Deciphering Molecular Determinants Underlying Penicillium digitatum’s Response to Biological and Chemical Antifungal Agents by Tandem Mass Tag (TMT)-Based High-Resolution LC-MS/MS
Abstract
:1. Introduction
2. Results
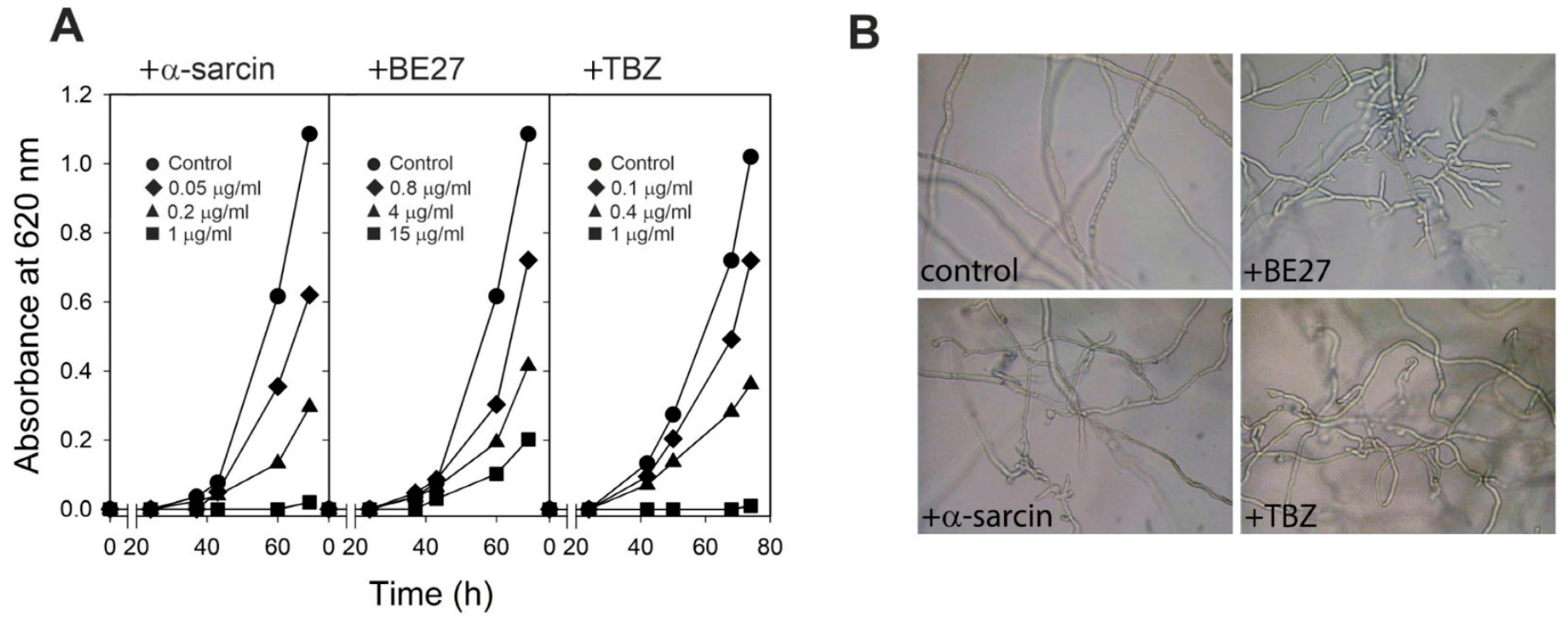
2.1. Effects of BE27, α-Sarcin and TBZ Treatments on the Growth and Morphogenesis of P. digitatum
2.2. Quantitative Proteomic Analysis of P. digitatum in Response to α-Sarcin, BE27 and TBZ Treatments
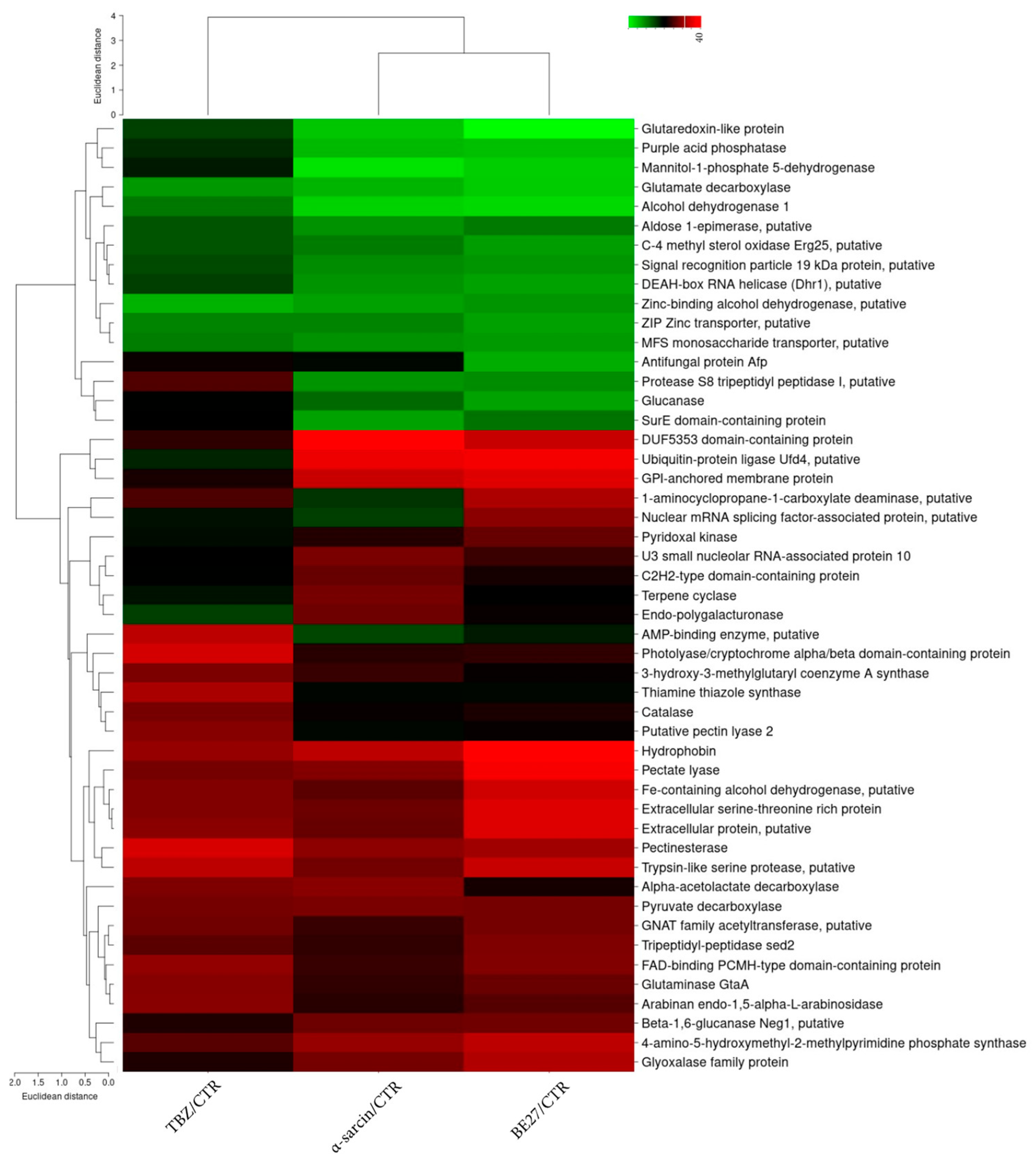
2.3. Categorization of Differentially Expressed Proteins
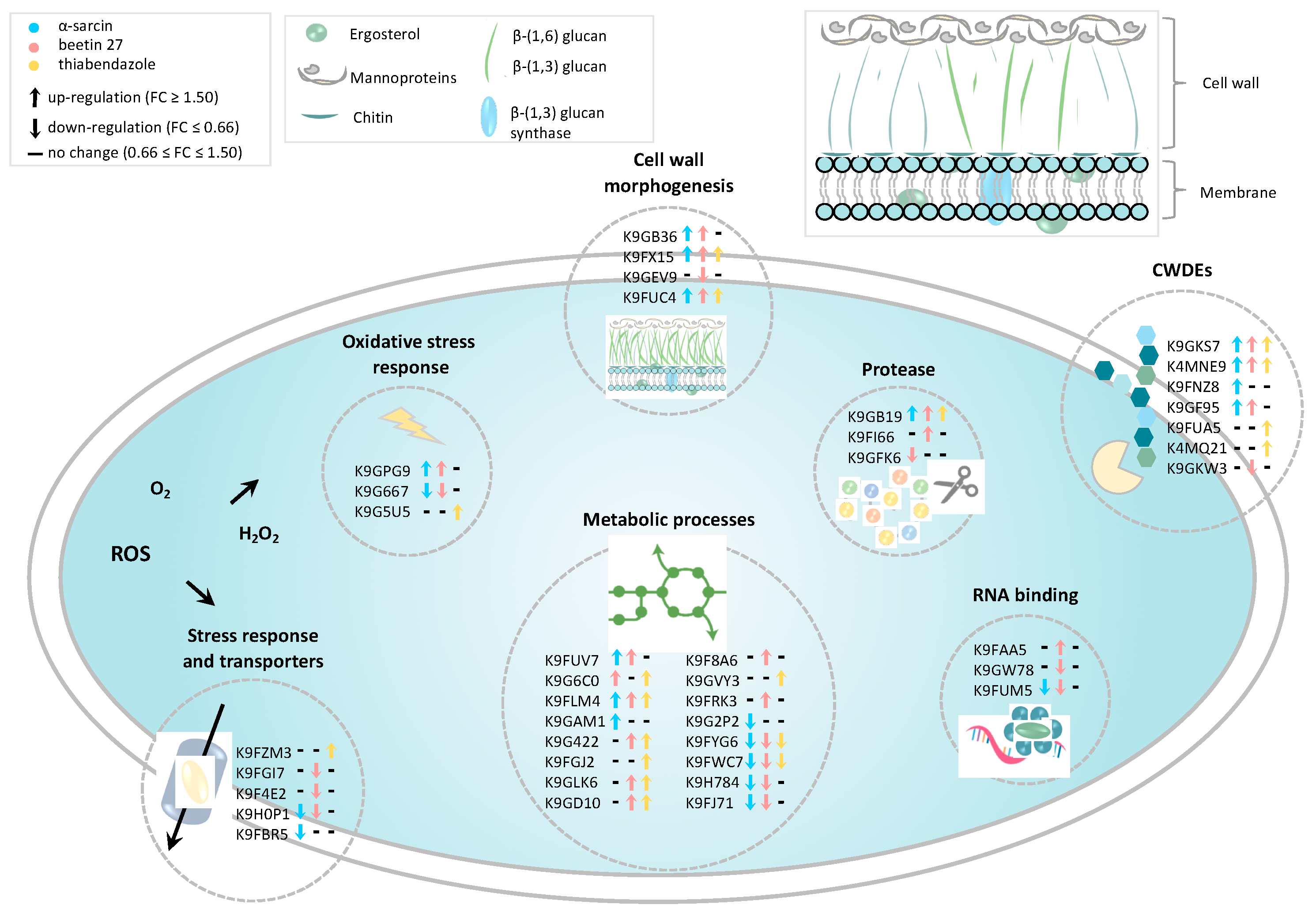
2.4. Cell Wall-Degrading Enzymes (CWDEs) and Fungal Morphogenesis
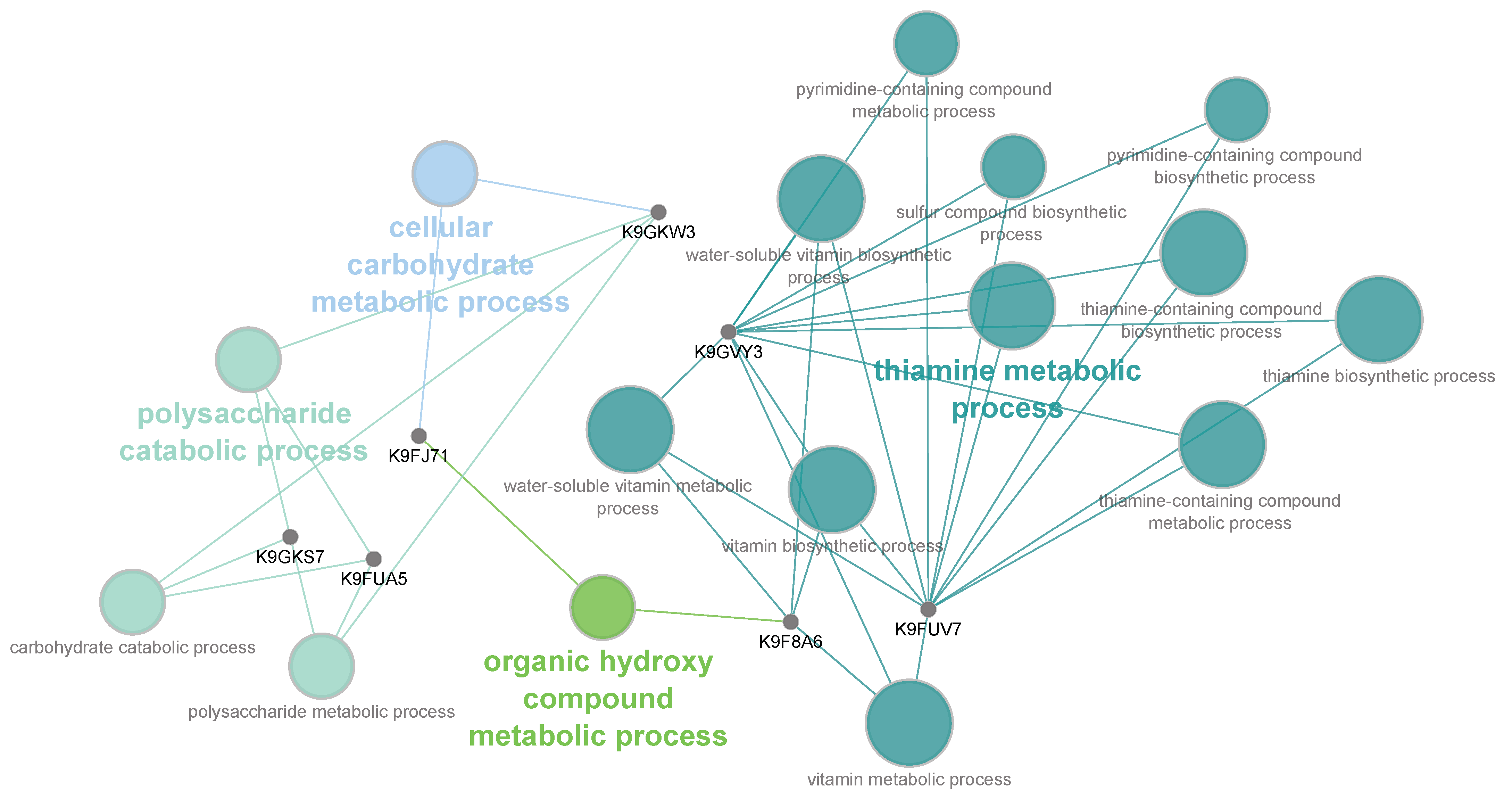
2.5. Antifungal Agents Affect Penicillium digitatum Metabolic Processes
2.6. Antioxidant and Detoxification Mechanisms and Stress Response
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Antifungal Treatments of P. digitatum
4.3. Antifungal Activity Measurements
4.4. Sample Preparation for Mass Spectrometry Analysis
4.5. High-Resolution NanoLC−Tandem Mass Spectrometry
4.6. Protein Identification and Quantitation
4.7. Bioinformatic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BE27 | Beetin 27 |
| CWDEs | Cell Wall-Degrading Enzymes |
| MS | Mass Spectrometry |
| RIPs | Ribosome-Inactivating Proteins |
| SRL | Sarcin Ricin Loop |
| TBZ | Thiabendazole |
| TMT | Tandem Mass Tag |
References
- Passam, H.C.; Karapanos, I.C.; Alexopoulos, A.A. The Biological Basis of Fruit Quality. In Breeding for Fruit Quality; Jenks, M.A., Bebeli, P.J., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2011; pp. 3–38. [Google Scholar]
- Talibi, I.; Boubaker, H.; Boudyach, E.H.; Ait Ben Aoumar, A. Alternative methods for the control of postharvest citrus diseases. J. Appl. Microbiol. 2014, 117, 1–17. [Google Scholar] [CrossRef]
- Tian, S.; Torres, R.; Ballester, A.R.; Li, B.; Vilanova, L.; González-Candelas, L. Molecular aspects in pathogen-fruit interactions: Virulence and resistance. Postharvest Biol. Technol. 2016, 122, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Lin, Y.; Cao, H.; Li, Z. Citrus Postharvest Green Mold: Recent Advances in Fungal Pathogenicity and Fruit Resistance. Microorganisms 2020, 8, 449. [Google Scholar] [CrossRef] [Green Version]
