Genetic Study of Four Candidate Holliday Junction Processing Proteins in the Thermophilic Crenarchaeon Sulfolobus acidocaldarius
Abstract
:1. Introduction
2. Results
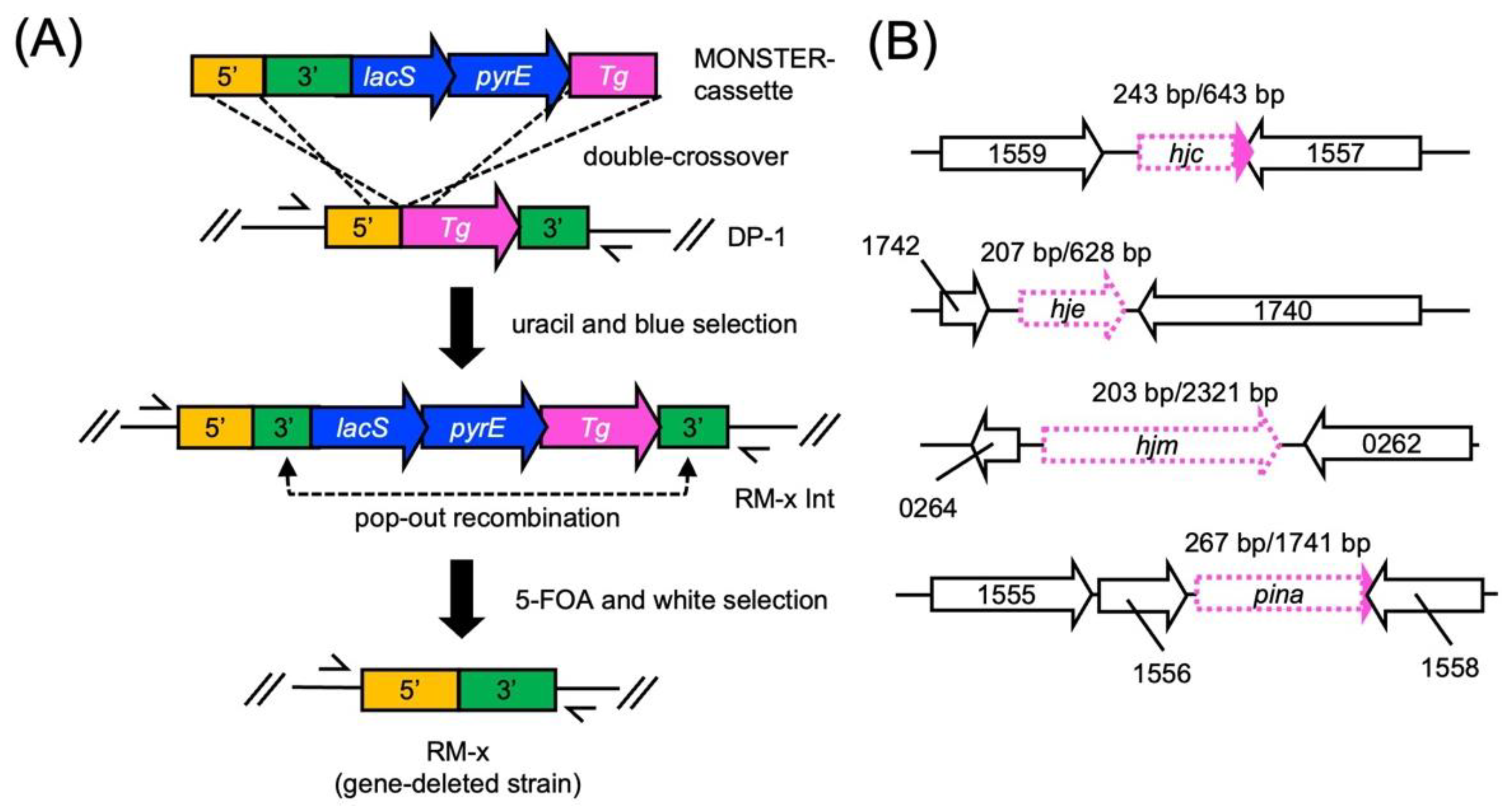
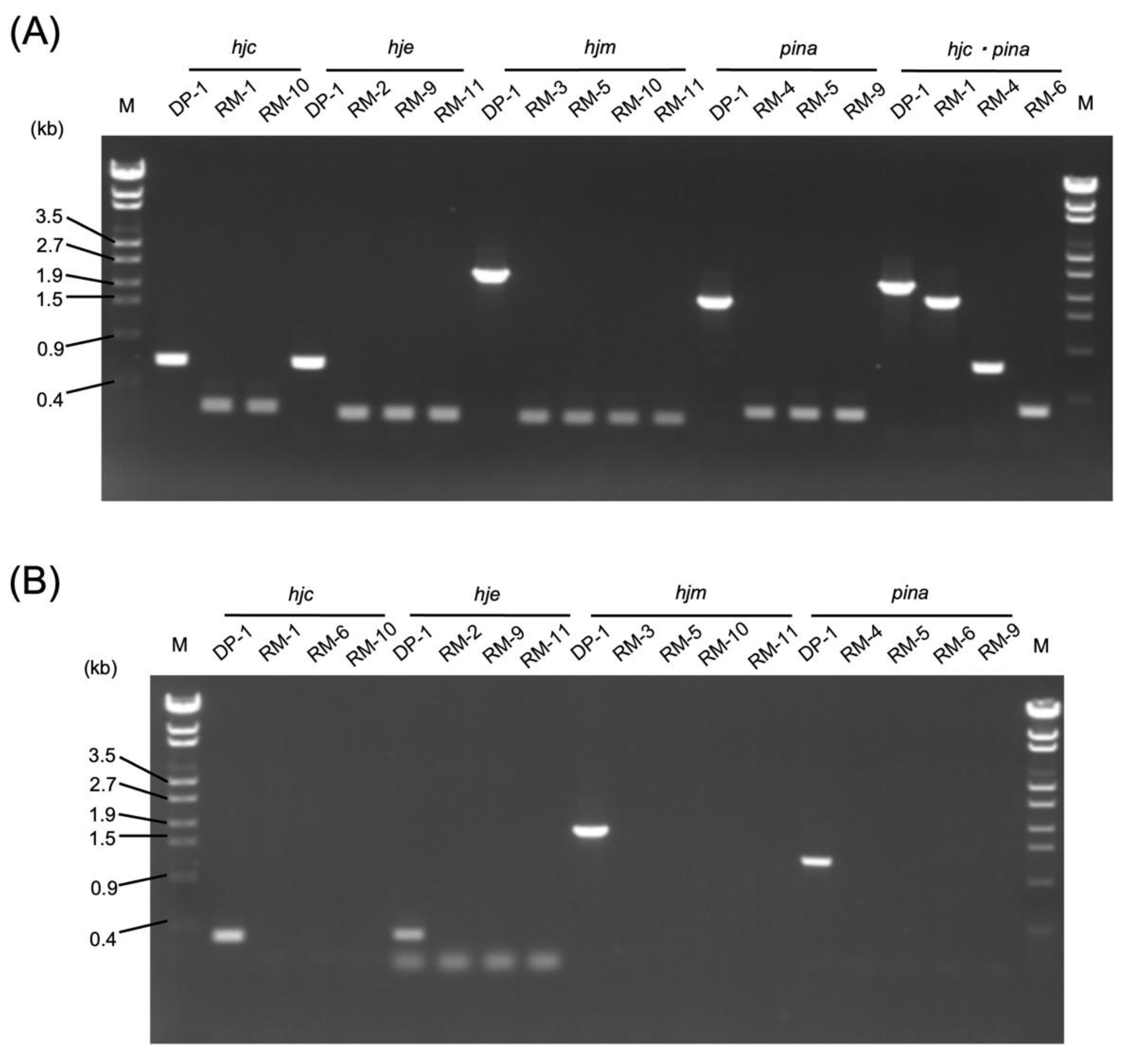
2.1. Construction of the hjc-, hje-, hjm-, and pina-Deleted Strains
2.2. Growth Characteristics of the Gene-Deleted Strains
2.3. Sensitivity of the Gene-Deleted Strains to Interstrand Crosslinker MMC
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. General DNA Manipulation
4.3. Construction of Gene-Deleted Strains
4.4. Growth Temperature Range Test
4.5. Growth Curve after Treatment with DNA-Damaging Agent
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- White, M.F.; Allers, T. DNA repair in the archaea-an emerging picture. FEMS Microbiol. Rev. 2018, 42, 514–526. [Google Scholar] [CrossRef]
- Grogan, D.W. Understanding DNA repair in hyperthermophilic archaea: Persistent gaps and other reasons to focus on the fork. Archaea 2015, 2015, 942605. [Google Scholar] [CrossRef]
- Fujikane, R.; Ishino, S.; Ishino, Y.; Forterre, P. Genetic analysis of DNA repair in the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Genet. Syst. 2010, 85, 243–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, S.; Kurosawa, N. Endonucleases responsible for DNA repair of helix-distorting DNA lesions in the thermophilic crenarchaeon Sulfolobus acidocaldarius in vivo. Extremophiles 2019, 23, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Muniyappa, K.; Shaner, S.L.; Tsang, S.S.; Radding, C.M. Mechanism of the concerted action of recA protein and helix- destabilizing proteins in homologous recombination. Proc. Natl. Acad. Sci. USA 1984, 81, 2757–2761. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, T.; Zaitseva, E.M.; Kowalczykowski, S.C. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997, 272, 7940–7945. [Google Scholar] [CrossRef] [Green Version]
- Seitz, E.M.; Brockman, J.P.; Sandler, S.J.; Clark, A.J.; Kowalczykowski, S.C. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 1998, 12, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Kerr, I.D.; Wadsworth, R.I.M.; Cubeddu, L.; Blankenfeldt, W.; Naismith, J.H.; White, M.F. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 2003, 22, 2561–2570. [Google Scholar] [CrossRef] [Green Version]
- Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 2008, 89, 285–307. [Google Scholar] [CrossRef] [Green Version]
