncogenic role | [131] | |
| RAB16 | miR-758 | Oncogenic role | [132] | ||
| ORC1 | miR-140-5p | Cervical cancer | Oncogenic role | [83] | |
| Fus | miR-200a | Oncogenic role | [82] | ||
| SIX1 | miR-889-3p | Oncogenic role | [84] | ||
| FOXP3 | miR-149-3p | Ovarian cancer | Oncogenic role | [85] | |
| BCL2L2 | miR-335 | Oncogenic role | [86] | ||
| ANLN | miR-200c-3p | Breast cancer | Doxorubicin resistance | [133] | |
| GINS2 | miR-23a-3p | Melanoma | Oncogenic role | [134] | |
| MYC | miR-29a | Acute myeloid leukemia | Oncogenic role | [135] | |
| Bcl-w | miR-497 | Extranodal natural killer/T-cell lymphoma | Oncogenic role | [136] | |
| Smad7 | miR-92b | HCC | Tumor suppressor | [87] | |
| SOX6, PTEN | miR-155-5p | Tumor suppressor | [88] | ||
| PDCD4 | miR-497-5p | Tumor suppressor | [89] | ||
| PDK1 | miR-139-5p | Oncogenic role | [137] | ||
| ZEB1/2 | miR-200b-3p | Oncogenic role | [104] | ||
| O-GlcNAc transferase | miR-424-5p | Oncogenic role | [138] | ||
| PIK3CA | miR-320a | Oncogenic role | [139] | ||
| P21 | miR-106b-5p | Renal cell carcinoma | Tumor suppressor | [140] | |
| CUL3 | miR-15a-5p | Acute kidney injury | Pathogenesis promotion | [141] | |
| PDCD4 | miR-142-5p | Acute kidney injury | Pathogenesis promotion | [142] | |
| YAP | miR-194-5p | Wilms tumor | Oncogenic role | [143] | |
| CDKN1A | miR-93-5p | Diabetic nephropathy | Promotion of renal interstitial fibrosis | [144] | |
| TLR4 | miR-217 | Membranous nephropathy | Promotion of podocyte apoptosis and disease development | [145] | |
| SOX-6 | miR-19b | Renal fibrosis | Apoptosis and inflammation promotion | [146] | |
| NOD2 | miR-320 | Atherosclerosis | Promotion of oxidative-LDL-induced cell injury | [147] | |
| TLR4 | miR-370-3p | Pneumonia | Proinflammatory role in LPS-induced injury | [92] | |
| CCL16 | miR-30b-5p | Pneumonia | Proinflammatory role in LPS-induced injury | [148] | |
| IRF2 | miR-204 | Respiratory distress syndrome (mice) | Promotion of LPS-induced acute respiratory distress syndrome | [149] | |
| EGR3 | miR-200c-3p | Chronic obstructive pulmonary disease | Apoptosis and inflammation promotion | [150] | |
| IL-12A | miR-21 | Primary graft dysfunction in lung injury | Induction of neutrophil extracellular trap formation and dysfunction progression | [151] | |
| HMGB3 | miR-101-3p | Bronchopulmonar dysplasia | BP dysplasia promotion | [152] | |
| TLR5 | miR-154-5p | Neuropathic pain development (rats) | Neuropathic pain progression | [153] | |
| SIRT1 | miR-30d-5p | Diabetes | Diabetic peripheral neuropathy attenuation | [154] | |
| Nav1.7 | miR-146a | Satellite glial cell activation and inflammatory pain (rats) | Proinflammatory role | [93] | |
| STAT3 | let-7c-5p | Rheumatoid arthritis | [155] | ||
| Smurf1 | miR-27a | Microglial cells (spinal cord injury, rats) | [94] | ||
| NFAT5 | miR-29c-3p | Epilepsy (rat model) | [156] | ||
| NLRP3 | miRNA-223-3p | Renal calculus (mouse model) | [157] | ||
| OPN | miR-376c-5p | Osteoarthritis | Promotion of inflammatory microenvironment and chondrocyte apoptosis | [95] | |
| CXCR4 | miR-211 | Promotion of chondrocyte apoptosis | [96] | ||
| MMP-13, ADAMTS5 | miR-1277-5p | Promotion of extracellular matrix degradation | [97] | ||
| DNMT3A | miR-149-5p | Promotion of osteoarthritis | [99] | ||
| STAT3 | miR-130a | Promotion of inflammation and extracellular matrix degradation | [112] | ||
| SIRT1 | miR-653-5p | Protective role | [158] | ||
| ZFPM2 | miR-203-3p | Fracture healing | Interferes with proliferation and differentiation of osteoblasts | [159] | |
| AHNAK | miR-17-5p | Cervical ossification of the Posterior longitudinal ligament | Promotion of osteogenic differentiation | [160] | |
| PTEN | miR-19 | Intervertebral disc degeneration | Autophagy induction | [161] | |
| BACE1 | miR-124 | Alzheimer’s disease | Contribution to disease progression | [162] | |
| Sp1 | miR-199a-3p | Parkinson’s disease | Contribution to disease progression | [163] | |
| BACH1 | miR-98 | Cerebral injury | Promotion of neuronal injury | [164] | |
| TIPARP | miR-455-3p | Promotion of neuronal injury | [165] | ||
| FOXO3 | miR-27a-3p | Promotion of neuronal injury | [166] | ||
| IKKβ | miR-96-5p | Aggravation of neuronal apoptosis | [167] | ||
| Itga5 or KLF4 | miR-92a | Alleviation of cerebral vascular injury | [168] | ||
| COL1A1 | miR-29b-3p | Skin fibroblasts (thermal injury) | Promotion of extracellular matrix synthesis, proliferation and migration for wound healing | [169] | |
| HMGB1 | miR-29b | Hepatic stellate cells (alcoholic liver fibrogenesis) | Enhancement ethanol-induced hepatic stellate cell autophagy and activation | [170] | |
| PDE4D | miR-130a-3p | Myocardial infarction | Promotion of myocardial cell apoptosis and inhibition of cell proliferation | [100] | |
| Notch1 | miR-449 | Myocardial infarction | Promotes myocardial cell apoptosis | [101] | |
| S100B | miR-330-3p | Cardiomyocyte hypertrophy | Antihypertrophy effect | [102] | |
| TLR2 | miR-101 | Cardiac hypertrophy | Promotes the progression of cardiac hypertrophy | [103] | |
| FOXP2 | miR-122-5p | Hypoxia-induced H9c2 cardiomyocyte injury | Attenuates hypoxia-induced H9c2 cardiomyocyte injury | [171] | |
| c-Fos | miR-150-5p | Septic myocardial injury | Induces pyroptosis | [172] | |
| PTEN | miR-17 | Stanford type A aortic dissection (TAAD) | Contribution to disease progression | [173] | |
| ELN | miR-29b-3p | Thoracic aortic aneurysm | Aggravation of aortic smooth muscle cell apoptosis | [174] | |
| Arl2 | miR-214-3p | Atrial fibrillation | Suppression of myocardial pyroptosis | [175] | |
| FTX | ZEB2,HOXB9, NOB1, YY1 | miR-215 | Colorectal cancer | Oncogenic role | [176] |
| RBPJ | miR-590-5p | Oncogenic role | [177] | ||
| ZFX | miR-144 | Gastric cancer | Oncogenic role | [178] | |
| SIVA1 | miR-215 | [179] | |||
| AEG-1 | miR-342-3p | Glioma | Oncogenic role | [180] | |
| ALG3 | miR-342-3p | Drug resistance in acute myeloid leukemia | Drug resistance | [181] | |
| c-Met | miR-186 | Bone marrow mesenchymal stem cells | Oncogenic role | [182] | |
| WIF1,PTEN, WNT5A | mir-374a | HCC | Tumor suppressor | [183] | |
| FOXA2 | miR-200a-3p | Lung cancer | Tumor suppressor | [100] | |
| SOX7 | miR-21-5p | Epileptiform hippocampal neurons (rat) | Apoptosis inhibition | [184] | |
| Bcl2l2 | miR-29b-1-5p | Cardiomyocytes (mouse) | Apoptosis inhibition | [185] | |
| Fmr1 | miR-410-3p | Myocardial ischemia/reperfusion injury | Alleviation of hypoxia/reoxygenation-induced cardiomyocyte injury | [186] | |
| JPX | Notch1 | miR-137 | Osteoclasts (osteoporosis) | Osteogenic differentiation inhibition | [187] |
| CCND2 | miR-145-5p | Lung cancer | Oncogenic role | [188] | |
| Twist1 | miR-33a-5p | Oncogenic role | [189] | ||
| CXCR6 | miR-197 | Gastric cancer | Oncogenic role | [190] | |
| CDH2 | miR-944 | Oral squamous cell carcinoma | Oncogenic role | [191] | |
| HIF-1alfa | miR-18a-5p | Intervertebral disc degeneration (human pulposus cells) | Apoptosis inhibition | [192] | |