- Lucas, J.A.; Hawkins, N.J.; Fraaije, B.A. Chapter Two—The Evolution of Fungicide Resistance. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 90, pp. 29–92. [Google Scholar]
- Liu, R.S.; Huang, H.; Yang, Q.; Liu, W.Y. Purification of alpha-sarcin and an antifungal protein from mold (Aspergillus giganteus) by chitin affinity chromatography. Protein Expr. Purif. 2002, 25, 50–58. [Google Scholar] [CrossRef]
- Theis, T.; Wedde, M.; Meyer, V.; Stahl, U. The antifungal protein from Aspergillus giganteus causes membrane permeabilization. Antimicrob. Agents Chemother. 2003, 47, 588–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szappanos, H.; Szigeti, G.P.; Pál, B.; Rusznák, Z.; Szucs, G.; Rajnavölgyi, E.; Balla, J.; Balla, G.; Nagy, E.; Leiter, E.; et al. The antifungal protein AFP secreted by Aspergillus giganteus does not cause detrimental effects on certain mammalian cells. Peptides 2006, 27, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.B.; Del Pozo, A.M.; Borja, M.; Segundo, B.S. Activity of the Antifungal Protein from Aspergillus giganteus Against Botrytis cinerea. Phytopathology 2003, 93, 1344–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citores, L.; Iglesias, R.; Ragucci, S.; Di Maro, A.; Ferreras, J.M. Antifungal Activity of α-Sarcin against Penicillium digitatum: Proposal of a New Role for Fungal Ribotoxins. ACS Chem. Biol. 2018, 13, 1978–1982. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Zhou, Y.K.; Ji, Z.L.; Chen, X.R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Citores, L.; Iglesias, R.; Ferreras, J.M. Antiviral Activity of Ribosome-Inactivating Proteins. Toxins 2021, 13, 80. [Google Scholar] [CrossRef]
- Citores, L.; Iglesias, R.; Gay, C.; Ferreras, J.M. Antifungal activity of the ribosome-inactivating protein BE27 from sugar beet (Beta vulgaris L.) against the green mould Penicillium digitatum. Mol. Plant Pathol. 2016, 17, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, J.; Zhu, Y.; Duan, Y.; Zhou, M. Mechanism of Action of the Benzimidazole Fungicide on Fusarium graminearum: Interfering with Polymerization of Monomeric Tubulin But Not Polymerized Microtubule. Phytopathology 2016, 106, 807–813. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhao, M.; Zhang, Z. Quantitative proteomics reveals the antifungal effect of canthin-6-one isolated from Ailanthus altissima against Fusarium oxysporum f. sp. cucumerinum in vitro. PLoS ONE 2021, 16, e0250712. [Google Scholar] [CrossRef] [PubMed]
