Changes in the Transcriptome Profiles of Human Amnion-Derived Mesenchymal Stromal/Stem Cells Induced by Three-Dimensional Culture: A Potential Priming Strategy to Improve Their Properties
Abstract
:1. Introduction
2. Results
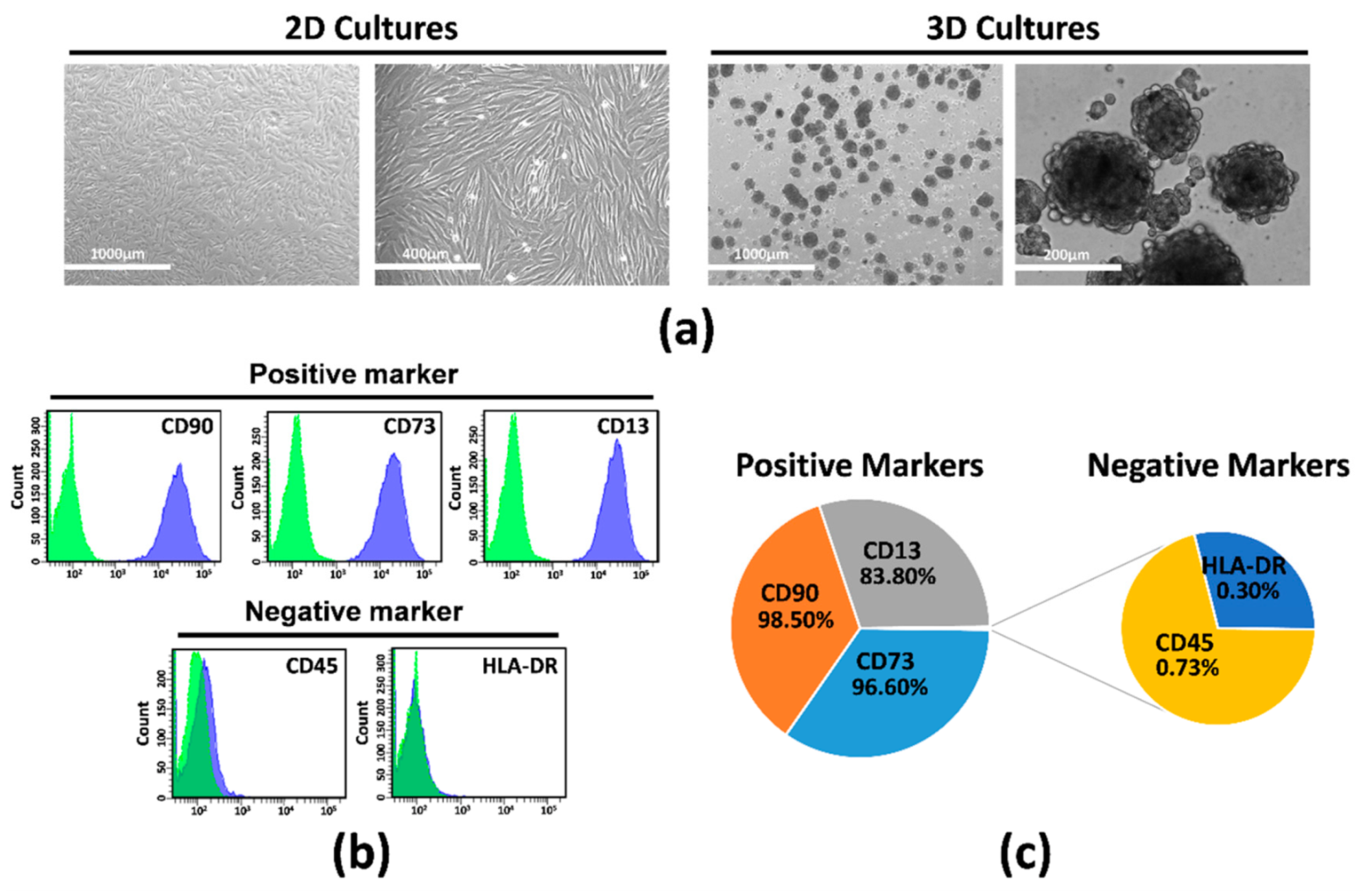
2.1. Isolation, Characterization, and Culture of hAMSCs
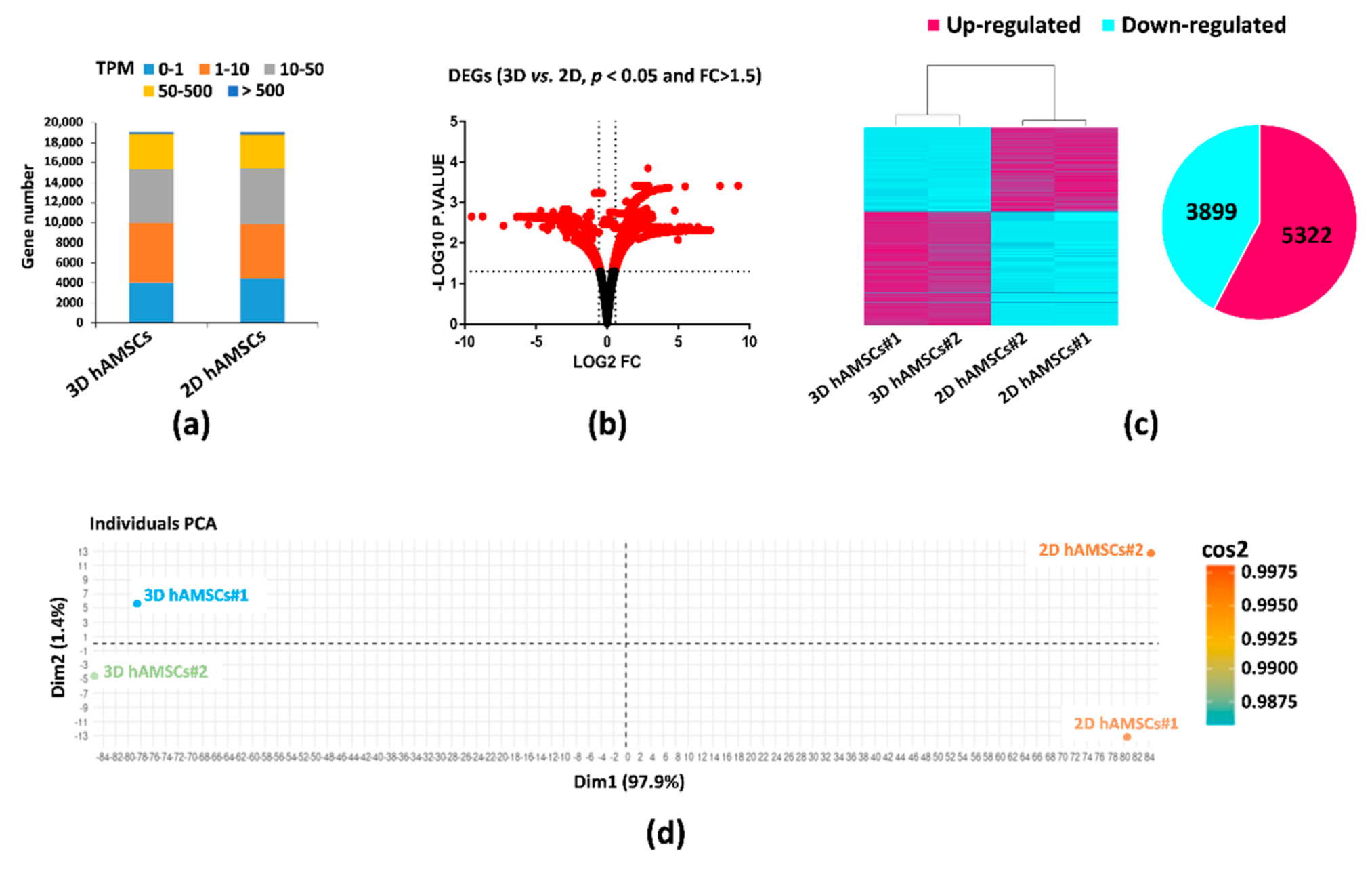
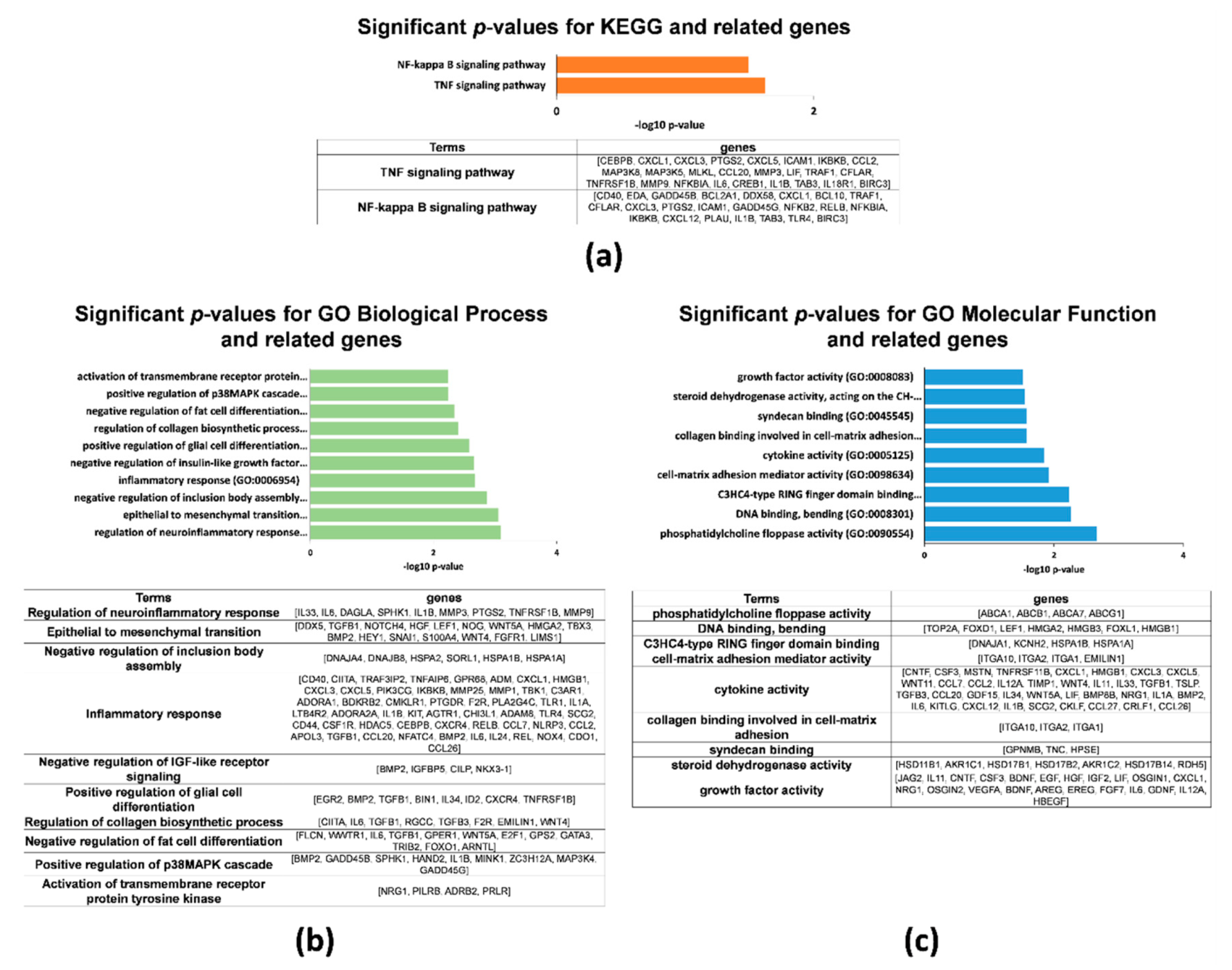
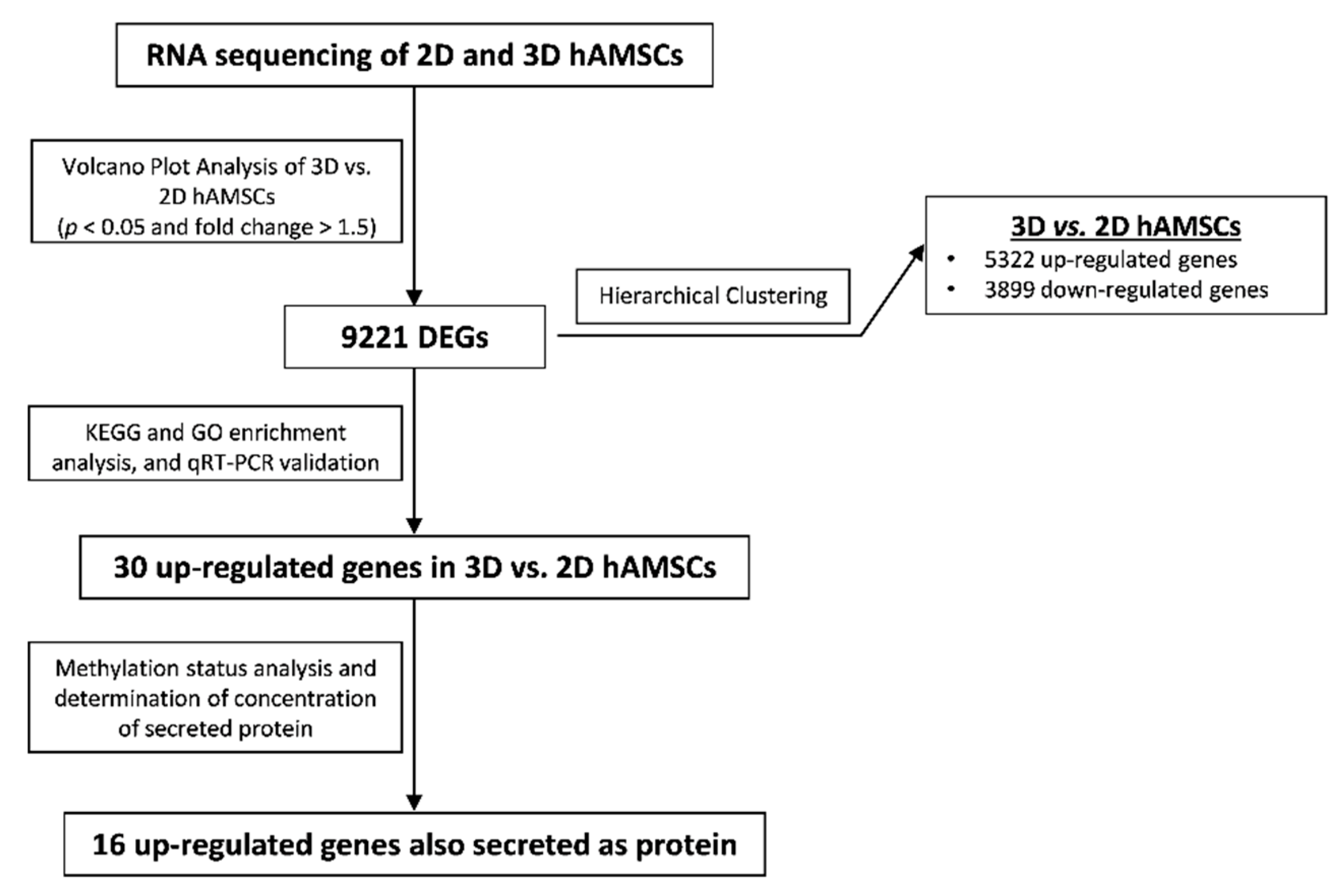
2.2. Gene Expression Profiles in 2D and 3D hAMSCs Revealed Enhanced Regenerative Properties of the hAMSC Spheroids
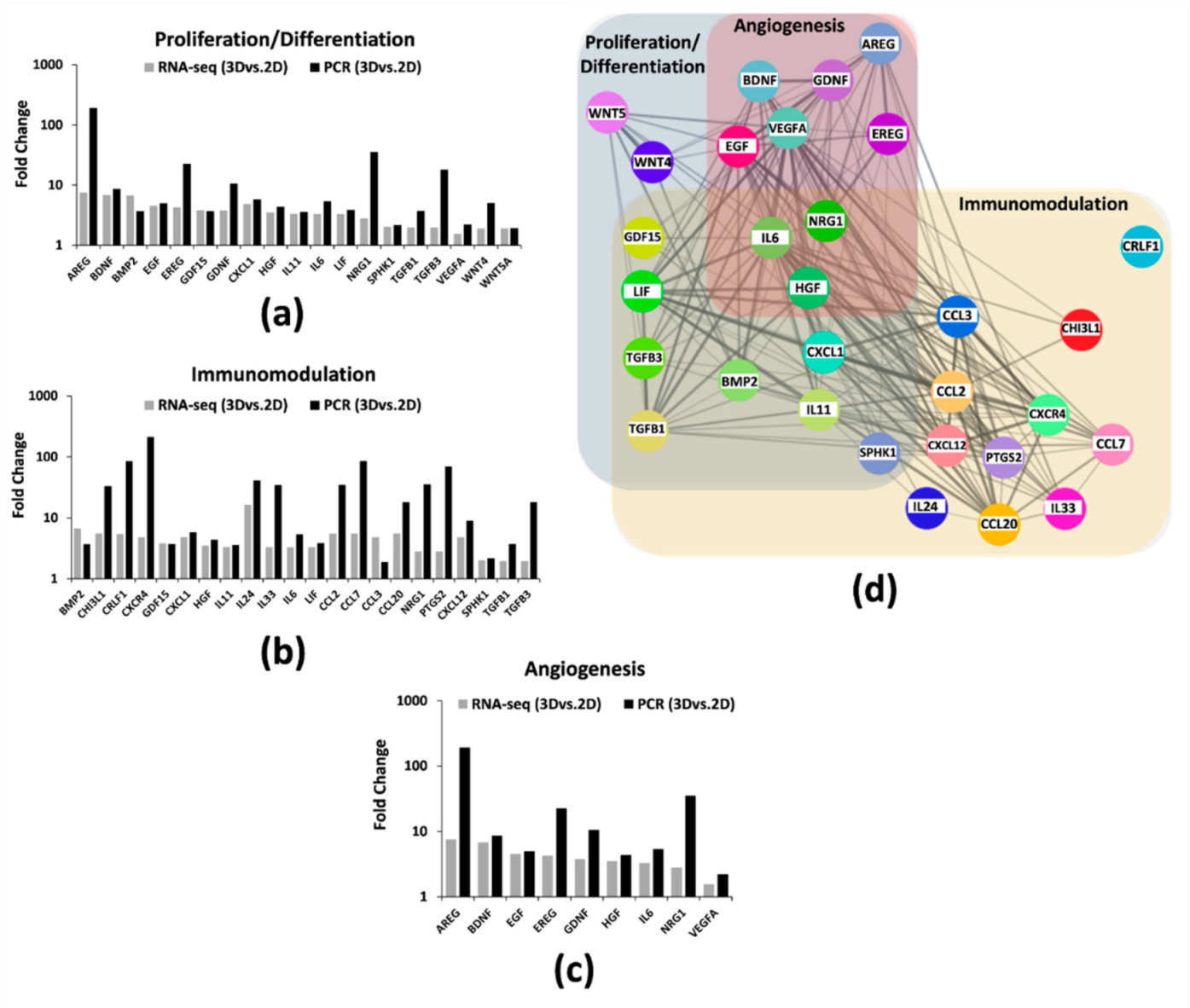
2.3. Spheroid Formation of hAMSCs Induced Changes in Methylation Status and Increased the Production of Bioactive Factors
3. Discussion
4. Materials and Methods
4.1. Isolation, Culture, and Phenotypic Characterization of Human Amnion-Derived Mesenchymal Stromal/Stem Cells
4.2. Mesenchymal Stromal/Stem Cell Spheroid Cultures
4.3. Conditioned Media Preparation
4.4. Gene Expression Profiling
4.5. Protein Expression Analysis
4.6. RNA-Seq, Library Construction, Sequencing, and Analysis
4.7. Bisulfite Genomic Sequencing Analysis of DNA Methylation
