CircRNAs as Potential Blood Biomarkers and Key Elements in Regulatory Networks in Gastric Cancer
Abstract
:1. Introduction
2. Results
2.1. Deregulated circRNAs in GC Tissue
2.2. CircRNAs Are Differentially Expressed in Blood and Are Potentially Less Invasive Biomarkers for Gastric Cancer
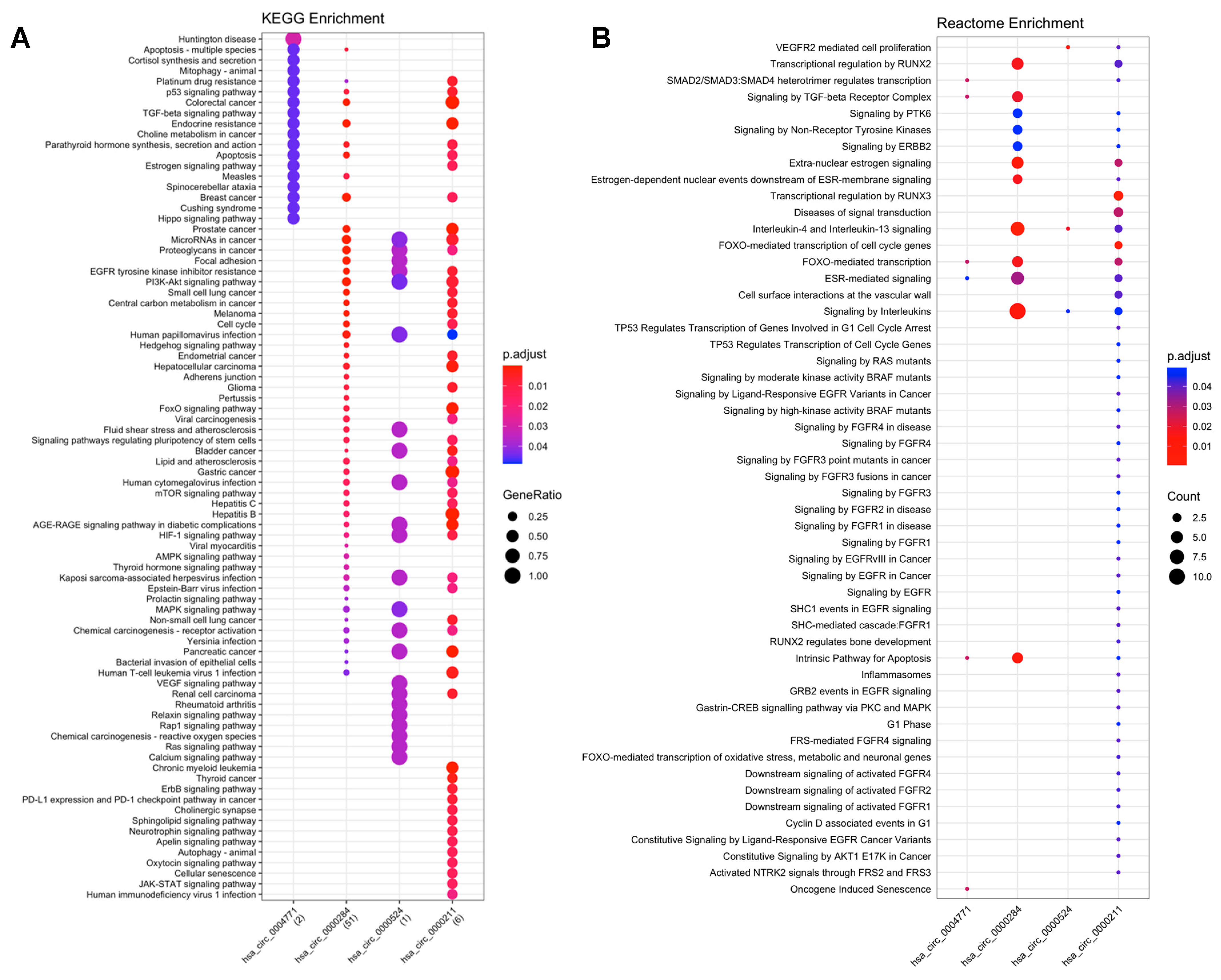
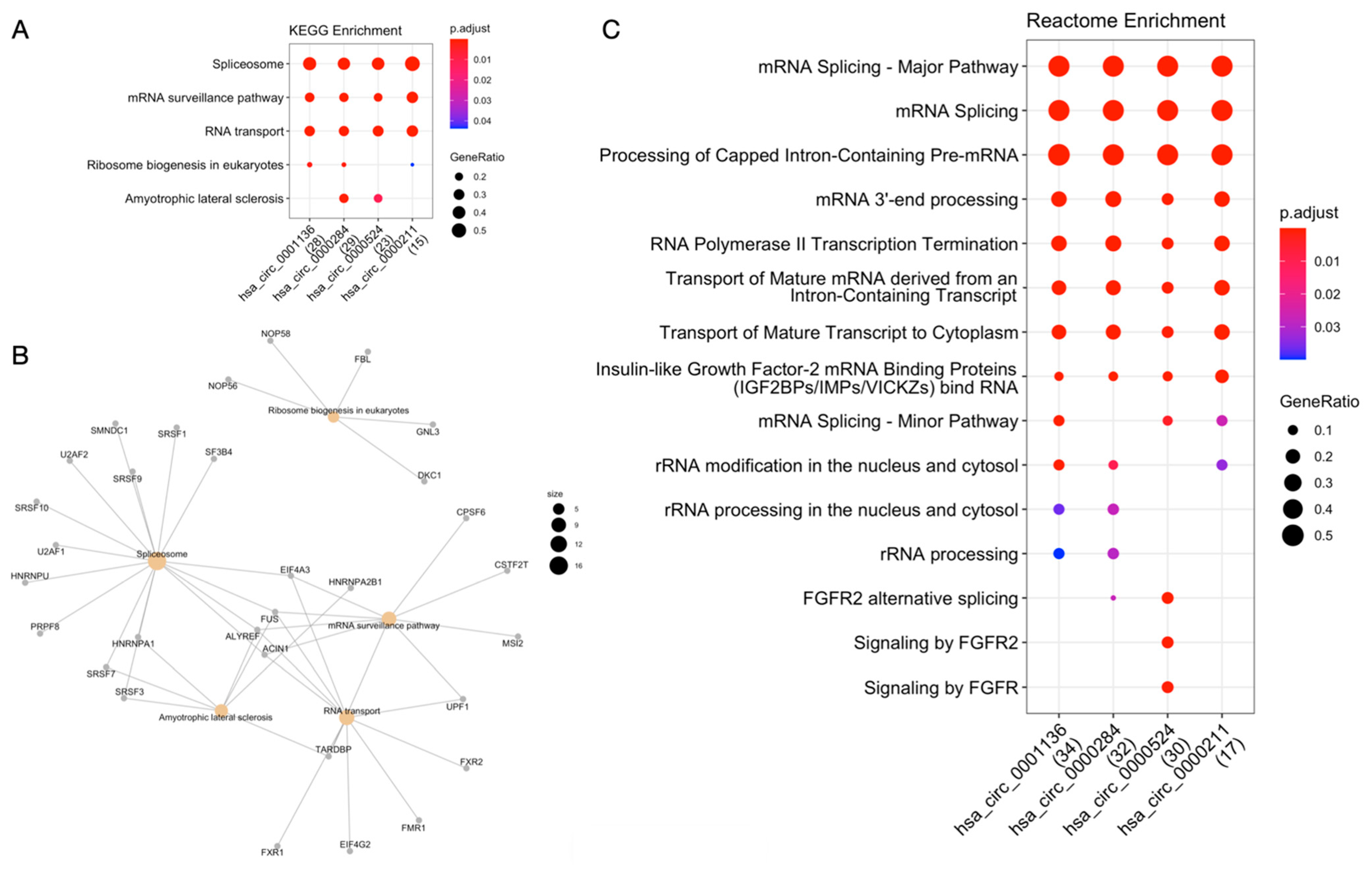
2.3. Functional Enrichment Analyses
2.3.1. CircRNA-miRNA-mRNA
2.3.2. CircRNAs-RBPs
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. RNA Isolation
4.3. Reverse Transcriptase Quantitative PCR (RT-qPCR)
4.4. Functional Enrichment Analyses
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Agency for Research on Cancer. Cancer Today. Available online: https://gco.iarc.fr/today/home (accessed on 12 February 2021).