- Aumer, T.; Voisin, S.N.; Knobloch, T.; Landon, C.; Bulet, P. Impact of an Antifungal Insect Defensin on the Proteome of the Phytopathogenic Fungus Botrytis cinerea. J. Proteome Res. 2020, 19, 1131–1146. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Ge, B.; Wang, J.; Liu, B.; Ma, J.; Wei, Q.; Zhang, K. iTRAQ-based proteomic analysis reveals the mechanisms of Botrytis cinerea controlled with Wuyiencin. BMC Microbiol. 2019, 19, 280. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Chen, C.; Xia, X.; Garba, B.; Shang, L.; Wang, Y. Proteomic analysis of the inhibitory effect of chitosan on Penicillium expansum. Food Sci. Technol. 2020, 40, 250–257. [Google Scholar] [CrossRef] [Green Version]
- Martins, I.; Varela, A.; Frija, L.M.T.; Estevão, M.A.S.; Planchon, S.; Renaut, J.; Afonso, C.A.M.; Silva Pereira, C. Proteomic Insights on the Metabolism of Penicillium janczewskii during the Biotransformation of the Plant Terpenoid Labdanolic Acid. Front. Bioeng. Biotechnol. 2017, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Zhou, T.; Ye, B.; Yan, Z.; Wang, X.; Lai, T. Uncovering proteomics changes of Penicillium expansum spores in response to decanal treatment by iTRAQ. J. Plant Pathol. 2020, 102, 721–730. [Google Scholar] [CrossRef]
- Lin, S.H.; Luo, P.; Yuan, E.; Zhu, X.; Zhang, B.; Wu, X. Physiological and Proteomic Analysis of Penicillium digitatum in Response to X33 Antifungal Extract Treatment. Front. Microbiol. 2020, 11, 584331. [Google Scholar] [CrossRef]
- Di Pietro, A.; Roncero, M.I.G.; Roldán, M.C.R. From Tools of Survival to Weapons of Destruction: The Role of Cell Wall-Degrading Enzymes in Plant Infection. In Plant Relationships. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research); Deising, H.B., Ed.; Springer: Berlin, Heidelberg, 2009; Volume 5. [Google Scholar]
