Telomere and Telomerase-Associated Proteins in Endometrial Carcinogenesis and Cancer-Associated Survival
Abstract
:1. Introduction
2. Results
2.1. Patient Population and Demographic Details
2.2. Rationale for Selecting Specific TTAPs
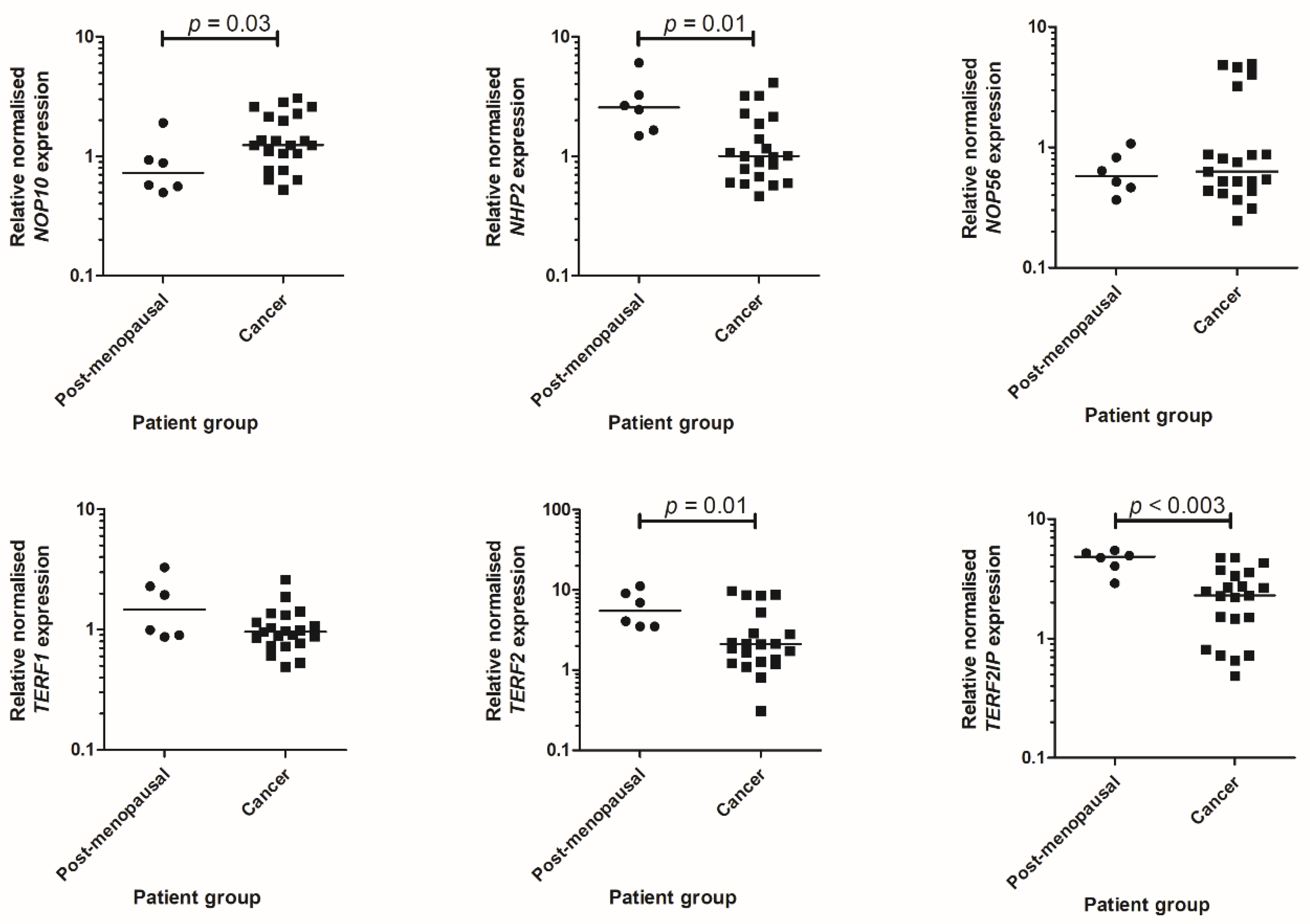
2.3. qPCR Data
2.4. Characterisation of Telomere and Telomerase Associated Protein Expression
2.5. Survival Analysis
2.6. Correlations
3. Discussion
3.1. NOP10
3.2. NHP2
3.3. NOP56
3.4. TERF1
3.5. TERF2 and TER2IP
3.6. Limitations
4. Materials and Methods
4.1. Patient Population and Sample Collection
4.2. qPCR
4.3. Immunohistochemistry
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TTAPs | Telomere and telomerase associated proteins |
| TCGA | The Cancer Genome Atlas |
| EC | Endometrial cancer |
| qPCR | Quantitative polymerase chain reaction |
| IHC | Immunohistochemistry |
References
- Amant, F.; Moerman, P.; Neven, P.; Timmerman, D.; Van Limbergen, E.; Vergote, I. Endometrial cancer. Lancet 2005, 366, 491–505. [Google Scholar] [CrossRef]
- Cancer Research UK Uterine Cancer Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/uterine-cancer (accessed on 10 October 2021).