3.2. ceRNETs Involving FTX
3.3. ceRNETs Involving JPX
4. Implications for X Chromosome Aneuploidy Syndromes
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. Classical and noncanonical functions of miRNAs in cancers. Trends Genet. 2021. [Google Scholar] [CrossRef]
- Alessio, E.; Bonadio, R.S.; Buson, L.; Chemello, F.; Cagnin, S. A Single Cell but Many Different Transcripts: A Journey into the World of Long Non-Coding RNAs. Int. J. Mol. Sci. 2020, 21, 302. [Google Scholar] [CrossRef] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, J.A.; Westhof, E. The dynamic landscapes of RNA architecture. Cell 2009, 136, 604–609. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hennelly, S.P.; Tung, C.S.; Sanbonmatsu, K.Y. Rise of the RNA machines: Exploring the structure of long non-coding RNAs. J. Mol. Biol. 2013, 425, 3731–3746. [Google Scholar] [CrossRef]
- Diederichs, S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014, 30, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Pathania, A.S.; Challagundla, K.B. Exosomal Long Non-coding RNAs: Emerging Players in the Tumor Microenvironment. Mol. Ther. Nucleic Acids. 2020, 23, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [Green Version]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics-challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 9, 1310. [Google Scholar] [CrossRef] [Green Version]
- Di Palo, A.; Siniscalchi, C.; Mosca, N.; Russo, A.; Potenza, N. A Novel ceRNA Regulatory Network Involving the Long Non-Coding Antisense RNA SPACA6P-AS, miR-125a and its mRNA Targets in Hepatocarcinoma Cells. Int. J. Mol. Sci. 2020, 21, 5068. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [Green Version]
- Marshall, E.A.; Stewart, G.L.; Sage, A.P.; Lam, W.L.; Brown, C.J. Beyond sequence homology: Cellular biology limits the potential of XIST to act as a miRNA sponge. PLoS ONE 2019, 14, e0221371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, A.K.L. The Whereabouts of microRNA Actions: Cytoplasm and Beyond. Trends Cell Biol. 2015, 25, 601–610. [Google Scholar] [CrossRef] [Green Version]
- Lyon, M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961, 190, 372–373. [Google Scholar] [CrossRef]
- Furlan, G.; Rougeulle, C. Function and evolution of the long noncoding RNA circuitry orchestrating X-chromosome inactivation in mammals. Wiley Interdiscip. Rev. RNA 2016, 7, 702–722. [Google Scholar] [CrossRef] [PubMed]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrel, L.; Willard, H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005, 434, 400–404. [Google Scholar] [CrossRef]
- Brooks, W.H. X chromosome inactivation and autoimmunity. Clin. Rev. Allergy Immunol. 2010, 39, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Berletch, J.B.; Nguyen, D.K.; Disteche, C.M. X chromosome regulation: Diverse patterns in development, tissues and disease. Nat. Rev. Genet. 2014, 15, 367–378. [Google Scholar] [CrossRef]
- Brown, C.J.; Lafreniere, R.G.; Powers, V.E.; Sebastio, G.; Ballabio, A.; Pettigrew, A.L.; Ledbetter, D.H.; Levy, E.; Craig, I.W.; Willard, H.F. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 1991, 349, 82–84. [Google Scholar] [CrossRef]
- Chureau, C.; Prissette, M.; Bourdet, A.; Barbe, V.; Cattolico, L.; Jones, L.; Eggen, A.; Avner, P.; Duret, L. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002, 12, 894–908. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.L.; Ogawa, Y. Understanding the Complex Circuitry of lncRNAs at the X-inactivation Center and Its Implications in Disease Conditions. Curr. Top. Microbiol. Immunol. 2016, 394, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Weakley, S.M.; Wang, H.; Yao, Q.; Chen, C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J. Surg. 2011, 35, 1751–1756. [Google Scholar] [CrossRef] [Green Version]
- Borsani, G.; Tonlorenzi, R.; Simmler, M.C.; Dandolo, L.; Arnaud, D.; Capra, V.; Grompe, M.; Pizzuti, A.; Muzny, D.; Lawrence, C.; et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature 1991, 351, 325–329. [Google Scholar] [CrossRef]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; Cooper, P.; Smith, S.; McCabe, V.M.; Norris, D.P.; Penny, G.D.; Patel, D.; Rastan, S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 1991, 351, 329–331. [Google Scholar] [CrossRef]
- Chaumeil, J.; Le Baccon, P.; Wutz, A.; Heard, E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006, 20, 2223–2237. [Google Scholar] [CrossRef] [Green Version]
- Clemson, C.M.; Hall, L.L.; Byron, M.; McNeil, J.; Lawrence, J.B. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl. Acad. Sci. USA 2006, 103, 7688–7693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Sousa, L.B.A.; Jonkers, I.; Syx, L.; Dunkel, I.; Chaumeil, J.; Picard, C.; Foret, B.; Chen, C.J.; Lis, J.T.; Heard, E.; et al. Kinetics of Xist-induced gene silencing can be predicted from combinations of epigenetic and genomic features. Genome Res. 2019, 29, 1087–1099. [Google Scholar] [CrossRef] [Green Version]
- Sado, T.; Brockdorff, N. Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013, 368, 20110325. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.J.; Hendrich, B.D.; Rupert, J.L.; Lafrenière, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992, 71, 527–542. [Google Scholar] [CrossRef]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef]
- Nesterova, T.B.; Slobodyanyuk, S.Y.; Elisaphenko, E.A.; Shevchenko, A.I.; Johnston, C.; Pavlova, M.E.; Rogozin, I.B.; Kolesnikov, N.N.; Brockdorff, N.; Zakian, S.M. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001, 11, 833–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wutz, A.; Rasmussen, T.P.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [Green Version]
- Nesterova, T.B.; Wei, G.; Coker, H.; Pintacuda, G.; Bowness, J.S.; Zhang, T.; Almeida, M.; Bloechl, B.; Moindrot, B.; Carter, E.J.; et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 2019, 10, 3129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colognori, D.; Sunwoo, H.; Kriz, A.J.; Wang, C.-Y.; Lee, J.T. Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X. Mol. Cell 2019, 74, 101–117.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678, Erratum in Cell 2012, 149, 1647–1648. [Google Scholar] [CrossRef] [Green Version]
- Pintacuda, G.; Wei, G.; Roustan, C.; Kirmizitas, B.A.; Solcan, N.; Cerase, A.; Castello, A.; Mohammed, S.; Moindrot, B.; Nesterova, T.B.; et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol. Cell 2017, 68, 955–969. [Google Scholar] [CrossRef] [Green Version]
- Jeon, Y.; Lee, J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 2011, 146, 119–133. [Google Scholar] [CrossRef] [Green Version]
- Almeida, M.; Pintacuda, G.; Masui, O.; Koseki, Y.; Gdula, M.; Cerase, A.; Brown, D.; Mould, A.; Innocent, C.; Nakayama, M.; et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 2017, 356, 1081–1084. [Google Scholar] [CrossRef]