- Lopez-Perez, M.; Ballester, A.R.; Gonzalez-Candelas, L. Identification and functional analysis of Penicillium digitatum genes putatively involved in virulence towards citrus fruit. Mol. Plant Pathol. 2015, 16, 262–275. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Qian, X.; Dhanasekaran, S.; Boateng, N.A.S.; Yan, X.; Zhu, H.; He, F.; Zhang, H. Study on the Infection Mechanism of Penicillium Digitatum on Postharvest Citrus (Citrus Reticulata Blanco) Based on Transcriptomics. Microorganisms 2019, 7, 672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Wang, Y.; Hu, X.; Lei, M.; Wang, M.; Zeng, J.; Li, H.; Liu, Z.; Zhou, T.; Yu, D. Involvement of LaeA in the regulation of conidia production and stress responses in Penicillium digitatum. J. Basic Microbiol. 2020, 60, 82–88. [Google Scholar] [CrossRef]
- Mutz, M.; Roemer, T. The GPI anchor pathway: A promising antifungal target? Future Med. Chem. 2016, 8, 1387–1391. [Google Scholar] [CrossRef] [Green Version]
- Ruan, R.; Wang, M.; Liu, X.; Sun, X.; Chung, K.R.; Li, H. Functional analysis of two sterol regulatory element binding proteins in Penicillium digitatum. PLoS ONE 2017, 12, e0176485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blosser, S.J.; Merriman, B.; Grahl, N.; Chung, D.; Cramer, R.A. Two C4-sterol methyl oxidases (Erg25) catalyse ergosterol intermediate demethylation and impact environmental stress adaptation in Aspergillus fumigatus. Microbiol. (Read. Engl.) 2014, 160, 2492–2506. [Google Scholar] [CrossRef] [Green Version]
- Costa, J.H.; Bazioli, J.M.; de Moraes Pontes, J.G.; Fill, T.P. Penicillium digitatum infection mechanisms in citrus: What do we know so far? Fungal Biol. 2019, 123, 584–593. [Google Scholar] [CrossRef]
- Macarisin, D.; Cohen, L.; Eick, A.; Rafael, G.; Belausov, E.; Wisniewski, M.; Droby, S. Penicillium digitatum Suppresses Production of Hydrogen Peroxide in Host Tissue During Infection of Citrus Fruit. Phytopathology 2007, 97, 1491–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, A.; Kumar, S.G.; Kim, S.W.; Hwang, H.J.; Baek, Y.M.; Lee, S.H.; Hwang, H.S.; Shon, Y.H.; Nam, K.S.; Yun, J.W. Proteomic analysis for inhibitory effect of chitosan oligosaccharides on 3T3-L1 adipocyte differentiation. Proteomics 2008, 8, 569–581. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; Zwiers, L.H.; De Waard, M.A. Secretion of Natural and Synthetic Toxic Compounds from Filamentous Fungi by Membrane Transporters of the ATP-binding Cassette and Major Facilitator Superfamily. Eur. J. Plant Pathol. 2002, 108, 719–734. [Google Scholar] [CrossRef]