- Fung-Kee-Fung, M.; Dodge, J.; Elit, L.; Lukka, H.; Chambers, A.; Oliver, T. Follow-up after primary therapy for endometrial cancer: A systematic review. Gynecol. Oncol. 2006, 101, 520–529. [Google Scholar] [CrossRef]
- Adishesh, M.; Hapangama, D.K. Enriching Personalized Endometrial Cancer Research with the Harmonization of Biobanking Standards. Cancers 2019, 11, 1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palm, W.; de Lange, T. How Shelterin Protects Mammalian Telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, E.R.; Blackburn, E.H. An overhanging 3’ terminus is a conserved feature of telomeres. Mol. Cell. Biol. 1989, 9. [Google Scholar] [CrossRef]
- Marión, R.M.; Montero, J.J.; López de Silanes, I.; Graña-Castro, O.; Martínez, P.; Schoeftner, S.; Palacios-Fábrega, J.A.; Blasco, M.A. TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2. eLife 2019, 8, e44656. [Google Scholar] [CrossRef]
- Adishesh, M.; Alnafakh, R.; Baird, D.M.; Jones, R.E.; Simon, S.; Button, L.; Kamal, A.M.; Kirwan, J.; Decruze, S.B.; Drury, J.; et al. Human Endometrial Carcinogenesis Is Associated with Significant Reduction in Long Non-Coding RNA, TERRA. Int. J. Mol. Sci. 2020, 21, 8686. [Google Scholar] [CrossRef]
- Osterhage, J.L.; Friedman, K.L. Chromosome End Maintenance by Telomerase. J. Biol. Chem. 2009, 284, 16061–16065. [Google Scholar] [CrossRef] [Green Version]
- Shammas, M.A. Telomeres, lifestyle, cancer, and aging. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 28–34. [Google Scholar] [CrossRef] [Green Version]
- Kong, C.M.; Lee, X.W.; Wang, X. Telomere shortening in human diseases. FEBS J. 2013, 280, 3180–3193. [Google Scholar] [CrossRef] [PubMed]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse Transcriptase Motifs in the Catalytic Subunit of Telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A Human Telomerase Holoenzyme Protein Required for Cajal Body Localization and Telomere Synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef] [Green Version]
- Blackburn, E.H.; Collins, K. Telomerase: An RNP Enzyme Synthesizes DNA. Cold Spring Harb. Perspect. Biol. 2010, 3, a003558. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, J.R.; Cheng, J.; Collins, K. A Box H/ACA Small Nucleolar RNA-Like Domain at the Human Telomerase RNA 3′ End. Mol. Cell. Biol. 1999, 19, 567–576. [Google Scholar] [CrossRef] [Green Version]
- Pogacić, V.; Dragon, F.; Filipowicz, W. Human H/ACA Small Nucleolar RNPs and Telomerase Share Evolutionarily Conserved Proteins NHP2 and NOP10. Mol. Cell. Biol. 2000, 20, 9028–9040. [Google Scholar] [CrossRef] [Green Version]
- Egan, E.D.; Collins, K. Specificity and Stoichiometry of Subunit Interactions in the Human Telomerase Holoenzyme Assembled In Vivo. Mol. Cell. Biol. 2010, 30, 2775–2786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier, U.T. The many facets of H/ACA ribonucleoproteins. Chromosoma 2005, 114, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Dez, C.; Henras, A.; Faucon, B.; Lafontaine, D.; Caizergues-Ferrer, M.; Henry, Y. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 2001, 29, 598–603. [Google Scholar] [CrossRef] [Green Version]
- Knox, A.A.; McKeegan, K.S.; Debieux, C.M.; Traynor, A.; Richardson, H.; Watkins, N.J. A Weak C’ Box Renders U3 snoRNA Levels Dependent on hU3-55K Binding. Mol. Cell. Biol. 2011, 31, 2404–2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautier, T.; Bergès, T.; Tollervey, D.; Hurt, E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 1997, 17, 7088–7098. [Google Scholar] [CrossRef] [Green Version]
- Lechertier, T.; Grob, A.; Hernandez-Verdun, D.; Roussel, P. Fibrillarin and Nop56 interact before being co-assembled in box C/D snoRNPs. Exp. Cell Res. 2009, 315, 928–942. [Google Scholar] [CrossRef]