4.8. Whole-Genome Bisulfite Sequencing Data Mapping and Quality Analysis
4.9. Pathway Enrichment Analysis
4.10. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| CM | Conditioned medium |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| hAMSCs | Amnion-derived mesenchymal stromal/stem cells |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MSCs | Mesenchymal stromal/stem cells |
| NGS | Next-generation sequencing |
| PCA | Principal component analysis |
| PPI | Protein–protein interactions |
| RNA-seq | RNA sequencing |
| TPM | Transcripts per kilobase million |
References
- Wosczyna, M.N.; Konishi, C.T.; Perez Carbajal, E.E.; Wang, T.T.; Walsh, R.A.; Gan, Q.; Wagner, M.W.; Rando, T.A. Mesenchymal Stromal Cells Are Required for Regeneration and Homeostatic Maintenance of Skeletal Muscle. Cell Rep. 2019, 27, 2029–2035.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagaradze, G.; Basalova, N.; Kirpatovsky, V.; Ohobotov, D.; Nimiritsky, P.; Grigorieva, O.; Popov, V.; Kamalov, A.; Tkachuk, V.; Efimenko, A. A magic kick for regeneration: Role of mesenchymal stromal cell secretome in spermatogonial stem cell niche recovery. Stem Cell Res. Ther. 2019, 10, 342. [Google Scholar] [CrossRef] [Green Version]