- Sanchez-Torres, P. Molecular Mechanisms Underlying Fungicide Resistance in Citrus Postharvest Green Mold. J Fungi 2021, 7, 783. [Google Scholar] [CrossRef]
- de Ramon-Carbonell, M.; Sanchez-Torres, P. Penicilliumdigitatum MFS transporters can display different roles during pathogen-fruit interaction. Int. J. Food Microbiol. 2021, 337, 108918. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, S.; Gandia, M.; Borics, A.; Marx, F.; Manzanares, P.; Marcos, J.F. Mapping and Identification of Antifungal Peptides in the Putative Antifungal Protein AfpB from the Filamentous Fungus Penicillium digitatum. Front Microbiol. 2017, 8, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrigues, S.; Gandia, M.; Popa, C.; Borics, A.; Marx, F.; Coca, M.; Marcos, J.F.; Manzanares, P. Efficient production and characterization of the novel and highly active antifungal protein AfpB from Penicillium digitatum. Sci. Rep. 2017, 7, 14663. [Google Scholar] [CrossRef] [Green Version]
- Marcet-Houben, M.; Ballester, A.R.; de la Fuente, B.; Harries, E.; Marcos, J.F.; Gonzalez-Candelas, L.; Gabaldon, T. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genom. 2012, 13, 646. [Google Scholar] [CrossRef] [Green Version]
- Meyer, V.; Jung, S. Antifungal Peptides of the AFP Family Revisited: Are These Cannibal Toxins? Microorganisms 2018, 6, 50. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, R.; Citores, L.; Di Maro, A.; Ferreras, J.M. Biological activities of the antiviral protein BE27 from sugar beet (Beta vulgaris L.). Planta 2015, 241, 421–433. [Google Scholar] [CrossRef]
- Iglesias, R.; Perez, Y.; de Torre, C.; Ferreras, J.M.; Antolin, P.; Jimenez, P.; Rojo, M.A.; Mendez, E.; Girbes, T. Molecular characterization and systemic induction of single-chain ribosome-inactivating proteins (RIPs) in sugar beet (Beta vulgaris) leaves. J. Exp. Bot. 2005, 56, 1675–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Matteo, F.; Pipicelli, F.; Kyrousi, C.; Tovecci, I.; Penna, E.; Crispino, M.; Chambery, A.; Russo, R.; Ayo-Martin, A.C.; Giordano, M.; et al. Cystatin B is essential for proliferation and interneuron migration in individuals with EPM1 epilepsy. EMBO Mol. Med. 2020, 12, e11419. [Google Scholar] [CrossRef]