- Cowling, V.H.; Turner, S.A.; Cole, M.D. Burkitt’s lymphoma-associated c-Myc mutations converge on a dramatically altered target gene response and implicate Nol5a/Nop56 in oncogenesis. Oncogene 2014, 33, 3519–3527. [Google Scholar] [CrossRef] [Green Version]
- STRING: Functional Protein Association Networks. Available online: https://string-db.org/ (accessed on 14 June 2020).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Ungar, L.; Yosef, N.; Sela, Y.; Sharan, R.; Ruppin, E.; Kupiec, M. A genome-wide screen for essential yeast genes that affect telomere length maintenance. Nucleic Acids Res. 2009, 37, 3840–3849. [Google Scholar] [CrossRef] [Green Version]
- Brien, T.P.; Kallakury, B.V.; Lowry, C.V.; A Ambros, R.; Muraca, P.J.; Malfetano, J.H.; Ross, J.S. Telomerase activity in benign endometrium and endometrial carcinoma. Cancer Res. 1997, 57, 2760–2764. [Google Scholar] [PubMed]
- Lehner, R.; Enomoto, T.; McGregor, J.A.; Shroyer, A.L.; Haugen, B.R.; Pugazhenthi, U.; Shroyer, K.R. Quantitative analysis of telomerase hTERT mRNA and telomerase activity in endometrioid adenocarcinoma and in normal endometrium. Gynecol. Oncol. 2002, 84, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; de Lange, T. Mammalian Telomeres Resemble Fragile Sites and Require TRF1 for Efficient Replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Steensel, B.; de Lange, T. Control of telomere length by the human telomeric protein TRF1. Nature 1997, 385, 740–743. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010, 11, 319–330. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Hapangama, D.K.; Kamal, A.; Saretzki, G. Implications of telomeres and telomerase in endometrial pathology. Hum. Reprod. Update 2016, 23, 166–187. [Google Scholar] [CrossRef] [Green Version]
- Alnafakh, R.A.A.; Adishesh, M.; Button, L.; Saretzki, G.; Hapangama, D.K. Telomerase and Telomeres in Endometrial Cancer. Front. Oncol. 2019, 9, 344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valentijn, A.J.; Saretzki, G.; Tempest, N.; Critchley, H.O.D.; Hapangama, D.K. Human endometrial epithelial telomerase is important for epithelial proliferation and glandular formation with potential implications in endometriosis. Hum. Reprod. 2015, 30, 2816–2828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, M.; Kyo, S.; Takakura, M.; Kanaya, T.; Sagawa, T.; Yamashita, K.; Okada, Y.; Hiyama, E.; Inoue, M. Expression of Telomerase Activity in Human Endometrium Is Localized to Epithelial Glandular Cells and Regulated in a Menstrual Phase-Dependent Manner Correlated with Cell Proliferation. Am. J. Pathol. 1998, 153, 1985–1991. [Google Scholar] [CrossRef]
- Kyo, S.; Takakura, M.; Kohama, T.; Inoue, M. Telomerase activity in human endometrium. Cancer Res. 1997, 57, 610–614. [Google Scholar]
- Gul, I.; Dundar, O.; Bodur, S.; Tunca, Y.; Tutuncu, L. The Status of Telomerase Enzyme Activity in Benign and Malignant Gynaecologic Pathologies. Balk. Med J. 2013, 30, 287–292. [Google Scholar] [CrossRef]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef] [Green Version]
- Bradfield, A.; Button, L.; Drury, J.; Green, D.C.; Hill, C.J.; Hapangama, D.K. Investigating the Role of Telomere and Telomerase Associated Genes and Proteins in Endometrial Cancer. Methods Protoc. 2020, 3, 63. [Google Scholar] [CrossRef]
- Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; Benz, C.C.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Alnafakh, R.; Saretzki, G.; Midgley, A.; Flynn, J.; Kamal, A.M.; Dobson, L.; Natarajan, P.; Stringfellow, H.; Martin-Hirsch, P.; Decruze, S.B.; et al. Aberrant Dyskerin Expression Is Related to Proliferation and Poor Survival in Endometrial Cancer. Cancers 2021, 13, 273. [Google Scholar] [CrossRef]
- The Human Protein Atlas. GAR1—ENDOMETRIAL CANCER—Interactive Survival Scatter Plot & Survival Analysis. Available online: https://www.proteinatlas.org/ENSG00000109534-GAR1/pathology/endometrial+cancer (accessed on 24 December 2021).