- Komori, K.; Sakae, S.; Shinagawa, H.; Morikawa, K.; Ishino, Y. A holliday junction resolvase from Pyrococcus furiosus: Functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in Bacteria, Eukarya, and Archaea. Proc. Natl. Acad. Sci. USA 1999, 96, 8873–8878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kvaratskhelia, M.; White, M.F. An archaeal Holliday junction resolving enzyme from Sulfolobus solfataricus exhibits unique properties. J. Mol. Biol. 2000, 295, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.P.; Bolt, E.L. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 2005, 33, 3678–3690. [Google Scholar] [CrossRef] [PubMed]
- Fujikane, R.; Komori, K.; Shinagawa, H.; Ishino, Y. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 2005, 280, 12351–12358. [Google Scholar] [CrossRef] [Green Version]
- White, M.F. Homologous recombination in the archaea: The means justify the ends. Biochem. Soc. Trans. 2011, 39, 15–19. [Google Scholar] [CrossRef]
- Zhai, B.; DuPrez, K.; Doukov, T.I.; Li, H.; Huang, M.; Shang, G.; Ni, J.; Gu, L.; Shen, Y.; Fan, L. Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration. J. Mol. Biol. 2017, 429, 1009–1029. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Lu, S.; Hou, G.; Ma, X.; Sheng, D.; Ni, J.; Shen, Y. Hjm/Hel308a DNA Helicase from Sulfolobus tokodaii promotes replication fork regression and interacts with Hjc endonuclease in vitro. J. Bacteriol. 2008, 190, 3006–3017. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Chu, M.; Li, Y.; Ni, J.; Sheng, D.; Hou, G.; She, Q.; Shen, Y. Dissection of the functional domains of an archaeal Holliday junction helicase. DNA Repair 2012, 11, 102–111. [Google Scholar] [CrossRef]
- van Wolferen, M.; Ma, X.; Albers, S.V. DNA processing proteins involved in the UV-induced stress response of sulfolobales. J. Bacteriol. 2015, 197, 2941–2951. [Google Scholar] [CrossRef] [Green Version]
- Buckley, R.J.; Kramm, K.; Cooper, C.D.O.; Grohmann, D.; Bolt, E.L. Mechanistic insights into Lhr helicase function in DNA repair. Biochem. J. 2020, 477, 2935–2947. [Google Scholar] [CrossRef]
- Zhai, B.; DuPrez, K.; Han, X.; Yuan, Z.; Ahmad, S.; Xu, C.; Gu, L.; Ni, J.; Fan, L.; Shen, Y. The archaeal ATPase PINA interacts with the helicase Hjm via its carboxyl terminal KH domain remodeling and processing replication fork and Holliday junction. Nucleic Acids Res. 2018, 46, 6627–6641. [Google Scholar] [CrossRef] [Green Version]