- Żylicz, J.J.; Bousard, A.; Žumer, K.; Dossin, F.; Mohammad, E.; da Rocha, S.T.; Schwalb, B.; Syx, L.; Dingli, F.; Loew, D.; et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 2019, 176, 182–197.e23. [Google Scholar] [CrossRef] [Green Version]
- Chow, J.; Heard, E. X inactivation and the complexities of silencing a sex chromosome. Curr. Opin. Cell Biol. 2009, 21, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Sun, S.; Lee, J.T. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 2010, 143, 390–403. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Del Rosario, B.C.; Szanto, A.; Ogawa, Y.; Jeon, Y.; Lee, J.T. Jpx RNA activates Xist by evicting CTCF. Cell 2013, 153, 1537–1551. [Google Scholar] [CrossRef] [Green Version]
- Carmona, S.; Lin, B.; Chou, T.; Arroyo, K.; Sun, S. LncRNA Jpx induces Xist expression in mice using both trans and cis mechanisms. PLoS Genet. 2018, 14, e1007378. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, I.; Barakat, T.S.; Achame, E.M.; Monkhorst, K.; Kenter, A.; Rentmeester, E.; Grosveld, F.; Grootegoed, J.A.; Gribnau, J. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell 2009, 139, 999–1011. [Google Scholar] [CrossRef] [Green Version]
- Barakat, T.S.; Loos, F.; van Staveren, S.; Myronova, E.; Ghazvini, M.; Grootegoed, J.A.; Gribnau, J. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol. Cell 2014, 53, 965–978. [Google Scholar] [CrossRef] [Green Version]
- Chureau, C.; Chantalat, S.; Romito, A.; Galvani, A.; Duret, L.; Avner, P.; Rougeulle, C. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 2011, 20, 705–718. [Google Scholar] [CrossRef] [Green Version]
- Furlan, G.; Gutierrez Hernandez, N.; Huret, C.; Galupa, R.; van Bemmel, J.G.; Romito, A.; Heard, E.; Morey, C.; Rougeulle, C. The Ftx Noncoding Locus Controls X Chromosome Inactivation Independently of Its RNA Products. Mol. Cell 2018, 70, 462–472.e8. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.T.; Lu, N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 1999, 99, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Sado, T.; Wang, Z.; Sasaki, H.; Li, E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 2001, 128, 1275–1286. [Google Scholar] [CrossRef]
- Migeon, B.R.; Chowdhury, A.K.; Dunston, J.A.; McIntosh, I. Identification of TSIX, encoding an RNA antisense to human XIST, reveals differences from its murine counterpart: Implications for X inactivation. Am. J. Hum. Genet. 2001, 69, 951–960. [Google Scholar] [CrossRef] [Green Version]
- Migeon, B.R.; Lee, C.H.; Chowdhury, A.K.; Carpenter, H. Species differences in TSIX/Tsix reveal the roles of these genes in X-chromosome inactivation. Am. J. Hum. Genet. 2002, 71, 286–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, P.; Page, D.R.; Avner, P.; Rougeulle, C. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev. 2006, 20, 2787–2792. [Google Scholar] [CrossRef] [Green Version]
- Navarro, P.; Chantalat, S.; Foglio, M.; Chureau, C.; Vigneau, S.; Clerc, P.; Avner, P.; Rougeulle, C. A role for non-coding Tsix transcription in partitioning chromatin domains within the mouse X-inactivation centre. Epigenetics Chromatin. 2009, 2, 8. [Google Scholar] [CrossRef] [Green Version]
- Karner, H.; Webb, C.H.; Carmona, S.; Liu, Y.; Lin, B.; Erhard, M.; Chan, D.; Baldi, P.; Spitale, R.C.; Sun, S. Functional Conservation of LncRNA JPX Despite Sequence and Structural Divergence. J. Mol. Biol. 2020, 432, 283–300. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, G.; Liu, Y. Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway. Gene 2018, 665, 141–148. [Google Scholar] [CrossRef]
- Liu, A.; Liu, L.; Lu, H. LncRNA XIST facilitates proliferation and epithelial-mesenchymal transition of colorectal cancer cells through targeting miR-486-5p and promoting neuropilin-2. J. Cell Physiol. 2019, 234, 13747–13761. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, R.; Yang, D.; Li, J.; Yan, X.; Jin, K.; Li, W.; Liu, X.; Zhao, J.; Shang, W.; et al. Knockdown of Long Non-Coding RNA XIST Inhibited Doxorubicin Resistance in Colorectal Cancer by Upregulation of miR-124 and Downregulation of SGK1. Cell Physiol. Biochem. 2018, 51, 113–128. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Zheng, S. The role of XBP-1-mediated unfolded protein response in colorectal cancer progression-a regulatory mechanism associated with lncRNA-miRNA-mRNA network. Cancer Cell Int. 2021, 21, 488. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Ju, H.Q.; Lu, Y.X.; Chen, L.Z.; Zeng, Z.L.; Zhang, D.S.; Luo, H.Y.; Wang, F.; Qiu, M.Z.; Wang, D.S.; et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J. Exp. Clin. Cancer Res. 2016, 35, 142. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wang, L.; Li, P.; Hu, F.; Cao, Y.; Tang, D.; Ye, G.; Li, H.; Wang, D. Silencing lncRNA XIST exhibits antiproliferative and proapoptotic effects on gastric cancer cells by up-regulating microRNA-132 and down-regulating PXN. Aging 2020, 12, 14469–14481. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wan, L.; Liu, Z.; Xu, G.; Wang, S.; Su, Z.; Zhang, Y.; Zhang, C.; Liu, X.; Lei, Z.; et al. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018, 418, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hong, L.; Yu, D.; Cao, T.; Zhou, Z.; He, S. LncRNA XIST promotes pancreatic cancer migration, invasion and EMT by sponging miR-429 to modulate ZEB1 expression. Int. J. Biochem. Cell Biol. 2019, 113, 17–26. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Y. LncRNA XIST enhanced TGF-β2 expression by targeting miR-141-3p to promote pancreatic cancer cells invasion. Biosci. Rep. 2019, 39, BSR20190332. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Chang, Q.; Zheng, B.; Xu, J.; Li, H.; Wang, R. LncRNA XIST promotes the epithelial to mesenchymal transition of retinoblastoma via sponging miR-101. Eur. J. Pharmacol. 2019, 843, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, X.; Zhao, Y.; Sun, G.; Zhang, J.; Gao, Y.; Liu, Q.; Zhang, W.; Zhu, H. Downregulation of lncRNA XIST Represses Tumor Growth and Boosts Radiosensitivity of Neuroblastoma via Modulation of the miR-375/L1CAM Axis. Neurochem. Res. 2020, 45, 2679–2690. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Luo, C.; Guo, Z. LncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J. Cell Biochem. 2020, 121, 2170–2183. [Google Scholar] [CrossRef]
- Yao, Y.; Ma, J.; Xue, Y.; Wang, P.; Li, Z.; Liu, J.; Chen, L.; Xi, Z.; Teng, H.; Wang, Z.; et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015, 359, 75–86. [Google Scholar] [CrossRef]
- Du, P.; Zhao, H.; Peng, R.; Liu, Q.; Yuan, J.; Peng, G.; Liao, Y. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci. Rep. 2017, 37, BSR20170696. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.P.; Li, H.Q.; Chen, J.X.; Kong, F.G.; Mo, Z.H.; Wang, J.Z.; Huang, K.M.; Li, X.N.; Yan, Y. Overexpression of XIST facilitates cell proliferation, invasion and suppresses cell apoptosis by reducing radio-sensitivity of glioma cells via miR-329-3p/CREB1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3190–3203. [Google Scholar] [CrossRef]