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Liu, F.; Pu, X.-Y.; Huang, S.-G.; Xiang, G.-M.; Jiang, D.-N.; Hou, G.; Huang, D.-N. Expression of hPOT1 in HeLa cells and the probability of gene variation of hpot1 Exon14 in endometrial cancer are much higher than in other cancers. Asian Pac. J. Cancer Prev. 2012, 13, 5659–5663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Lange, T. What I got wrong about shelterin. J. Biol. Chem. 2018, 293, 10453–10456. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, P.C.; Panero, J.; Stanganelli, C.; Palau Nagore, V.; Stella, F.; Bezares, R.; Slavutsky, I. Dysregulation of H/ACA ribonucleoprotein components in chronic lymphocytic leukemia. PLoS ONE 2017, 12, e0179883. [Google Scholar] [CrossRef] [Green Version]
- Elsharawy, K.A.; Althobiti, M.; Mohammed, O.J.; Aljohani, A.I.; Toss, M.S.; Green, A.R.; Rakha, E.A. Nucleolar protein 10 (NOP10) predicts poor prognosis in invasive breast cancer. Breast Cancer Res. Treat. 2020, 185, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Kim, S.S.; Yoo, N.J.; Lee, S.H. Expressional analysis of NOLA1, NOLA2, NOLA3 and DKC1, the core proteins in H/ACA riboproteins, in gastric and colorectal cancers. Pathology 2012, 44, 576–577. [Google Scholar] [CrossRef] [PubMed]
- Witkowska, A.; Gumprecht, J.; Głogowska-Ligus, J.; Wystrychowski, G.; Owczarek, A.; Stachowicz, M.; Bocianowska, A.; Nowakowska-Zajdel, E.; Mazurek, U. Expression profile of significant immortalization genes in colon cancer. Int. J. Mol. Med. 2010, 25, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Sorosky, J.I. Endometrial Cancer. Obstet. Gynecol. 2008, 111, 436–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomica, D.; Ramić, S.; Danolić, D.; Knezević, F.; Kolak, T.; Balja, M.P.; Alvir, I.; Mamić, I.; Puljiz, M. A correlation between the expression of estrogen receptors and progesterone receptors in cancer cells and in the myometrium and prognostic factors in endometrial cancer. Coll. Antropol. 2014, 38, 129–134. [Google Scholar] [PubMed]
- Kamal, A.; Tempest, N.; Parkes, C.; Alnafakh, R.; Makrydima, S.; Adishesh, M.; Hapangama, D.K. Hormones and endometrial carcinogenesis. Horm. Mol. Biol. Clin. Investig. 2016, 25, 129–148. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Catez, F.; Berger, C.M.; Perrial, E.; Plesa, A.; Thomas, X.; Mattei, E.; Hayette, S.; Saintigny, P.; Bouvet, P.; et al. Expression Profiling of Ribosome Biogenesis Factors Reveals Nucleolin as a Novel Potential Marker to Predict Outcome in AML Patients. PLoS ONE 2017, 12, e0170160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Human Protein Atlas. NOP56—Pathology. Available online: http://www.proteinatlas.org (accessed on 14 June 2020).