- Miceli, V.; Bulati, M.; Iannolo, G.; Zito, G.; Gallo, A.; Conaldi, P.G. Therapeutic Properties of Mesenchymal Stromal/Stem Cells: The Need of Cell Priming for Cell-Free Therapies in Regenerative Medicine. Int. J. Mol. Sci. 2021, 22, 763. [Google Scholar] [CrossRef]
- Lu, Z.; Chen, Y.; Dunstan, C.; Roohani-Esfahani, S.; Zreiqat, H. Priming Adipose Stem Cells with Tumor Necrosis Factor-Alpha Preconditioning Potentiates Their Exosome Efficacy for Bone Regeneration. Tissue Eng. Part A 2017, 23, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Jeong, H.J.; Lee, S.K.; Kim, S.J. Hypoxic Conditioned Medium from Human Adipose-Derived Stem Cells Promotes Mouse Liver Regeneration through JAK/STAT3 Signaling. Stem Cells Transl. Med. 2016, 5, 816–825. [Google Scholar] [CrossRef]
- Bulati, M.; Miceli, V.; Gallo, A.; Amico, G.; Carcione, C.; Pampalone, M.; Conaldi, P.G. The Immunomodulatory Properties of the Human Amnion-Derived Mesenchymal Stromal/Stem Cells Are Induced by INF-gamma Produced by Activated Lymphomonocytes and Are Mediated by Cell-To-Cell Contact and Soluble Factors. Front. Immunol. 2020, 11, 54. [Google Scholar] [CrossRef] [Green Version]