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Chia, N.Y.; Tan, P. Molecular classification of gastric cancer. Ann. Oncol. 2016, 27, 763–769. [Google Scholar] [CrossRef]
- Hudler, P. Challenges of deciphering gastric cancer heterogeneity. World J. Gastroenterol. 2015, 21, 10510–10527. [Google Scholar] [CrossRef]
- Necula, L.; Matei, L.; Dragu, D.; Neagu, A.I.; Mambet, C.; Nedeianu, S.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 2019, 25, 2029–2044. [Google Scholar] [CrossRef]
- Li, P.-F.; Chen, S.-C.; Xia, T.; Jiang, X.-M.; Shao, Y.-F.; Xiao, B.-X.; Guo, J.-M. Non-coding RNAs and gastric cancer. World J. Gastroenterol. 2014, 20, 5411–5419. [Google Scholar] [CrossRef]
- Matsuoka, T.; Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 2018, 24, 2818–2832. [Google Scholar] [CrossRef]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef] [Green Version]
- Vicens, Q.; Westhof, E. Biogenesis of Circular RNAs. Cell 2014, 159, 13–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilusz, J.E. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA 2018, 9, e1478. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, Biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Li, P.; Chen, S.; Chen, H.; Mo, X.; Li, T.; Shao, Y.; Xiao, B.; Guo, J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta 2015, 444, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, T.; Zhao, Q.; Xiao, B.; Guo, J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta 2017, 466, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Li, J.; Lu, R.; Li, T.; Yang, Y.; Xiao, B.; Guo, J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017, 6, 1173–1180. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Vidal, A.; Ribeiro-Dos-Santos, A.M.; Vinasco-Sandoval, T.; Magalhães, L.; Pinto, P.; Anaissi, A.K.M.; Demachki, S.; De Assumpção, P.P.; Dos Santos, S.E.B. The comprehensive expression analysis of circular RNAs in gastric cancer and its association with field cancerization. Sci. Rep. 2017, 7, 14551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assumpção, M.B.; Moreira, F.C.; Hamoy, I.G.; Magalhães, L.; Vidal, A.; Pereira, A.L.; Burbano, R.; Khayat, A.; Silva, A.; Santos, S.; et al. High-Throughput miRNA Sequencing Reveals a Field Effect in Gastric Cancer and Suggests an Epigenetic Network Mechanism. Bioinform. Biol. Insights 2015, 9, BBI-S24066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, A.; Moreira, F.; Vinasco-Sandoval, T.; Cunha, A.; Vidal, A.; Ribeiro-Dos-Santos, A.M.; Pinto, P.; Magalhães, L.; Assumpção, M.; Demachki, S.; et al. miRNome Reveals New Insights Into the Molecular Biology of Field Cancerization in Gastric Cancer. Front. Genet. 2019, 10, 592. [Google Scholar] [CrossRef]
- Pereira, A.L.; Magalhães, L.; Moreira, F.C.; Reis-Das-Mercês, L.; Vidal, A.F.; Ribeiro-Dos-Santos, A.M.; Demachki, S.; Anaissi, A.K.M.; Burbano, R.M.R.; Albuquerque, P.; et al. Epigenetic Field Cancerization in Gastric Cancer: MicroRNAs as Promising Biomarkers. J. Cancer 2019, 10, 1560–1569. [Google Scholar] [CrossRef]
- Shao, Y.; Chen, L.; Lu, R.; Zhang, X.; Xiao, B.; Ye, G.; Guo, J. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumor Biol. 2017, 39, 1010428317699125. [Google Scholar] [CrossRef] [Green Version]
- Tian, M.; Chen, R.; Li, T.; Xiao, B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J. Clin. Lab. Anal. 2017, 32, e22281. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Ding, H.; Yang, L.; Yu, Y.; Zhou, J.; Yan, Z.; Guo, J. Reduced expression of circRNA hsa_circ_0067582 in human gastric cancer and its potential diagnostic values. J. Clin. Lab. Anal. 2020, 34, e23080. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, J.; Long, Z.; Li, G. Circular RNAs in Gastric Cancer: Potential Biomarkers and Therapeutic Targets. BioMed Res. Int. 2020, 2020, 2790679. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Xu, Y.; Hu, J.; Zhang, S.; Li, M.; Xu, L. A novel circular RNA, hsa-circ-0000211, Promotes lung adenocarcinoma migration and invasion through sponging of hsa-miR-622 and modulating HIF1-α expression. Biochem. Biophys. Res. Commun. 2020, 521, 395–401. [Google Scholar] [CrossRef]