- Russo, R.; Matrone, N.; Belli, V.; Ciardiello, D.; Valletta, M.; Esposito, S.; Pedone, P.V.; Ciardiello, F.; Troiani, T.; Chambery, A. Macrophage Migration Inhibitory Factor Is a Molecular Determinant of the Anti-EGFR Monoclonal Antibody Cetuximab Resistance in Human Colorectal Cancer Cells. Cancers 2019, 11, 1430. [Google Scholar] [CrossRef] [Green Version]
- Napolitano, F.; De Rosa, A.; Russo, R.; Di Maio, A.; Garofalo, M.; Federici, M.; Migliarini, S.; Ledonne, A.; Rizzo, F.R.; Avallone, L.; et al. The striatal-enriched protein Rhes is a critical modulator of cocaine-induced molecular and behavioral responses. Sci. Rep. 2019, 9, 15294. [Google Scholar] [CrossRef] [Green Version]
- Valletta, M.; Russo, R.; Baglivo, I.; Russo, V.; Ragucci, S.; Sandomenico, A.; Iaccarino, E.; Ruvo, M.; De Feis, I.; Angelini, C.; et al. Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF. Int. J. Mol. Sci. 2020, 21, 8950. [Google Scholar] [CrossRef] [PubMed]

| Classification | AC | Description | α-Sarcin vs. CTR | BE27 vs. CTR | TBZ vs. CTR | Cov | #Pep | #PSM |
|---|---|---|---|---|---|---|---|---|
| Cell wall morphogenesis | K9GB36 | GPI-anchored membrane protein | 2.06 | 2.22 | 1.22 | 21 | 4 | 10 |
| K9FX15 | Hydrophobin | 1.99 | 2.73 | 1.76 | 69 | 5 | 66 | |
| K9GEV9 | C-4 methyl sterol oxidase Erg25, putative | 0.71 | 0.64 | 0.82 | 11 | 3 | 5 | |
| K9FUC4 | Extracellular serine-threonine rich protein | 1.55 | 2.21 | 1.67 | 7 | 4 | 10 | |
| CWDEs | K9GKS7 | Pectinesterase | 1.72 | 1.81 | 2.17 | 15 | 5 | 13 |
| K4MNE9 | Pectate lyase | 1.67 | 2.4 | 1.59 | 42 | 8 | 45 | |
| K9FNZ8 | Endo-polygalacturonase | 1.57 | 1.17 | 0.88 | 19 | 6 | 21 | |
| K9GF95 | Beta-1,6-glucanase Neg1, putative | 1.55 | 1.57 | 1.24 | 13 | 6 | 25 | |
| K9FUA5 | Arabinan endo-1,5-alpha-L-arabinosidase | 1.28 | 1.46 | 1.69 | 6 | 2 | 3 | |
| K4MQ21 | Putative pectin lyase 2 | 1.06 | 1.16 | 1.68 | 20 | 4 | 11 | |
| K9GKW3 | Glucanase | 0.76 | 0.62 | 1.11 | 4 | 2 | 3 | |
| Metabolic processes | K9FUV7 | 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate synthase | 1.76 | 2 | 1.46 | 60 | 18 | 241 |
| K9G6C0 | Alpha-acetolactate decarboxylase | 1.7 | 1.22 | 1.64 | 49 | 12 | 63 | |
| K9FLM4 | Pyruvate decarboxylase | 1.62 | 1.61 | 1.6 | 69 | 30 | 699 | |
| K9GAM1 | Terpene cyclase | 1.6 | 1.11 | 1.02 | 41 | 12 | 60 | |