- Luo, C.; Quan, Z.; Zhong, B.; Zhang, M.; Zhou, B.; Wang, S.; Luo, X.; Tang, C. lncRNA XIST promotes glioma proliferation and metastasis through miR-133a/SOX4. Exp. Ther. Med. 2020, 19, 1641–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Deng, H.; Zhao, Y.; Li, C.; Liang, Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J. Exp. Clin. Cancer Res. 2018, 37, 279. [Google Scholar] [CrossRef]
- Song, P.; Ye, L.F.; Zhang, C.; Peng, T.; Zhou, X.H. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene 2016, 592, 8–14. [Google Scholar] [CrossRef]
- Zhu, H.; Zheng, T.; Yu, J.; Zhou, L.; Wang, L. LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed. Pharmacother. 2018, 105, 789–797. [Google Scholar] [CrossRef]
- Chen, X.; Xiong, D.; Ye, L.; Wang, K.; Huang, L.; Mei, S.; Wu, J.; Chen, S.; Lai, X.; Zheng, L.; et al. Up-regulated lncRNA XIST contributes to progression of cervical cancer via regulating miR-140-5p and ORC1. Cancer Cell Int. 2019, 19, 45. [Google Scholar] [CrossRef]
- Liu, X.; Xie, S.; Zhang, J.; Kang, Y. Long Noncoding RNA XIST Contributes to Cervical Cancer Development Through Targeting miR-889-3p/SIX1 Axis. Cancer Biother. Radiopharm. 2020, 35, 640–649. [Google Scholar] [CrossRef]
- Jiang, R.; Zhang, H.; Zhou, J.; Wang, J.; Xu, Y.; Zhang, H.; Gu, Y.; Fu, F.; Shen, Y.; Zhang, G.; et al. Inhibition of long non-coding RNA XIST upregulates microRNA-149-3p to repress ovarian cancer cell progression. Cell Death Dis. 2021, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Wang, N.; Duan, G. Long non-coding RNA XIST regulates ovarian cancer progression via modulating miR-335/BCL2L2 axis. World J. Surg. Oncol. 2021, 19, 165. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.K.; Yang, Y.T.; Ma, X.; Han, B.; Wang, Z.S.; Zhao, Q.Y.; Wu, L.Q.; Qu, Z.Q. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016, 7, e2203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.Q.; Huang, Z.M.; Chen, X.; Wu, F.; Wu, W. XIST Induced by JPX Suppresses Hepatocellular Carcinoma by Sponging miR-155-5p. Yonsei Med. J. 2018, 59, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, Z.; Huang, S.; Zhao, Q.; Huang, C.; Tang, Y.; Sun, C.; Zhang, Z.; Wang, L.; Chen, H.; et al. lncRNA XIST regulates proliferation and migration of hepatocellular carcinoma cells by acting as miR-497-5p molecular sponge and targeting PDCD4. Cancer Cell Int. 2019, 19, 198. [Google Scholar] [CrossRef]
- Guy, J.; Peters, M.G. Liver disease in women: The influence of gender on epidemiology, natural history, and patient outcomes. Gastroenterol. Hepatol. 2013, 9, 633–639. [Google Scholar]
- Ruggieri, A.; Barbati, C.; Malorni, W. Cellular and molecular mechanisms involved in hepatocellular carcinoma gender disparity. Int. J. Cancer 2010, 127, 499–504. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Y.; Gao, G.; Zhou, Z. Knockdown XIST alleviates LPS-induced WI-38 cell apoptosis and inflammation injury via targeting miR-370-3p/TLR4 in acute pneumonia. Cell Biochem. Funct. 2019, 37, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ma, M.; Yu, H.; Yu, H. Inhibition of lncRNA X inactivate-specific transcript ameliorates inflammatory pain by suppressing satellite glial cell activation and inflammation by acting as a sponge of miR-146a to inhibit Nav 1.7. J. Cell Biochem. 2018, 119, 9888–9898. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Lu, F.; Su, Q.; Liu, Z.; Xia, X.; Yan, Z.; Zhou, F.; Qin, R. Knockdown of long noncoding RNA XIST mitigates the apoptosis and inflammatory injury of microglia cells after spinal cord injury through miR-27a/Smurf1 axis. Neurosci. Lett. 2020, 715, 134649. [Google Scholar] [CrossRef]
- Li, L.; Lv, G.; Wang, B.; Kuang, L. XIST/miR-376c-5p/OPN axis modulates the influence of proinflammatory M1 macrophages on osteoarthritis chondrocyte apoptosis. J. Cell Physiol. 2020, 235, 281–293. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, L.; Zheng, Y.; Liu, W.; Xu., Y. Kaempferol attenuates the effects of XIST/miR-130a/STAT3 on inflammation and extracellular matrix degradation in osteoarthritis. Future Med. Chem. 2021, 13, 1451–1464. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lv, G.; Wang, B.; Kuang, L. The role of lncRNA XIST/miR-211 axis in modulating the proliferation and apoptosis of osteoarthritis chondrocytes through CXCR4 and MAPK signaling. Biochem. Biophys. Res. Commun. 2018, 503, 2555–2562. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Y.; Wang, Y.; Huang, X.; Zhao, W.; Zhao, Z. Long non-coding RNA XIST promotes extracellular matrix degradation by functioning as a competing endogenous RNA of miR-1277-5p in osteoarthritis. Int. J. Mol. Med. 2019, 44, 630–642. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, K.; Tang, C.; Shi, Z.; Jing, K.; Zheng, J. Long non-coding RNA XIST contributes to osteoarthritis progression via miR-149-5p/DNMT3A axis. Biomed. Pharmacother. 2020, 128, 110349. [Google Scholar] [CrossRef]
- Zhou, T.; Qin, G.; Yang, L.; Xiang, D.; Li, S. LncRNA XIST regulates myocardial infarction by targeting miR-130a-3p. J. Cell Physiol. 2019, 234, 8659–8667. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, H.Y.; Han, Y.L.; Wang, L.; Zhai, D.D.; Ma, T.; Zhang, M.J.; Liang, C.Z.; Shen, Y. Silence of lncRNA XIST represses myocardial cell apoptosis in rats with acute myocardial infarction through regulating miR-449. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8566–8572. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, X.; Chen, L.; Chen, W.; Zhang, Y.; Chen, J.; Wu, X.; Zhao, Y.; Wu, X.; Sun, G. The long noncoding RNA XIST protects cardiomyocyte hypertrophy by targeting miR-330-3p. Biochem. Biophys. Res. Commun. 2018, 505, 807–815. [Google Scholar] [CrossRef]
- Xiao, L.; Gu, Y.; Sun, Y.; Chen, J.; Wang, X.; Zhang, Y.; Gao, L.; Li, L. The long noncoding RNA XIST regulates cardiac hypertrophy by targeting miR-101. J. Cell Physiol. 2019, 234, 13680–13692. [Google Scholar] [CrossRef]
- Liu, L.; Jiang, H.; Pan, H.; Zhu, X. LncRNA XIST promotes liver cancer progression by acting as a molecular sponge of miR-200b-3p to regulate ZEB1/2 expression. J. Int. Med. Res. 2021, 49, 3000605211016211. [Google Scholar] [CrossRef]
- Li, W.; He, Y.; Cheng, Z. Long noncoding RNA XIST knockdown suppresses the growth of colorectal cancer cells via regulating microRNA-338-3p/PAX5 axis. Eur. J. Cancer Prev. 2021, 30, 132–142. [Google Scholar] [CrossRef]
- Yang, L.G.; Cao, M.Z.; Zhang, J.; Li, X.Y.; Sun, Q.L. LncRNA XIST modulates HIF-1A/AXL signaling pathway by inhibiting miR-93-5p in colorectal cancer. Mol. Genet. Genom. Med. 2020, 8, e1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Wang, Z.; Yu, Q.; Shen, J.; He, W.; Zhou, D.; Yu, Q.; Fan, J.; Gao, S.; Duan, L. Atractylenolide II reverses the influence of lncRNA XIST/miR-30a-5p/ROR1 axis on chemo-resistance of colorectal cancer cells. J. Cell Mol. Med. 2019, 23, 3151–3165. [Google Scholar] [CrossRef]