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Li, X.; Wang, W.; Wang, J.; Malovannaya, A.; Xi, Y.; Li, W.; Guerra, R.; Hawke, D.H.; Qin, J.; Chen, J. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol. Syst. Biol. 2015, 11, 775. [Google Scholar] [CrossRef]
- Wan, C.; Borgeson, B.; Phanse, S.; Tu, F.; Drew, K.; Clark, G.W.; Xiong, X.; Kagan, O.; Kwan, J.; Berzginov, A.; et al. Panorama of ancient metazoan macromolecular complexes. Nature 2015, 525, 339–344. [Google Scholar] [CrossRef] [Green Version]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kustatscher, G.; Grabowski, P.; Schrader, T.A.; Passmore, J.B.; Schrader, M.; Rappsilber, J. Co-regulation map of the human proteome enables identification of protein functions. Nat. Biotechnol. 2019, 37, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Sun, L.; Zhang, C.; Zhou, X.L. Expression of telomeric repeat binding factor 1 protein in nonsmall cell lung cancer with human telomerase reverse transcriptase positive. Int. J. Surg. Pathol. 2008, 16, 414–418. [Google Scholar] [CrossRef]
- Lin, X.; Gu, J.; Lu, C.; Spitz, M.R.; Wu, X. Expression of Telomere-Associated Genes as Prognostic Markers for Overall Survival in Patients with Non–Small Cell Lung Cancer. Clin. Cancer Res. 2006, 12, 5720–5725. [Google Scholar] [CrossRef] [Green Version]
- Yamada, M.; Tsuji, N.; Nakamura, M.; Moriai, R.; Kobayashi, D.; Yagihashi, A.; Watanabe, N. Down-regulation of TRF1, TRF2 and TIN2 genes is important to maintain telomeric DNA for gastric cancers. Anticancer Res. 2003, 22, 3303–3307. [Google Scholar]
- Miyachi, K.; Fujita, M.; Tanaka, N.; Sasaki, K.; Sunagawa, M. Correlation between telomerase activity and telomeric-repeat binding factors in gastric cancer. J. Exp. Clin. Cancer Res. 2002, 21, 269–275. [Google Scholar]
- Bejarano, L.; Schuhmacher, A.J.; Méndez, M.; Megias, D.; Blanco-Aparicio, C.; Martínez, S.; Pastor, J.; Squatrito, M.; Blasco, M.A. Inhibition of TRF1 Telomere Protein Impairs Tumor Initiation and Progression in Glioblastoma Mouse Models and Patient-Derived Xenografts. Cancer Cell 2017, 32, 590–607.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Wu, X. Abnormal function of telomere protein TRF2 induces cell mutation and the effects of environmental tumor-promoting factors (Review). Oncol. Rep. 2021, 46, 1–20. [Google Scholar] [CrossRef]
- Wu, M.; Lin, Z.; Li, X.; Xin, X.; An, J.; Zheng, Q.; Yang, Y.; Lu, D. HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Sci. Rep. 2016, 6, 36045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özden, S.; Tiber, P.M.; Ozgen, Z.; Ozyurt, H.; Serakinci, N.; Orun, O. Expression of TRF2 and its prognostic relevance in advanced stage cervical cancer patients. Biol. Res. 2014, 47, 61. [Google Scholar] [CrossRef] [Green Version]