| K9G422 | Fe-containing alcohol dehydrogenase, putative | 1.47 | 2.12 | 1.66 | 17 | 8 | 52 | |
| K9FGJ2 | 3-hydroxy-3-methylglutaryl coenzyme A synthase | 1.33 | 1.16 | 1.63 | 28 | 12 | 62 | |
| K9GLK6 | GNAT family acetyltransferase, putative | 1.33 | 1.6 | 1.58 | 25 | 4 | 18 | |
| K9GD10 | Glutaminase GtaA | 1.3 | 1.54 | 1.68 | 12 | 5 | 9 | |
| K9F8A6 | Pyridoxal kinase | 1.26 | 1.53 | 1.05 | 9 | 2 | 4 | |
| K9GVY3 | Thiamine thiazole synthase | 1.07 | 1.06 | 1.88 | 27 | 9 | 35 | |
| K9FRK3 | 1-aminocyclopropane-1-carboxylate deaminase, putative | 0.92 | 1.9 | 1.41 | 18 | 5 | 9 | |
| K9G2P2 | Aldose 1-epimerase, putative | 0.66 | 0.72 | 0.82 | 6 | 3 | 3 | |
| K9FYG6 | Zinc-binding alcohol dehydrogenase, putative | 0.63 | 0.65 | 0.59 | 37 | 10 | 48 | |
| K9FWC7 | Glutamate decarboxylase | 0.59 | 0.54 | 0.64 | 52 | 25 | 125 | |
| K9H784 | Alcohol dehydrogenase 1 | 0.53 | 0.51 | 0.72 | 50 | 15 | 124 | |
| K9FJ71 | Mannitol-1-phosphate 5-dehydrogenase | 0.49 | 0.53 | 1 | 25 | 8 | 12 | |
| Other | K9FX00 | DUF5353 domain-containing protein | 2.79 | 2.06 | 1.3 | 10 | 2 | 3 |
| K9GTK7 | Ubiquitin-protein ligase Ufd4, putative | 2.32 | 2.39 | 0.97 | 2 | 3 | 5 | |
| K9G185 | U3 small nucleolar RNA-associated protein 10 | 1.63 | 1.35 | 1.12 | 1 | 2 | 3 | |
| K9G4M0 | Extracellular protein, putative | 1.53 | 2.21 | 1.71 | 6 | 2 | 3 | |
| K9GB66 | FAD-binding PCMH-type domain-containing protein | 1.33 | 1.66 | 1.74 | 41 | 17 | 115 | |
| K9GEC9 | AMP-binding enzyme, putative | 0.86 | 1 | 1.99 | 4 | 2 | 3 | |
| K9F863 | Purple acid phosphatase | 0.57 | 0.56 | 0.95 | 16 | 5 | 10 | |
| K9FDZ3 | C2H2-type domain-containing protein | 1.53 | 1.21 | 1.11 | 11 | 5 | 9 | |
| Protease | K9GB19 | Trypsin-like serine protease, putative | 1.59 | 2.05 | 1.98 | 27 | 6 | 73 |
| K9FI66 | Tripeptidyl-peptidase sed2 | 1.3 | 1.64 | 1.47 | 29 | 10 | 37 | |
| K9GFK6 | Protease S8 tripeptidyl peptidase I, putative | 0.66 | 0.67 | 1.44 | 3 | 3 | 4 | |
| Oxidative stress response | K9GPG9 | Glyoxalase family protein | 1.6 | 1.92 | 1.25 | 6 | 3 | 5 |
| K9G667 | Glutaredoxin-like protein | 0.55 | 0.45 | 0.88 | 28 | 4 | 11 | |
| K9G5U5 | Catalase | 1.16 | 1.23 | 1.6 | 27 | 15 | 35 | |
| RNA binding | K9FAA5 | Nuclear mRNA splicing factor-associated protein, putative | 0.89 | 1.7 | 1.03 | 3 | 2 | 3 |