- Cheng, J.; Zhuo, H.; Xu, M.; Wang, L.; Xu, H.; Peng, J.; Hou, J.; Lin, L.; Cai, J. Regulatory network of circRNA–miRNA–mRNA contributes to the histological classification and disease progression in gastric cancer. J. Transl. Med. 2018, 16, 216. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Li, L. Down-Regulation of circNRIP1 Promotes the Apoptosis and Inhibits the Migration and Invasion of Gastric Cancer Cells by miR-182/ROCK1 Axis. OncoTargets Ther. 2020, 13, 6279–6288. [Google Scholar] [CrossRef]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef] [PubMed]
- De Vita, F.; Di Martino, N.; Fabozzi, A.; Laterza, M.M.; Ventriglia, J.; Savastano, B.; Petrillo, A.; Gambardella, V.; Sforza, V.; Marano, L.; et al. Clinical management of advanced gastric cancer: The role of new molecular drugs. World J. Gastroenterol. 2014, 20, 14537–14558. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Shi, Y.; Liu, M.; Sun, J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 175. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, J.; Zhang, L.-Y.; Wang, L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3713–3718. [Google Scholar] [PubMed]
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018, 9, 417. [Google Scholar] [CrossRef]
- Wei, J.; Xu, H.; Wei, W.; Wang, Z.; Zhang, Q.; De, W.; Shu, Y. circHIPK3 Promotes Cell Proliferation and Migration of Gastric Cancer by Sponging miR-107 and Regulating BDNF Expression. OncoTargets Ther. 2020, 13, 1613–1624. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Che, X.; Qu, X.; Li, X.; Lu, W.; Wu, J.; Wang, Y.; Hou, K.; Li, C.; Zhang, X.; et al. CircHIPK3 Promotes Metastasis of Gastric Cancer via miR-653-5p/miR-338-3p-NRP1 Axis Under a Long-Term Hypoxic Microenvironment. Front. Oncol. 2020, 10, 1612. [Google Scholar] [CrossRef]
- Liu, W.-G.; Xu, Q. Upregulation of CircHIPK3 Promotes the Progression of Gastric Cancer via Wnt/β-Catenin Pathway and Indi-cates a Poor Prognosis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7905–7912. [Google Scholar] [CrossRef]
- Li, Q.; Tian, Y.; Liang, Y.; Li, C. CircHIPK3/miR-876-5p/PIK3R1 axis regulates regulation proliferation, migration, invasion, and glutaminolysis in gastric cancer cells. Cancer Cell Int. 2020, 20, 391. [Google Scholar] [CrossRef]
- Sun, Z.; Xu, Q.; Ma, Y.; Yang, S.; Shi, J. Circ_0000524/miR-500a-5p/CXCL16 axis promotes podocyte apoptosis in membranous nephropathy. Eur. J. Clin. Investig. 2021, 51, e13414. [Google Scholar] [CrossRef]
- Wang, B.; Chen, H.; Zhang, C.; Yang, T.; Zhao, Q.; Yan, Y.; Zhang, Y.; Xu, F. Effects ofhsa_circRBM23on Hepatocellular Carcinoma Cell Viability and Migration as Produced by Regulating miR-138 Expression. Cancer Biother. Radiopharm. 2018, 33, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, G.; He, H.; Zheng, Z.; Li, X.; Lin, R.; Xu, D. Differential expression of circular RNAs in bone marrow-derived exosomes from essential thrombocythemia patients. Cell Biol. Int. 2020, 45, 869–881. [Google Scholar] [CrossRef]
- Tang, G.; Xie, W.; Qin, C.; Zhen, Y.; Wang, Y.; Chen, F.; Du, Z.; Wu, Z.; Zhang, B.; Shen, Z.; et al. Expression of circular RNA circASXL1 correlates with TNM classification and predicts overall survival in bladder cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 8495–8502. [Google Scholar]
- Li, T.; Shao, Y.; Fu, L.; Xie, Y.I.; Zhu, L.; Sun, W.; Yu, R.; Xiao, B.; Guo, J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J. Mol. Med. 2018, 96, 85–96. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, Y.; Xu, L.; Qu, C.; Zhang, L.; Xiao, X.; Chen, W.; Li, K.; Liang, Q.; Wu, H. circ-NRIP1 Promotes Glycolysis and Tumor Progression by Regulating miR-186-5p/MYH9 Axis in Gastric Cancer. Cancer Manag. Res. 2020, 12, 5945–5956. [Google Scholar] [CrossRef]