- Teo, H.; Ghosh, S.; Luesch, H.; Ghosh, A.; Wong, E.T.; Malik, N.; Orth, A.; De Jesus, P.; Perry, A.S.; Oliver, J.D.; et al. Telomere-independent Rap1 is an IKK adaptor and regulates NF-κB-dependent gene expression. Nat. Cell Biol. 2010, 12, 758–767. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, C.; Wang, L.; An, G.; Tian, Z.; Meng, L.; Qu, L.; Lian, S.; Shou, C. Repressor activator protein 1–promoted colorectal cell migration is associated with the regulation of Vimentin. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Adishesh, M.; Fyson, A.; Decruze, S.B.; Kirwan, J.; ENITEC Consortium; Werner, H.M.J.; Hapangama, D.K. Harmonisation of biobanking standards in endometrial cancer research. Br. J. Cancer 2017, 117, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Alnafakh, R.; Choi, F.; Bradfield, A.; Adishesh, M.; Saretzki, G.; Hapangama, D.K. Endometriosis Is Associated with a Significant Increase in hTERC and Altered Telomere/Telomerase Associated Genes in the Eutopic Endometrium, an Ex-Vivo and In Silico Study. Biomedicines 2020, 8, 588. [Google Scholar] [CrossRef]
- Maclean, A.; Bunni, E.; Makrydima, S.; Withington, A.; Kamal, A.M.; Valentijn, A.J.; Hapangama, D.K. Fallopian tube epithelial cells express androgen receptor and have a distinct hormonal responsiveness when compared with endometrial epithelium. Hum. Reprod. 2020, 35, 2097–2106. [Google Scholar] [CrossRef]
- Tempest, N.; Jansen, M.; Baker, A.M.; Hill, C.J.; Hale, M.; Magee, D.; Treanor, D.; Wright, N.A.; Hapangama, D.K. Histological 3D reconstruction and in vivo lineage tracing of the human endometrium. J. Pathol. 2020, 251, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Hapangama, D.K.; Drury, J.; Da Silva, L.; Al-Lamee, H.; Earp, A.; Valentijn, A.J.; Edirisinghe, D.P.; Murray, P.A.; Fazleabas, A.T.; Gargett, C.E. Abnormally located SSEA1+/SOX9+ endometrial epithelial cells with a basalis-like phenotype in the eutopic functionalis layer may play a role in the pathogenesis of endometriosis. Hum. Reprod. 2019, 34, 56–68. [Google Scholar] [CrossRef]
- Kamal, A.M.; Bulmer, J.N.; DeCruze, S.B.; Stringfellow, H.F.; Martin-Hirsch, P.; Hapangama, D.K. Androgen receptors are acquired by healthy postmenopausal endometrial epithelium and their subsequent loss in endometrial cancer is associated with poor survival. Br. J. Cancer 2016, 114, 688–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamal, A.; Valentijn, A.; Barraclough, R.; Rudland, P.; Rahmatalla, N.; Martin-Hirsch, P.; Stringfellow, H.; Decruze, S.B.; Hapangama, D.K. High AGR2 protein is a feature of low grade endometrial cancer cells. Oncotarget 2018, 9, 31459–31472. [Google Scholar] [CrossRef] [PubMed]
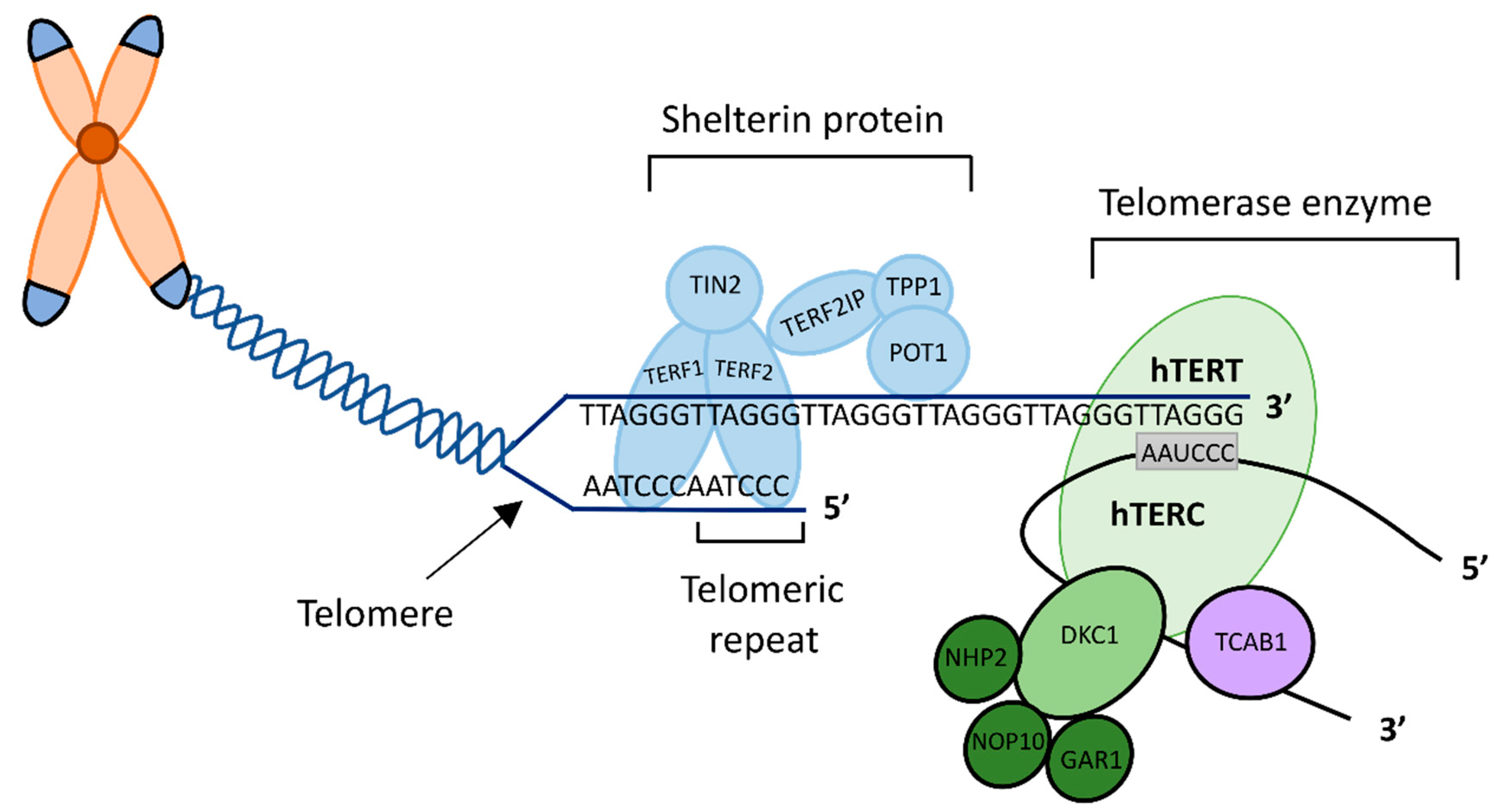



| Patient Group | Number of Patients | Age Median (Range) | BMI Median (Range) | Parity Median (Range) | Smokers Number (%) | HRT Number (%) | Death Number (%) | Recurrence Number (%) |