- Zeng, Z.L.; Lu, J.H.; Wang, Y.; Sheng, H.; Wang, Y.N.; Chen, Z.H.; Wu, Q.N.; Zheng, J.B.; Chen, Y.X.; Yang, D.D.; et al. The lncRNA XIST/miR-125b-2-3p axis modulates cell proliferation and chemotherapeutic sensitivity via targeting Wee1 in colorectal cancer. Cancer Med. 2021, 10, 2423–2441. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Li, J.; Zhou, X.; Cui, L.; Wang, Y. The lncRNA XIST promotes proliferation, migration and invasion of gastric cancer cells by targeting miR-337. Arab J. Gastroenterol. 2020, 21, 199–206. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, G.; Cheng, Z.; Dai, L.; Jia, L.; Jing, X.; Wang, H.; Zhang, R.; Liu, M.; Jiang, T.; et al. Knockdown of LncRNA-XIST Suppresses Proliferation and TGF-β1-Induced EMT in NSCLC Through the Notch-1 Pathway by Regulation of miR-137. Genet. Test Mol. Biomark. 2018, 22, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yao, L.; Zhang, M.; Jiang, J.; Yang, M.; Wang, Y. Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging 2019, 11, 7830–7846. [Google Scholar] [CrossRef]
- Wang, J.; Cai, H.; Dai, Z.; Wang, G. Down-regulation of lncRNA XIST inhibits cell proliferation via regulating miR-744/RING1 axis in non-small cell lung cancer. Clin. Sci. 2019, 133, 1567–1579, Erratum in Clin. Sci. 2019, 133, 1825. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Xing, W.; Cheng, J.; Yu, Y. Knockdown of lncRNA XIST Suppresses Cell Tumorigenicity in Human Non-Small Cell Lung Cancer by Regulating miR-142-5p/PAX6 Axis. Onco Targets Ther. 2020, 13, 4919–4929. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zu, Y.; Fu, X.; Deng, Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol. Rep. 2017, 38, 3347–3354. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; Chen, B.; Wei, X.; Peng, J.; Ma, K.; Duan, S.; He, J. Long non-coding RNA XIST expedites lung adenocarcinoma progression through upregulating MDM2 expression via binding to miR-363-3p. Thorac. Cancer 2020, 11, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.J.; Pan, Y.H.; Wang, D.W.; You, D. Long non-coding RNA XIST promotes cell proliferation of pancreatic cancer through miR-137 and Notch1 pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12161–12170. [Google Scholar] [CrossRef]
- Hu, C.; Liu, S.; Han, M.; Wang, Y.; Xu, C. Knockdown of lncRNA XIST inhibits retinoblastoma progression by modulating the miR-124/STAT3 axis. Biomed. Pharmacother. 2018, 107, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, D.; Sheng, Y.; Guo, H.; Meng, F.; Song, T. XIST promotes cell proliferation and invasion by regulating miR-140-5p and SOX4 in retinoblastoma. World J. Surg. Oncol. 2020, 18, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Fu, Z.; Gao, X.; Wang, R.; Li, Q. Long non-coding RNA XIST promotes retinoblastoma cell proliferation, migration, and invasion by modulating microRNA-191-5p/brain derived neurotrophic factor. Bioengineered 2021, 12, 1587–1598. [Google Scholar] [CrossRef]
- Yang, L.L.; Li, Q.; Zhang, X.; Cao, T. Long non-coding RNA XIST confers aggressive progression via miR-361-3p/STX17 in retinoblastoma cells. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10433–10444. [Google Scholar] [CrossRef]
- Zhou, K.; Li, S.; Du, G.; Fan, Y.; Wu, P.; Sun, H.; Zhang, T. LncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. OncoTargets Ther. 2019, 12, 7095–7109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.L.; Han, P.H.; Pan, D.Q.; Chen, G.; Lu, X.H.; Li, J. LncRNA XIST regulates cell proliferation, migration and invasion of glioblastoma via regulating miR-448 and ROCK1. J. Biol. Regul. Homeost. Agents 2020, 34, 2049–2058. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.L.; Liang, Y.; Cao, Y.; Liu, L.; Li, J.; Shi, G.Q. LncRNA XIST Promotes Migration and Invasion of Papillary Thyroid Cancer Cell by Modulating MiR-101-3p/CLDN1 Axis. Biochem. Genet. 2021, 59, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Tan, S.; Song, L.; Song, L.; Wang, Y. LncRNA XIST knockdown suppresses the malignancy of human nasopharyngeal carcinoma through XIST/miRNA-148a-3p/ADAM17 pathway in vitro and in vivo. Biomed. Pharmacother. 2020, 121, 109620. [Google Scholar] [CrossRef]
- Zhao, C.H.; Bai, X.F.; Hu, X.H. Knockdown of lncRNA XIST inhibits hypoxia-induced glycolysis, migration and invasion through regulating miR-381-3p/NEK5 axis in nasopharyngeal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2505–2517. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, M.; Qu, H. lncRNA XIST regulates cell proliferation, migration and invasion via regulating miR-30b and RECK in nasopharyngeal carcinoma. Oncol. Lett. 2021, 21, 256. [Google Scholar] [CrossRef]
- Xiao, D.; Cui, X.; Wang, X. Long noncoding RNA XIST increases the aggressiveness of laryngeal squamous cell carcinoma by regulating miR-124-3p/EZH2. Exp. Cell Res. 2019, 381, 172–178. [Google Scholar] [CrossRef]
- Cui, C.L.; Li, Y.N.; Cui, X.Y.; Wu, X. lncRNA XIST promotes the progression of laryngeal squamous cell carcinoma by sponging miR-144 to regulate IRS1 expression. Oncol. Rep. 2020, 43, 525–535. [Google Scholar] [CrossRef]
- Liu, C.; Lu, Z.; Liu, H.; Zhuang, S.; Guo, P. LncRNA XIST promotes the progression of laryngeal squamous cell carcinoma via sponging miR-125b-5p to modulate TRIB2. Biosci. Rep. 2020, 40, BSR20193172. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Hu, X.; Wu, Y.; Cong, L.; He, X.; Lu, J.; Feng, J.; Liu, D. Long non-coding RNA XIST promotes the development of esophageal cancer by sponging miR-494 to regulate CDK6 expression. Biomed. Pharmacother. 2019, 109, 2228–2236. [Google Scholar] [CrossRef]
- Sun, X.; Wei, B.; Peng, Z.H.; Fu, Q.L.; Wang, C.J.; Zheng, J.C.; Sun, J.C. Knockdown of lncRNA XIST suppresses osteosarcoma progression by inactivating AKT/mTOR signaling pathway by sponging miR-375-3p. Int. J. Clin. Exp. Pathol. 2019, 12, 1507–1517. [Google Scholar]
- Liu, W.; Long, Q.; Zhang, L.; Zeng, D.; Hu, B.; Zhang, W.; Liu, S.; Deng, S.; Chen, L. Long non-coding RNA X-inactive specific transcript promotes osteosarcoma metastasis via modulating microRNA-758/Rab16. Ann. Transl. Med. 2021, 9, 841. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, F.; Xiang, Z.; Huang, T.; Zhou, W.B. LncRNA XIST promotes chemoresistance of breast cancer cells to doxorubicin by sponging miR-200c-3p to upregulate ANLN. Clin. Exp. Pharmacol. Physiol. 2020, 47, 1464–1472. [Google Scholar] [CrossRef]
- Hao, Y.Q.; Liu, K.W.; Zhang, X.; Kang, S.X.; Zhang, K.; Han, W.; Li, L.; Li, Z.H. GINS2 was regulated by lncRNA XIST/miR-23a-3p to mediate proliferation and apoptosis in A375 cells. Mol. Cell Biochem. 2021, 476, 1455–1465. [Google Scholar] [CrossRef]