- Chinnici, C.M.; Russelli, G.; Bulati, M.; Miceli, V.; Gallo, A.; Busa, R.; Tinnirello, R.; Conaldi, P.G.; Iannolo, G. Mesenchymal stromal cell secretome in liver failure: Perspectives on COVID-19 infection treatment. World J. Gastroenterol. 2021, 27, 1905–1919. [Google Scholar] [CrossRef]
- Lo Nigro, A.; Gallo, A.; Bulati, M.; Vitale, G.; Paini, D.S.; Pampalone, M.; Galvagno, D.; Conaldi, P.G.; Miceli, V. Amnion-Derived Mesenchymal Stromal/Stem Cell Paracrine Signals Potentiate Human Liver Organoid Differentiation: Translational Implications for Liver Regeneration. Front. Med. 2021, 8, 746298. [Google Scholar] [CrossRef] [PubMed]
- Maacha, S.; Sidahmed, H.; Jacob, S.; Gentilcore, G.; Calzone, R.; Grivel, J.C.; Cugno, C. Paracrine Mechanisms of Mesenchymal Stromal Cells in Angiogenesis. Stem Cells Int. 2020, 2020, 4356359. [Google Scholar] [CrossRef]
- Miceli, V.; Bertani, A.; Chinnici, C.M.; Bulati, M.; Pampalone, M.; Amico, G.; Carcione, C.; Schmelzer, E.; Gerlach, J.C.; Conaldi, P.G. Conditioned Medium from Human Amnion-Derived Mesenchymal Stromal/Stem Cells Attenuating the Effects of Cold Ischemia-Reperfusion Injury in an in vitro Model Using Human Alveolar Epithelial Cells. Int. J. Mol. Sci. 2021, 22, 510. [Google Scholar] [CrossRef] [PubMed]