| K9GW78 | Signal recognition particle 19 kDa protein, putative | 0.67 | 0.65 | 0.85 | 7 | 2 | 4 | |
| K9FUM5 | DEAH-box RNA helicase (Dhr1), putative | 0.65 | 0.62 | 0.89 | 2 | 3 | 4 | |
| Stress response and transporters | K9FZM3 | Photolyase/cryptochrome alpha/beta domain-containing protein | 1.27 | 1.3 | 2.13 | 3 | 2 | 3 |
| K9FGI7 | Antifungal protein Afp | 1.06 | 0.6 | 1.18 | 23 | 3 | 9 | |
| K9F4E2 | ZIP Zinc transporter, putative | 0.7 | 0.63 | 0.69 | 7 | 3 | 5 | |
| K9H0P1 | MFS monosaccharide transporter, putative | 0.66 | 0.64 | 0.71 | 5 | 3 | 10 | |
| K9FBR5 | SurE domain-containing protein | 0.62 | 0.74 | 1.12 | 26 | 6 | 20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Citores, L.; Valletta, M.; Singh, V.P.; Pedone, P.V.; Iglesias, R.; Ferreras, J.M.; Chambery, A.; Russo, R. Deciphering Molecular Determinants Underlying Penicillium digitatum’s Response to Biological and Chemical Antifungal Agents by Tandem Mass Tag (TMT)-Based High-Resolution LC-MS/MS. Int. J. Mol. Sci. 2022, 23, 680. https://doi.org/10.3390/ijms23020680
Citores L, Valletta M, Singh VP, Pedone PV, Iglesias R, Ferreras JM, Chambery A, Russo R. Deciphering Molecular Determinants Underlying Penicillium digitatum’s Response to Biological and Chemical Antifungal Agents by Tandem Mass Tag (TMT)-Based High-Resolution LC-MS/MS. International Journal of Molecular Sciences. 2022; 23(2):680. https://doi.org/10.3390/ijms23020680
Chicago/Turabian StyleCitores, Lucía, Mariangela Valletta, Vikram Pratap Singh, Paolo Vincenzo Pedone, Rosario Iglesias, José Miguel Ferreras, Angela Chambery, and Rosita Russo. 2022. "Deciphering Molecular Determinants Underlying Penicillium digitatum’s Response to Biological and Chemical Antifungal Agents by Tandem Mass Tag (TMT)-Based High-Resolution LC-MS/MS" International Journal of Molecular Sciences 23, no. 2: 680. https://doi.org/10.3390/ijms23020680
APA StyleCitores, L., Valletta, M., Singh, V. P., Pedone, P. V., Iglesias, R., Ferreras, J. M., Chambery, A., & Russo, R. (2022). Deciphering Molecular Determinants Underlying Penicillium digitatum’s Response to Biological and Chemical Antifungal Agents by Tandem Mass Tag (TMT)-Based High-Resolution LC-MS/MS. International Journal of Molecular Sciences, 23(2), 680. https://doi.org/10.3390/ijms23020680