- Wang, C.; Li, L.; Li, M.; Wang, W.; Liu, Y.; Wang, S. Silencing long non-coding RNA XIST suppresses drug resistance in acute myeloid leukemia through down-regulation of MYC by elevating microRNA-29a expression. Mol. Med. 2020, 26, 114. [Google Scholar] [CrossRef]
- Liu, Q.; Ran, R.; Wu, Z.; Li, X.; Zeng, Q.; Xia, R.; Wang, Y. Long Non-coding RNA X-Inactive Specific Transcript Mediates Cell Proliferation and Intrusion by Modulating the miR-497/Bcl-w Axis in Extranodal Natural Killer/T-cell Lymphoma. Front. Cell Dev. Biol. 2020, 8, 599070. [Google Scholar] [CrossRef]
- Mo, Y.; Lu, Y.; Wang, P.; Huang, S.; He, L.; Li, D.; Li, F.; Huang, J.; Lin, X.; Li, X.; et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour. Biol. 2017, 39, 1010428317690999. [Google Scholar] [CrossRef] [Green Version]
- Ning, D.; Chen, J.; Du, P.; Liu, Q.; Cheng, Q.; Li, X.; Zhang, B.; Chen, X.; Jiang, L. The crosstalk network of XIST/miR-424-5p/OGT mediates RAF1 glycosylation and participates in the progression of liver cancer. Liver Int. 2021, 41, 1933–1944. [Google Scholar] [CrossRef]
- Yang, L.; Xie, F.; Xu, W.; Xu, T.; Ni, Y.; Tao, X.; Zang, Y.; Jin, J. Long non-coding RNA XIST accelerates hepatic carcinoma progression by targeting the microRNA-320a/PIK3CA axis. Oncol. Lett. 2021, 22, 801. [Google Scholar] [CrossRef]
- Sun, K.; Jia, Z.; Duan, R.; Yan, Z.; Jin, Z.; Yan, L.; Li, Q.; Yang, J. Long non-coding RNA XIST regulates miR-106b-5p/P21 axis to suppress tumor progression in renal cell carcinoma. Biochem. Biophys. Res. Commun. 2019, 510, 416–420. [Google Scholar] [CrossRef]
- Xu, G.; Mo, L.; Wu, C.; Shen, X.; Dong, H.; Yu, L.; Pan, P.; Pan, K. The miR-15a-5p-XIST-CUL3 regulatory axis is important for sepsis-induced acute kidney injury. Ren. Fail. 2019, 41, 955–966. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.; Li, W.; Ji, T.; Li, X.; Qu, X.; Feng, L.; Zhu, Y.; Qi, Y.; Zhu, C.; Bai, S. Downregulation of XIST ameliorates acute kidney injury by sponging miR-142-5p and targeting PDCD4. J. Cell Physiol. 2020, 235, 8852–8863. [Google Scholar] [CrossRef]
- He, X.; Luo, X.; Dong, J.; Deng, X.; Liu, F.; Wei, G. Long Non-Coding RNA XIST Promotes Wilms Tumor Progression Through the miR-194-5p/YAP Axis. Cancer Manag. Res. 2021, 13, 3171–3180. [Google Scholar] [CrossRef]
- Yang, J.; Shen, Y.; Yang, X.; Long, Y.; Chen, S.; Lin, X.; Dong, R.; Yuan, J. Silencing of long noncoding RNA XIST protects against renal interstitial fibrosis in diabetic nephropathy via microRNA-93-5p-mediated inhibition of CDKN1A. Am. J. Physiol. Renal. Physiol. 2019, 317, F1350–F1358. [Google Scholar] [CrossRef]
- Jin, L.W.; Pan, M.; Ye, H.Y.; Zheng, Y.; Chen, Y.; Huang, W.W.; Xu, X.Y.; Zheng, S.B. Down-regulation of the long non-coding RNA XIST ameliorates podocyte apoptosis in membranous nephropathy via the miR-217-TLR4 pathway. Exp. Physiol. 2019, 104, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Xia, W.P.; Chen, X.; Ru, F.; He, Y.; Liu, P.H.; Gan, Y.; Zhang, B.; Li, Y.; Dai, G.Y.; Jiang, Z.X.; et al. Knockdown of lncRNA XIST inhibited apoptosis and inflammation in renal fibrosis via microRNA-19b-mediated downregulation of SOX6. Mol. Immunol. 2021, 139, 87–96. [Google Scholar] [CrossRef]
- Xu, X.; Ma, C.; Liu, C.; Duan, Z.; Zhang, L. Knockdown of long noncoding RNA XIST alleviates oxidative low-density lipoprotein-mediated endothelial cells injury through modulation of miR-320/NOD2 axis. Biochem. Biophys. Res. Commun. 2018, 503, 586–592. [Google Scholar] [CrossRef]
- Xu, J.; Li, H.; Lv, Y.; Zhang, C.; Chen, Y.; Yu, D. Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway. Open Life Sci. 2021, 16, 108–127. [Google Scholar] [CrossRef]
- Wang, S.; Cao, F.; Gu, X.; Chen, J.; Xu, R.; Huang, Y.; Ying, L. LncRNA XIST, as a ceRNA of miR-204, aggravates lipopolysaccharide-induced acute respiratory distress syndrome in mice by upregulating IRF2. Int. J. Clin. Exp. Pathol. 2019, 12, 2425–2434. [Google Scholar]
- Chen, P.; Jiang, P.; Chen, J.; Yang, Y.; Guo, X. XIST promotes apoptosis and the inflammatory response in CSE-stimulated cells via the miR-200c-3p/EGR3 axis. BMC Pulm. Med. 2021, 21, 215. [Google Scholar] [CrossRef]
- Li, J.; Wei, L.; Han, Z.; Chen, Z.; Zhang, Q. Long non-coding RNA X-inactive specific transcript silencing ameliorates primary graft dysfunction following lung transplantation through microRNA-21-dependent mechanism. EBioMedicine 2020, 52, 102600. [Google Scholar] [CrossRef] [Green Version]
- Yuan, W.; Liu, X.; Zeng, L.; Liu, H.; Cai, B.; Huang, Y.; Tao, X.; Mo, L.; Zhao, L.; Gao, C. Silencing of Long Non-Coding RNA X Inactive Specific Transcript (Xist) Contributes to Suppression of Bronchopulmonary Dysplasia Induced by Hyperoxia in Newborn Mice via microRNA-101-3p and the transforming growth factor-beta 1 (TGF-β1)/Smad3 Axis. Med. Sci. Monit. 2020, 26, e922424. [Google Scholar] [CrossRef]
- Wei, M.; Li, L.; Zhang, Y.; Zhang, Z.J.; Liu, H.L.; Bao, H.G. LncRNA X inactive specific transcript contributes to neuropathic pain development by sponging miR-154-5p via inducing toll-like receptor 5 in CCI rat models. J. Cell Biochem. 2018, 120, 1271–1281. [Google Scholar] [CrossRef]
- Liu, B.Y.; Li, L.; Bai, L.W.; Xu, C.S. Long Non-coding RNA XIST Attenuates Diabetic Peripheral Neuropathy by Inducing Autophagy Through MicroRNA-30d-5p/sirtuin1 Axis. Front. Mol. Biosci. 2021, 8, 655157. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Xiu, D.H.; Jiang, J.L.; Liu, G.F. Long non-coding RNA XIST binding to let-7c-5p contributes to rheumatoid arthritis through its effects on proliferation and differentiation of osteoblasts via regulation of STAT3. J. Clin. Lab. Anal. 2020, 34, e23496. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, H.; Chen, Z.; Hu, X.; Wu, T.; Liu, W. Long Noncoding RNA X-Inactive-Specific Transcript Promotes the Secretion of Inflammatory Cytokines in LPS Stimulated Astrocyte Cell Via Sponging miR-29c-3p and Regulating Nuclear Factor of Activated T cell 5 Expression. Front. Endocrinol. 2021, 12, 573143. [Google Scholar] [CrossRef]
- Lv, P.; Liu, H.; Ye, T.; Yang, X.; Duan, C.; Yao, X.; Li, B.; Tang, K.; Chen, Z.; Liu, J. XIST Inhibition Attenuates Calcium Oxalate Nephrocalcinosis-Induced Renal Inflammation and Oxidative Injury via the miR-223/NLRP3 Pathway. Oxid. Med. Cell Longev. 2021, 2021, 1676152. [Google Scholar] [CrossRef]
- Lian, L.P.; Xi, X.Y. Long non-coding RNA XIST protects chondrocytes ATDC5 and CHON-001 from IL-1β-induced injury via regulating miR-653-5p/SIRT1 axis. J. Biol. Regul. Homeost. Agents 2020, 34, 379–391. [Google Scholar] [CrossRef]
- Niu, S.; Xiang, F.; Jia, H. Downregulation of lncRNA XIST promotes proliferation and differentiation, limits apoptosis of osteoblasts through regulating miR-203-3p/ZFPM2 axis. Connect. Tissue Res. 2020, 24, 381–392. [Google Scholar] [CrossRef]
- Chen, W.; Li, S.; Zhang, F. Role of lncRNA XIST/microRNA-19/PTEN network in autophagy of nucleus pulposus cells in intervertebral disc degeneration via the PI3K/Akt signaling pathway. Cell Cycle 2021, 20, 1629–1641. [Google Scholar] [CrossRef]