- Kvaratskhelia, M.; White, M.F. Two holliday junction resolving enzymes in Sulfolobus solfataricus. J. Mol. Biol. 2000, 297, 923–932. [Google Scholar] [CrossRef]
- Huang, Q.; Li, Y.; Zeng, C.; Song, T.; Yan, Z.; Ni, J.; She, Q.; Shen, Y. Genetic analysis of the Holliday junction resolvases Hje and Hjc in Sulfolobus islandicus. Extremophiles 2015, 19, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kurosawa, N. Development of the Multiple Gene Knockout System with One-Step PCR in Thermoacidophilic Crenarchaeon Sulfolobus acidocaldarius. Archaea 2017, 2017, 7459310. [Google Scholar] [CrossRef] [Green Version]
- Branzei, D.; Foiani, M. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 2005, 17, 568–575. [Google Scholar] [CrossRef]
- Michel, B.; Boubakri, H.; Baharoglu, Z.; LeMasson, M.; Lestini, R. Recombination proteins and rescue of arrested replication forks. DNA Repair 2007, 6, 967–980. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Tian, B.; Li, S.; Ao, X.; Dalgaard, K.; Gökce, S.; Liang, Y.; She, Q. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem. Soc. Trans. 2013, 41, 405–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lestini, R.; Duan, Z.; Allers, T. The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA Repair 2010, 9, 994–1002. [Google Scholar] [CrossRef]
- Sakai, H.D.; Kurosawa, N. Sulfodiicoccus acidiphilus gen. nov., sp. nov., a sulfur-inhibited thermoacidophilic archaeon belonging to the order Sulfolobales isolated from a terrestrial acidic hot spring. Int. J. Syst. Evol. Microbiol. 2017, 67, 1880–1886. [Google Scholar] [CrossRef]
- Zhang, C.; Phillips, A.P.R.; Wipfler, R.L.; Olsen, G.J.; Whitaker, R.J. The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat. Commun. 2018, 9, 4908. [Google Scholar] [CrossRef] [Green Version]
- Sakai, H.D.; Kurosawa, N. Saccharolobus caldissimus gen. nov., sp. nov., a facultatively anaerobic iron-reducing hyperthermophilic archaeon isolated from an acidic terrestrial hot spring, and reclassification of Sulfolobus solfataricus as Saccharolobus solfataricus comb. nov. and Sulfolobus shibatae as Saccharolobus shibatae comb. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 1271–1278. [Google Scholar] [CrossRef]