|---|---|---|---|---|---|---|---|---|
| Pre-Menopausal | 13 | 41 (32–57) | 26.7 (18.9–40.5) | 2 (1–6) | 6 (46%) | n/a | n/a | n/a |
| Post-Menopausal | 27 | 62 (51–85) | 24.9 (17.9–39.6) | 3 (0–5) | 6 (22%) | 1 (4%) | n/a | n/a |
| Cancer | 63 | 68 (37–78) | 28.75 (20.2–54.4) | 2 (0–7) | 3 (5%) | 7 (11%) | 30 (48%) | 30 (48%) |
| Ki67 Correlations | NOP10 | NHP2 | NOP56 | TERF1 | TERF2 | TERF2IP |
|---|---|---|---|---|---|---|
| Number | 51 | 48 | 48 | 36 | 70 | 73 |
| Spearman r | −0.09 | −0.45 | 0.04 | −0.33 | −0.23 | −0.42 |
| 95% confidence interval | −0.36 to 0.20 | −0.66 to −0.19 | −0.25 to 0.33 | −0.60 to 0.01 | −0.45 to 0.01 | −0.60 to−0.21 |
| p value (two-tailed) | 0.54 | 0.001 | 0.77 | 0.05 | 0.06 | <0.0002 |
| NOP10 | NHP2 | NOP56 | TERF1 | TERF2 | TERF2IP | |
|---|---|---|---|---|---|---|
| NOP10 | r | 0.16 | 0.24 | −0.14 | 0.15 | −0.24 |
| p | 0.22 | 0.07 | 0.35 | 0.31 | 0.08 | |
| n | 59 | 59 | 46 | 51 | 54 | |
| NHP2 | 0.16 | r | −0.12 | 0.34 | 0.24 | 0.27 |
| 0.22 | p | 0.39 | 0.02 | 0.11 | 0.05 | |
| 59 | n | 56 | 46 | 48 | 53 | |
| NOP56 | 0.24 | −0.12 | r | 0.13 | 0.53 | 0.24 |
| 0.07 | 0.39 | p | 0.41 | <0.0002 | 0.09 | |
| 59 | 56 | n | 43 | 46 | 52 | |
| TERF1 | −0.14 | 0.34 | 0.13 | r | 0.65 | 0.48 |
| 0.35 | 0.02 | 0.41 | p | <0.0001 | 0.002 | |
| 46 | 46 | 43 | n | 36 | 38 | |
| TERF2 | 0.15 | 0.27 | 0.53 | 0.65 | r | 0.46 |
| 0.31 | 0.05 | <0.0002 | <0.0001 | p | <0.0001 | |
| 51 | 48 | 46 | 36 | n | 76 | |
| TERF2IP | −0.24 | 0.27 | 0.24 | 0.48 | 0.46 | r |
| 0.08 | 0.05 | 0.09 | 0.002 | <0.0001 | p | |
| 54 | 53 | 52 | 38 | 76 | n |
| PR | ERα | ERβ | AR | |
|---|---|---|---|---|
| NOP10 | ||||
| n | 36 | 34 | 34 | 34 |
| Spearman r | −0.03 | 0.25 | 0.06 | −0.08 |
| 95% confidence interval | −0.36 to 0.32 | −0.10 to 0.55 | −0.29 to 0.40 | −0.41 to 0.28 |
| p value (two-tailed) | 0.88 | 0.15 | 0.72 | 0.66 |
| NHP2 | ||||
| n | 33 | 31 | 31 | 31 |
| Spearman r | 0.51 | 0.09 | 0.17 | 0.40 |
| 95% confidence interval | 0.20 to 0.73 | −0.28 to 0.44 | −0.20 to 0.51 | 0.04 to 0.67 |
| p value (two-tailed) | 0.002 | 0.63 | 0.35 | 0.03 |
| NOP56 | ||||
| n | 37 | 35 | 35 | 35 |
| Spearman r | −0.30 | −0.36 | −0.12 | −0.41 |
| 95% confidence interval | −0.57 to 0.04 | −0.62 to −0.02 | −0.44 to 0.23 | −0.66 to −0.08 |
| p value (two-tailed) | 0.07 | 0.03 | 0.49 | 0.02 |
| TERF1 | ||||
| n | 27 | 25 | 25 | 25 |
| Spearman r | −0.03 | −0.02 | −0.39 | −0.10 |
| 95% confidence interval | −0.41 to 0.37 | −0.42 to 0.39 | −0.69 to 0.01 | −0.49 to 0.32 |
| p value (two-tailed) | 0.90 | 0.91 | 0.05 | 0.62 |
| TERF2 | ||||
| n | 44 | 42 | 42 | 42 |
| Spearman r | −0.14 | 0.01 | −0.05 | −0.07 |
| 95% confidence interval | −0.43 to 0.17 | −0.30 to 0.32 | −0.36 to 0.26 | −0.37 to 0.25 |
| p value (two-tailed) | 0.37 | 0.92 | 0.73 | 0.68 |