- Liao, X.; Tang, D.; Yang, H.; Chen, Y.; Chen, D.; Jia, L.; Yang, L.; Chen, X. Long Non-coding RNA XIST May Influence Cervical Ossification of the Posterior Longitudinal Ligament Through Regulation of miR-17-5P/AHNAK/BMP2 Signaling Pathway. Calcif. Tissue Int. 2019, 105, 670–680. [Google Scholar] [CrossRef]
- Yue, D.; Guanqun, G.; Jingxin, L.; Sen, S.; Shuang, L.; Yan, S.; Minxue, Z.; Ping, Y.; Chong, L.; Zhuobo, Z.; et al. Silencing of long noncoding RNA XIST attenuated Alzheimer’s disease-related BACE1 alteration through miR-124. Cell Biol. Int. 2020, 44, 630–636. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, M.M.; Liu, M.; Tan, Z.G.; Qin, Q.L.; Jiang, Y.G. LncRNA XIST sponges miR-199a-3p to modulate the Sp1/LRRK2 signal pathway to accelerate Parkinson’s disease progression. Aging 2021, 13, 4115–4137. [Google Scholar] [CrossRef]
- Weng, S.; Wang, S.; Jiang, J. Long Noncoding RNA X-Inactive Specific Transcript Regulates Neuronal Cell Apoptosis in Ischemic Stroke Through miR-98/BACH1 Axis. DNA Cell Biol. 2021, 40, 979–987. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Ma, C.; Zhou, T.; Lu, C.; Ding, L.; Li, L. LncRNA XIST Promoted OGD-Induced Neuronal Injury Through Modulating/miR-455-3p/TIPARP Axis. Neurochem. Res. 2021, 46, 1447–1456. [Google Scholar] [CrossRef]
- Zhang, H.; Xia, J.; Hu, Q.; Xu, L.; Cao, H.; Wang, X.; Cao, M. Long non-coding RNA XIST promotes cerebral ischemia/reperfusion injury by modulating miR-27a-3p/FOXO3 signaling. Mol. Med. Rep. 2021, 24, 566. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, J.K.; Ma, J. Regulation of the long noncoding RNA XIST on the inflammatory polarization of microglia in cerebral infarction. Exp. Ther. Med. 2021, 22, 924. [Google Scholar] [CrossRef]
- Wang, C.; Dong, J.; Sun, J.; Huang, S.; Wu, F.; Zhang, X.; Pang, D.; Fu, Y.; Li, L. Silencing of lncRNA XIST impairs angiogenesis and exacerbates cerebral vascular injury after ischemic stroke. Mol. Ther. Nucleic Acids 2021, 26, 148–160. [Google Scholar] [CrossRef]
- Cao, W.; Feng, Y. LncRNA XIST promotes extracellular matrix synthesis, proliferation and migration by targeting miR-29b-3p/COL1A1 in human skin fibroblasts after thermal injury. Biol. Res. 2019, 52, 52. [Google Scholar] [CrossRef]
- Xie, Z.Y.; Wang, F.F.; Xiao, Z.H.; Liu, S.F.; Lai, Y.L.; Tang, S.L. Long noncoding RNA XIST enhances ethanol-induced hepatic stellate cells autophagy and activation via miR-29b/HMGB1 axis. IUBMB Life 2019, 71, 1962–1972. [Google Scholar] [CrossRef]
- Peng, H.; Luo, Y.; Ying, Y. lncRNA XIST attenuates hypoxia-induced H9c2 cardiomyocyte injury by targeting the miR-122-5p/FOXP2 axis. Mol. Cell. Probes 2020, 50, 101500. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.L.; Qin, L.J. The lncRNA XIST/miR-150-5p/c-Fos axis regulates sepsis-induced myocardial injury via TXNIP-modulated pyroptosis. Lab. Investig. 2021, 101, 1118–1129. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, H.; Mai, C.; Qi, Y. Long Noncoding RNA XIST/miR-17/PTEN Axis Modulates the Proliferation and Apoptosis of Vascular Smooth Muscle Cells to Affect Stanford Type A Aortic Dissection. J. Cardiovasc. Pharmacol. 2020, 76, 53–62. [Google Scholar] [CrossRef]
- Liang, K.; Cui, M.; Fu, X.; Ma, J.; Zhang, K.; Zhang, D.; Zhai, S. LncRNA Xist induces arterial smooth muscle cell apoptosis in thoracic aortic aneurysm through miR-29b-3p/Eln pathway. Biomed. Pharmacother. 2021, 137, 111163. [Google Scholar] [CrossRef]
- Yan, B.; Liu, T.; Yao, C.; Liu, X.; Du, Q.; Pan, L. LncRNA XIST shuttled by adipose tissue-derived mesenchymal stem cell-derived extracellular vesicles suppresses myocardial pyroptosis in atrial fibrillation by disrupting miR-214-3p-mediated Arl2 inhibition. Lab. Investig. 2021, 101, 1427–1438. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, J.; Chen, X.; Xu, X.; Cao, G.; Li, H.; Wu, T. LncRNA FTX sponges miR-215 and inhibits phosphorylation of vimentin for promoting colorectal cancer progression. Gene Ther. 2018, 25, 321–330. [Google Scholar] [CrossRef]
- Chen, G.Q.; Liao, Z.M.; Liu, J.; Li, F.; Huang, D.; Zhou, Y.D. LncRNA FTX Promotes Colorectal Cancer Cells Migration and Invasion by miRNA-590-5p/RBPJ Axis. Biochem. Genet. 2021, 59, 560–573. [Google Scholar] [CrossRef]
- Li, H.; Yao, G.; Zhai, J.; Hu, D.; Fan, Y. LncRNA FTX Promotes Proliferation and Invasion of Gastric Cancer via miR-144/ZFX Axis. Onco Targets Ther. 2019, 12, 11701–11713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Wang, X.S.; Tang, B.; Li, P.A.; Wen, Y.; Yu, P.W. Long non-coding RNA FTX promotes gastric cancer progression by targeting miR-215. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3037–3048. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Bi, Y.; Li, J.; Peng, F.; Li, H.; Li, C.; Wang, L.; Ren, F.; Xie, C.; Wang, P.; et al. Long noncoding RNA FTX is upregulated in gliomas and promotes proliferation and invasion of glioma cells by negatively regulating miR-342-3p. Lab Investig. 2017, 97, 447–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Ma, X.; Liu, Q.; Xiao, Y.; Pan, S.; Jia, L. Aberrant mannosylation profile and FTX/miR-342/ALG3-axis contribute to development of drug resistance in acute myeloid leukemia. Cell Death Dis. 2018, 9, 688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Li, X.; Shi, Y.; Chen, H. The long noncoding RNA FTX promotes a malignant phenotype in bone marrow mesenchymal stem cells via the miR-186/c-Met axis. Biomed. Pharmacother. 2020, 131, 110666. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yuan, J.H.; Huang, J.F.; Yang, F.; Wang, T.T.; Ma, J.Z.; Zhang, L.; Zhou, C.C.; Wang, F.; Yu, J.; et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 2016, 35, 5422–5434. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Giri, V.; Cui, Y.; Yin, M.; Xian, Z.; Li, J. LncRNA FTX inhibits hippocampal neuron apoptosis by regulating miR-21-5p/SOX7 axis in a rat model of temporal lobe epilepsy. Biochem. Biophys. Res. Commun. 2019, 512, 79–86. [Google Scholar] [CrossRef]
- Long, B.; Li, N.; Xu, X.X.; Li, X.X.; Xu, X.J.; Guo, D.; Zhang, D.; Wu, Z.H.; Zhang, S.Y. Long noncoding RNA FTX regulates cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2l2. Biochem. Biophys. Res. Commun. 2018, 495, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, L.; Zhang, Y.Z.; Yang, H.Y.; Wang, Y.Y. Long non-coding RNA FTX alleviates hypoxia/reoxygenation-induced cardiomyocyte injury via miR-410-3p/Fmr1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 396–408. [Google Scholar] [CrossRef]
- Yu, Y.; Yao, P.; Wang, Z.; Xie, W. Down-regulation of FTX promotes the differentiation of osteoclasts in osteoporosis through the Notch1 signaling pathway by targeting miR-137. BMC Musculoskelet. Disord. 2020, 21, 456. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Ren, J.; Luo, M.; You, Z.; Fang, Y.; Han, Y.; Li, G.; Liu, H. Long non-coding RNA JPX correlates with poor prognosis and tumor progression in non-small-cell lung cancer by interacting with miR-145-5p and CCND2. Carcinogenesis 2020, 41, 634–645. [Google Scholar] [CrossRef]