- Nuzzi, R.; Caselgrandi, P.; Vercelli, A. Effect of Mesenchymal Stem Cell-Derived Exosomes on Retinal Injury: A Review of Current Findings. Stem Cells Int. 2020, 2020, 8883616. [Google Scholar] [CrossRef]
- Qian, X.; An, N.; Ren, Y.; Yang, C.; Zhang, X.; Li, L. Immunosuppressive Effects of Mesenchymal Stem Cells-derived Exosomes. Stem Cell Rev. Rep. 2021, 17, 411–427. [Google Scholar] [CrossRef]
- Sun, H.; Pratt, R.E.; Hodgkinson, C.P.; Dzau, V.J. Sequential paracrine mechanisms are necessary for the therapeutic benefits of stem cell therapy. Am. J. Physiol. Cell Physiol. 2020, 319, C1141–C1150. [Google Scholar] [CrossRef]
- Wang, M.; Yan, L.; Li, Q.; Yang, Y.; Turrentine, M.; March, K.; Wang, I.W. Mesenchymal stem cell secretions improve donor heart function following ex vivo cold storage. J. Thorac. Cardiovasc. Surg. 2020. [Google Scholar] [CrossRef] [PubMed]
- Fricova, D.; Korchak, J.A.; Zubair, A.C. Challenges and translational considerations of mesenchymal stem/stromal cell therapy for Parkinson’s disease. NPJ Regen. Med. 2020, 5, 20. [Google Scholar] [CrossRef]