- Woodman, I.L.; Bolt, E.L. Molecular biology of Hel308 helicase in archaea. Biochem. Soc. Trans. 2009, 37, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kurosawa, N. Robust growth of archaeal cells lacking a canonical single-stranded DNA-binding protein. FEMS Microbiol. Lett. 2019, 366, fnz124. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kurosawa, N. Disruption of the gene encoding restriction endonuclease SuaI and development of a host–vector system for the thermoacidophilic archaeon Sulfolobus acidocaldarius. Extremophiles 2016, 20, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.W. Isolation of Sulfolobus acidocaldarius mutants. In Archaea: A Laboratory Manual—Thermophiles; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1995; pp. 125–132. [Google Scholar]
- Kurosawa, N.; Itoh, Y.H.; Iwai, T.; Sugai, A.; Uda, I.; Kimura, N.; Horiuchi, T.; Itoh, T. Sulfurisphaera ohwakuensis gen. nov., sp. nov., a novel extremely thermophilic acidophile of the order Sulfolobales. Int. J. Syst. Bacteriol. 1998, 85507, 451–456. [Google Scholar] [CrossRef] [Green Version]


| Strains or DNAs | Relevant Characteristic(s) | Source or Reference |
|---|---|---|
| Strains | ||
| S. acidocaldarius | ||
| DP-1 | SK-1 with Δphr (ΔpyrE ΔsuaI Δphr) | [24] |
| RM-1 | DP-1 with Δhjc (ΔpyrE ΔsuaI Δphr Δhjc) | This study |
| RM-2 | DP-1 with Δhje (ΔpyrE ΔsuaI Δphr Δhje) | This study |
| RM-3 | DP-1 with Δhjm (ΔpyrE ΔsuaI Δphr Δhjm) | This study |
| RM-4 | DP-1 with Δpina (ΔpyrE ΔsuaI Δphr Δpina) | This study |
| RM-5 | RM-4 with Δhjm (ΔpyrE ΔsuaI Δphr Δpina Δhjm) | This study |
| RM-6 | RM-4 with Δhjc (ΔpyrE ΔsuaI Δphr Δpina Δhjc) | This study |
| RM-9 | RM-4 withΔhje (ΔpyrE ΔsuaI Δphr Δpina Δhje) | This study |
| RM-10 | RM-3 with Δhjc (ΔpyrE ΔsuaI Δphr Δhjm Δhjc) | This study |
| RM-11 | RM-3 with Δhje (ΔpyrE ΔsuaI Δphr Δhjm Δhje) | This study |
| Plasmid DNA | ||
| placSpyrE | Plasmid DNA carrying 0.8 kb of 5′ and 3′ homologous regions of suaI locus at both ends of pyrE-lacS dual marker | [24] |
| PCR products | ||
| MONSTER-hjc | Linear DNA containing the 38-bp 5′ and 30-bp 3′ sequences of the hjc flanking regions, and a 38-bp region of hjc as the Tg-arm at both ends of pyrE-lacS dual marker, respectively | This study |
| MONSTER-hje | Linear DNA containing the 38-bp 5′ and 30-bp 3′ sequences of the hje flanking regions, and a 38-bp region of hje as the Tg-arm at both ends of pyrE-lacS dual marker, respectively | This study |
| MONSTER-hjm | Linear DNA containing the 38-bp 5′ and 30-bp 3′ sequences of the hjm flanking regions, and a 38-bp region of hjm as the Tg-arm at both ends of pyrE-lacS dual marker, respectively | This study |