- Xu, G.; Li, M.; Wu, J.; Qin, C.; Tao, Y.; He, H. Circular RNA circNRIP1 Sponges microRNA-138-5p to Maintain Hypoxia-Induced Resistance to 5-Fluorouracil Through HIF-1α-Dependent Glucose Metabolism in Gastric Carcinoma. Cancer Manag. Res. 2020, 12, 2789–2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, A.; Takanashi, K. FUS interacts with nuclear matrix-associated protein SAFB1 as well as Matrin3 to regulate splicing and ligand-mediated transcription. Sci. Rep. 2016, 6, 35195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Hakimi, M.-A.; Baillat, D.; Chen, X.; Farber, M.J.; Klein-Szanto, A.J.P.; Cooch, N.S.; Godwin, A.K.; Shiekhattar, R. Linking Transcriptional Elongation and Messenger RNA Export to Metastatic Breast Cancers. Cancer Res. 2005, 65, 3011–3016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelle, L.; Cloutier, A.; Toutant, J.; Shkreta, L.; Thibault, P.; Durand, M.; Garneau, D.; Gendron, D.; Lapointe, E.; Couture, S.; et al. Proteins Associated with the Exon Junction Complex Also Control the Alternative Splicing of Apoptotic Regulators. Mol. Cell. Biol. 2012, 32, 954–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballut, L.; Marchadier, B.; Baguet, A.; Tomasetto, C.; Séraphin, B.; Le Hir, H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005, 12, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, M.; Keriel, A.; Staub, A.; Egly, J.-M. Kinase Activity and Phosphorylation of the Largest Subunit of TFIIF Transcription Factor. J. Biol. Chem. 1999, 274, 22387–22392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paronetto, M.P.; Achsel, T.; Massiello, A.; Chalfant, C.E.; Sette, C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 2007, 176, 929–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberstrass, F.C.; Auweter, S.D.; Erat, M.; Hargous, Y.; Henning, A.; Wenter, P.; Reymond, L.; Amir-Ahmady, B.; Pitsch, S.; Black, D.L.; et al. Structure of PTB Bound to RNA: Specific Binding and Implications for Splicing Regulation. Science 2005, 309, 2054–2057. [Google Scholar] [CrossRef]
- Norris, J.D.; Fan, D.; Sherk, A.; McDonnell, D.P. A Negative Coregulator for the Human ER. Mol. Endocrinol. 2002, 16, 459–468. [Google Scholar] [CrossRef]
- Mancarella, C.; Scotlandi, K. IGF2BP3 From Physiology to Cancer: Novel Discoveries, Unsolved Issues, and Future Perspectives. Front. Cell Dev. Biol. 2020, 7, 363. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. BioSyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reis-das-Mercês, L.; Vinasco-Sandoval, T.; Pompeu, R.; Ramos, A.C.; Anaissi, A.K.M.; Demachki, S.; de Assumpção, P.P.; Vidal, A.F.; Ribeiro-dos-Santos, Â.; Magalhães, L. CircRNAs as Potential Blood Biomarkers and Key Elements in Regulatory Networks in Gastric Cancer. Int. J. Mol. Sci. 2022, 23, 650. https://doi.org/10.3390/ijms23020650
Reis-das-Mercês L, Vinasco-Sandoval T, Pompeu R, Ramos AC, Anaissi AKM, Demachki S, de Assumpção PP, Vidal AF, Ribeiro-dos-Santos Â, Magalhães L. CircRNAs as Potential Blood Biomarkers and Key Elements in Regulatory Networks in Gastric Cancer. International Journal of Molecular Sciences. 2022; 23(2):650. https://doi.org/10.3390/ijms23020650
Chicago/Turabian StyleReis-das-Mercês, Laís, Tatiana Vinasco-Sandoval, Rafael Pompeu, Aline Cruz Ramos, Ana K. M. Anaissi, Samia Demachki, Paulo Pimentel de Assumpção, Amanda F. Vidal, Ândrea Ribeiro-dos-Santos, and Leandro Magalhães. 2022. "CircRNAs as Potential Blood Biomarkers and Key Elements in Regulatory Networks in Gastric Cancer" International Journal of Molecular Sciences 23, no. 2: 650. https://doi.org/10.3390/ijms23020650
APA StyleReis-das-Mercês, L., Vinasco-Sandoval, T., Pompeu, R., Ramos, A. C., Anaissi, A. K. M., Demachki, S., de Assumpção, P. P., Vidal, A. F., Ribeiro-dos-Santos, Â., & Magalhães, L. (2022). CircRNAs as Potential Blood Biomarkers and Key Elements in Regulatory Networks in Gastric Cancer. International Journal of Molecular Sciences, 23(2), 650. https://doi.org/10.3390/ijms23020650