- Lukomska, B.; Stanaszek, L.; Zuba-Surma, E.; Legosz, P.; Sarzynska, S.; Drela, K. Challenges and Controversies in Human Mesenchymal Stem Cell Therapy. Stem Cells Int. 2019, 2019, 9628536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squillaro, T.; Peluso, G.; Galderisi, U. Clinical Trials with Mesenchymal Stem Cells: An Update. Cell Transplant. 2016, 25, 829–848. [Google Scholar] [CrossRef] [Green Version]
- Tyndall, A. Successes and failures of stem cell transplantation in autoimmune diseases. Hematol. 2010 Am. Soc. Hematol. Educ. Program Book 2011, 2011, 280–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmelzer, E.; Miceli, V.; Chinnici, C.M.; Bertani, A.; Gerlach, J.C. Effects of Mesenchymal Stem Cell Coculture on Human Lung Small Airway Epithelial Cells. BioMed Res. Int. 2020, 2020, 9847579. [Google Scholar] [CrossRef] [PubMed]
- Frith, J.E.; Thomson, B.; Genever, P.G. Dynamic three-dimensional culture methods enhance mesenchymal stem cell properties and increase therapeutic potential. Tissue Eng. Part C Methods 2010, 16, 735–749. [Google Scholar] [CrossRef]