| MONSTER-pina | Linear DNA containing the 38-bp 5′ and 30-bp 3′ sequences of the pina flanking regions, and a 38-bp region of pina as the Tg-arm at both ends of pyrE-lacS dual marker, respectively | This study |
| Primers | Sequence (5′-3′) * |
|---|---|
| MONSTER-hjc-F | ttagagatataattctgagaggaaaacctaaatcctaagtaaggtatgttgagggaaaaataagtaaaTGTTTTTCTCTATATCAATCTC |
| MONSTER-hjc-R | cctttagttttattactcacataataaaataaaaacacACTCCTAGATCTAAAACTAAAG |
| MONSTER-hje-F | cactcctttttaaggcttatcagacaattttggtgcaatatttatttttcctgttagcgagatgtaagTGTTTTTCTCTATATCAATCTC |
| MONSTER-hje-R | agttccctttcagcactttttccaatgtctctattcatACTCCTAGATCTAAAACTAAAG |
| MONSTER-hjm-F | cttataaatcctacaaaataatgggtaacgtttaggatttacttaaacggtaaagtgacatttaaggaTGTTTTTCTCTATATCAATCTC |
| MONSTER-hjm-R | ctatcgacaggtaaatcttctacggtaatttcttccatACTCCTAGATCTAAAACTAAAG |
| MONSTER-pina-F | gtatgaaataaaatcctattcagaagggtattttagtctatgagaaaaagtacaagataaagataagaTGTTTTTCTCTATATCAATCTC |
| MONSTER-pina-R | agagcagacttatctggaagtaaatctcttgcaggcaaACTCCTAGATCTAAAACTAAAG |
| Hjc-out-F | gcaaatactatcaaagaagg |
| Hjc-out-R | tgtttaataaaaaagttgtctc |
| Hje-out-F | taggaagcaaataaatctatc |
| Hje-out-R | aaagagttaggaactcattg |
| Hjm-out-F | aaaggaaaagcttattaatgg |
| Hjm-out-R | tctatacgacttttcttacc |
| PINA-out-F | ttcatcctgaattatcagag |
| PINA-out-R | attatgttgcggatttagag |
| Hjc-in-F | tgagagatatcttgtttcaag |
| Hjc-in-R | tgacttaattgtctctaaatcc |
| Hje-in-F | ctgtggtatgtagttctctagg |
| Hje-in-R | cctagagaactacataccacag |
| Hjm-in-F | ttatggcagaattaggtatg |
| Hjm-in-R | actctgaccaactctaacaac |
| PINA-in-F | tttcgaagtacattgagaac |
| PINA-in-R | ttctccaaaaacgtatatctc |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuda, R.; Suzuki, S.; Kurosawa, N. Genetic Study of Four Candidate Holliday Junction Processing Proteins in the Thermophilic Crenarchaeon Sulfolobus acidocaldarius. Int. J. Mol. Sci. 2022, 23, 707. https://doi.org/10.3390/ijms23020707
Matsuda R, Suzuki S, Kurosawa N. Genetic Study of Four Candidate Holliday Junction Processing Proteins in the Thermophilic Crenarchaeon Sulfolobus acidocaldarius. International Journal of Molecular Sciences. 2022; 23(2):707. https://doi.org/10.3390/ijms23020707
Chicago/Turabian StyleMatsuda, Ryo, Shoji Suzuki, and Norio Kurosawa. 2022. "Genetic Study of Four Candidate Holliday Junction Processing Proteins in the Thermophilic Crenarchaeon Sulfolobus acidocaldarius" International Journal of Molecular Sciences 23, no. 2: 707. https://doi.org/10.3390/ijms23020707