| TERF2IP | ||||
| n | 49 | 47 | 47 | 47 |
| Spearman r | −0.03 | 0.07 | −0.18 | 0.21 |
| 95% confidence interval | −0.32 to 0.26 | −0.23 to 0.36 | −0.45 to 0.12 | −0.09 to 0.47 |
| p value (two-tailed) | 0.84 | 0.65 | 0.23 | 0.16 |
| Gene Name | Sequences (5′–3′)/Unique Assay ID | Amplicon Length |
|---|---|---|
| IPO8 | Forward: AGGATCAGAGGACAGCACTGCA Reverse: AGGTGAAGCCTCCCTGTTGTTC | 102 |
| PPIA | Forward: AGACAAGGTCCCAAAGAC Reverse: ACCACCTGACACATAAA | 118 |
| MRPL19 | Forward: CAGGAAGAGGACTTGGAGCTAC Reverse: GCTATCATCCAGCCGTTTCTCTA | 137 |
| NHP2 | qHsaCED0037860 | 164 |
| NOP10 | qHsaCID0012562 | 83 |
| NOP56 | qHsaCEP0025699 | 89 |
| TERF1 | qHsaCID0022415 | 100 |
| TERF2 | qHsaCID0015690 | 123 |
| TERF2IP | qHsaCID007952 | 123 |
| Antibody | Clone | Type and Host Species | Supplier | Dilution | Incubation |
|---|---|---|---|---|---|
| NOP10 | EPR8856 | Rabbit monoclonal | Abcam (Cambridge, UK) | 1/2000 | O/N 4 °C |
| NHP2 | H-9 | Mouse monoclonal | Santa Cruz Biotechnology(Insight Bio, Wembley, UK) | 1/8000 | O/N 4 °C |
| 1/2000 | 30 min RT | ||||
| NOP56 | CL2603 | Mouse monoclonal | Abcam (Cambridge, UK) | 1/1000 | O/N 4 °C |
| TERF1 | TRF-78 | Mouse monoclonal | Santa Cruz Biotechnology(Insight Bio, Wembley, UK) | 1/50 | O/N 4 °C |
| TERF2 | B5 | Mouse monoclonal | Santa Cruz Biotechnology(Insight Bio, Wembley, UK) | 1/200 | O/N 4 °C |
| TERF2IP | 4C8/1 | Mouse monoclonal | Santa Cruz Biotechnology(Insight Bio, Wembley, UK) | 1/100 | O/N 4 °C |
| Ki67 | MM1 | Mouse monoclonal | Leica (Milton Keynes, UK) | 1/200 | O/N 4 °C |
| ERα | 6F11 | Mouse monoclonal | Leica (Milton Keynes, UK) | 1/50 | 2 h RT |
| ERβ | PPG5/10 | Mouse monoclonal | Abcam (Cambridge, UK) | 1/50 | O/N 4 °C |
| PR | Pgr636 | Mouse monoclonal | DAKO (Ely, UK) | 1/1000 | 1 h RT |
| AR | AR441 | Mouse monoclonal | DAKO (Ely, UK) | 1/75 | O/N 4 °C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Button, L.; Rogers, B.; Thomas, E.; Bradfield, A.; Alnafakh, R.; Drury, J.; Hapangama, D.K. Telomere and Telomerase-Associated Proteins in Endometrial Carcinogenesis and Cancer-Associated Survival. Int. J. Mol. Sci. 2022, 23, 626. https://doi.org/10.3390/ijms23020626
Button L, Rogers B, Thomas E, Bradfield A, Alnafakh R, Drury J, Hapangama DK. Telomere and Telomerase-Associated Proteins in Endometrial Carcinogenesis and Cancer-Associated Survival. International Journal of Molecular Sciences. 2022; 23(2):626. https://doi.org/10.3390/ijms23020626
Chicago/Turabian StyleButton, Lucy, Bryony Rogers, Emily Thomas, Alice Bradfield, Rafah Alnafakh, Josephine Drury, and Dharani K. Hapangama. 2022. "Telomere and Telomerase-Associated Proteins in Endometrial Carcinogenesis and Cancer-Associated Survival" International Journal of Molecular Sciences 23, no. 2: 626. https://doi.org/10.3390/ijms23020626