- Kouroupis, D.; Correa, D. Increased Mesenchymal Stem Cell Functionalization in Three-Dimensional Manufacturing Settings for Enhanced Therapeutic Applications. Front. Bioeng. Biotechnol. 2021, 9, 621748. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.G.; Randau, T.M.; Hilgers, C.; Haddouti, E.M.; Masson, W.; Gravius, S.; Burger, C.; Wirtz, D.C.; Schildberg, F.A. Molecular and Functional Phenotypes of Human Bone Marrow-Derived Mesenchymal Stromal Cells Depend on Harvesting Techniques. Int. J. Mol. Sci. 2020, 21, 4382. [Google Scholar] [CrossRef] [PubMed]
- Burja, B.; Barlic, A.; Erman, A.; Mrak-Poljsak, K.; Tomsic, M.; Sodin-Semrl, S.; Lakota, K. Human mesenchymal stromal cells from different tissues exhibit unique responses to different inflammatory stimuli. Curr. Res. Transl. Med. 2020, 68, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Tesarova, L.; Jaresova, K.; Simara, P.; Koutna, I. Umbilical Cord-Derived Mesenchymal Stem Cells Are Able to Use bFGF Treatment and Represent a Superb Tool for Immunosuppressive Clinical Applications. Int. J. Mol. Sci. 2020, 21, 5366. [Google Scholar] [CrossRef]