- Pan, J.; Fang, S.; Tian, H.; Zhou, C.; Zhao, X.; Tian, H.; He, J.; Shen, W.; Meng, X.; Jin, X.; et al. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol. Cancer 2020, 19, 9. [Google Scholar] [CrossRef]
- Han, X.; Liu, Z. Long non-coding RNA JPX promotes gastric cancer progression by regulating CXCR6 and autophagy via inhibiting miR-197. Mol. Med. Rep. 2021, 23, 60. [Google Scholar] [CrossRef]
- Yao, Y.; Chen, S.; Lu, N.; Yin, Y.; Liu, Z. LncRNA JPX overexpressed in oral squamous cell carcinoma drives malignancy via miR-944/CDH2 axis. Oral Dis. 2021, 27, 924–933. [Google Scholar] [CrossRef]
- Yang, H.; Wang, G.; Liu, J.; Lin, M.; Chen, J.; Fang, Y.; Li, Y.; Cai, W.; Zhan, D. LncRNA JPX regulates proliferation and apoptosis of nucleus pulposus cells by targeting the miR-18a-5p/HIF-1α/Hippo-YAP pathway. Biochem. Biophys. Res. Commun. 2021, 566, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Z.H.; Zhang, B.; Chew, X.H.; Chan, J.J.; Teh, V.; Yang, H.; Kappei, D.; Tay, Y. Systematic Analysis of Intronic miRNAs Reveals Cooperativity within the Multicomponent FTX Locus to Promote Colon Cancer Development. Cancer Res. 2021, 81, 1308–1320. [Google Scholar] [CrossRef]
- Jin, S.; He, J.; Zhou, Y.; Wu, D.; Li, J.; Gao, W. LncRNA FTX activates FOXA2 expression to inhibit non-small-cell lung cancer proliferation and metastasis. J. Cell Mol. Med. 2020, 24, 4839–4849. [Google Scholar] [CrossRef] [Green Version]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [Green Version]
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef]
- Guo, X.; Su, B.; Zhou, Z.; Sha, J. Rapid evolution of mammalian X-linked testis microRNAs. BMC Genom. 2009, 10, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Palo, A.; Siniscalchi, C.; Salerno, M.; Russo, A.; Gravholt, C.H.; Potenza, N. What microRNAs could tell us about the human X chromosome. Cell Mol. Life Sci. 2021, 77, 4069–4080. [Google Scholar] [CrossRef]
- Berglund, A.; Stochholm, K.; Gravholt, C.H. The epidemiology of sex chromosome abnormalities. Am. J. Med. Genet C Semin. Med. Genet. 2020, 184, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.E.; Prakash, S.K.; Andersen, N.H.; Viuff, M.H.; Levitsky, L.L.; Rivera-Davila, M.; Crenshaw, M.L.; Hansen, L.; Colvin, M.K.; Hayes, F.J.; et al. Recognition and management of adults with Turner syndrome: From the transition of adolescence through the senior years. Am. J. Med. Genet. A 2019, 179, 1987–2033. [Google Scholar] [CrossRef] [Green Version]
- Skakkebaek, A.; Viuff, M.; Nielsen, M.M.; Gravholt, C.H. Epigenetics and genomics in Klinefelter syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Stochholm, K.; Juul, S.; Gravholt, C.H. Mortality and incidence in women with 47, XXX and variants. Am. J. Med. Genet. A 2010, 152, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Rao, E.; Weiss, B.; Fukami, M.; Rump, A.; Niesler, B.; Mertz, A.; Muroya, K.; Binder, G.; Kirsch, S.; Winkelmann, M.; et al. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat. Genet. 1997, 16, 54–63. [Google Scholar] [CrossRef]
- Ellison, J.W.; Wardak, Z.; Young, M.F.; Gehron Robey, P.; Laig-Webster, M.; Chiong, W. PHOG, a candidate gene for involvement in the short stature of Turner syndrome. Hum. Mol. Genet. 1997, 6, 1341–1347. [Google Scholar] [CrossRef] [Green Version]
- Ottesen, A.M.; Aksglaede, L.; Garn, I.; Tartaglia, N.; Tassone, F.; Gravholt, C.H.; Bojesen, A.; Sørensen, K.; Jørgensen, N.; Rajpert-De Meyts, E.; et al. Increased number of sex chromosomes affects height in a nonlinear fashion: A study of 305 patients with sex chromosome aneuploidy. Am. J. Med. Genet. A 2010, 152, 1206–1212. [Google Scholar] [CrossRef] [Green Version]
- Skakkebæk, A.; Nielsen, M.M.; Trolle, C.; Vang, S.; Hornshøj, H.; Hedegaard, J.; Wallentin, M.; Bojesen, A.; Hertz, J.M.; Fedder, J.; et al. DNA hypermethylation and differential gene expression associated with Klinefelter syndrome. Sci. Rep. 2018, 8, 13740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trolle, C.; Nielsen, M.M.; Skakkebæk, A.; Lamy, P.; Vang, S.; Hedegaard, J.; Nordentoft, I.; Ørntoft, T.F.; Pedersen, J.S.; Gravholt, C.H. Widespread DNA hypomethylation and differential gene expression in Turner syndrome. Sci. Rep. 2016, 6, 34220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, M.M.; Trolle, C.; Vang, S.; Hornshøj, H.; Skakkebaek, A.; Hedegaard, J.; Nordentoft, I.; Pedersen, J.S.; Gravholt, C.H. Epigenetic and transcriptomic consequences of excess X-chromosome material in 47, XXX syndrome-A comparison with Turner syndrome and 46,XX females. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 279–293. [Google Scholar] [CrossRef]
- Raznahan, A.; Parikshak, N.N.; Chandran, V.; Blumenthal, J.D.; Clasen, L.S.; Alexander-Bloch, A.F.; Zinn, A.R.; Wangsa, D.; Wise, J.; Murphy, D.G.M.; et al. Sex-chromosome dosage effects on gene expression in humans. Proc. Natl. Acad. Sci. USA 2018, 115, 7398–7403. [Google Scholar] [CrossRef] [Green Version]
- Schouten, P.C.; Vollebergh, M.A.; Opdam, M.; Jonkers, M.; Loden, M.; Wesseling, J.; Hauptmann, M.; Linn, S.C. High XIST and Low 53BP1 Expression Predict Poor Outcome after High-Dose Alkylating Chemotherapy in Patients with a BRCA1-like Breast Cancer. Mol. Cancer Ther. 2016, 15, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Bojesen, A.; Juul, S.; Birkebaek, N.H.; Gravholt, C.H. Morbidity in Klinefelter syndrome: A Danish register study based on hospital discharge diagnoses. J. Clin. Endocrinol. Metab. 2006, 91, 1254–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viuff, M.H.; Stochholm, K.; Lin, A.; Berglund, A.; Juul, S.; Gravholt, C.H. Cancer occurrence in Turner syndrome and the effect of sex hormone substitution therapy. Eur. J. Endocrinol. 2021, 184, 79–88. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siniscalchi, C.; Di Palo, A.; Russo, A.; Potenza, N. The lncRNAs at X Chromosome Inactivation Center: Not Just a Matter of Sex Dosage Compensation. Int. J. Mol. Sci. 2022, 23, 611. https://doi.org/10.3390/ijms23020611
Siniscalchi C, Di Palo A, Russo A, Potenza N. The lncRNAs at X Chromosome Inactivation Center: Not Just a Matter of Sex Dosage Compensation. International Journal of Molecular Sciences. 2022; 23(2):611. https://doi.org/10.3390/ijms23020611
Chicago/Turabian StyleSiniscalchi, Chiara, Armando Di Palo, Aniello Russo, and Nicoletta Potenza. 2022. "The lncRNAs at X Chromosome Inactivation Center: Not Just a Matter of Sex Dosage Compensation" International Journal of Molecular Sciences 23, no. 2: 611. https://doi.org/10.3390/ijms23020611
APA StyleSiniscalchi, C., Di Palo, A., Russo, A., & Potenza, N. (2022). The lncRNAs at X Chromosome Inactivation Center: Not Just a Matter of Sex Dosage Compensation. International Journal of Molecular Sciences, 23(2), 611. https://doi.org/10.3390/ijms23020611