- Parolini, O.; Alviano, F.; Bagnara, G.P.; Bilic, G.; Buhring, H.J.; Evangelista, M.; Hennerbichler, S.; Liu, B.; Magatti, M.; Mao, N.; et al. Concise review: Isolation and characterization of cells from human term placenta: Outcome of the first international Workshop on Placenta Derived Stem Cells. Stem Cells 2008, 26, 300–311. [Google Scholar] [CrossRef] [Green Version]
- Magatti, M.; Masserdotti, A.; Bonassi Signoroni, P.; Vertua, E.; Stefani, F.R.; Silini, A.R.; Parolini, O. B Lymphocytes as Targets of the Immunomodulatory Properties of Human Amniotic Mesenchymal Stromal Cells. Front. Immunol. 2020, 11, 1156. [Google Scholar] [CrossRef]
- Miceli, V.; Pampalone, M.; Vella, S.; Carreca, A.P.; Amico, G.; Conaldi, P.G. Comparison of Immunosuppressive and Angiogenic Properties of Human Amnion-Derived Mesenchymal Stem Cells between 2D and 3D Culture Systems. Stem Cells Int. 2019, 2019, 7486279. [Google Scholar] [CrossRef]
- Poggi, A.; Zocchi, M.R. Immunomodulatory Properties of Mesenchymal Stromal Cells: Still Unresolved “Yin and Yang”. Curr. Stem Cell Res. Ther. 2019, 14, 344–350. [Google Scholar] [CrossRef]
- Fan, X.L.; Zhang, Y.; Li, X.; Fu, Q.L. Mechanisms underlying the protective effects of mesenchymal stem cell-based therapy. Cell. Mol. Life Sci. 2020, 77, 2771–2794. [Google Scholar] [CrossRef] [Green Version]
- Tao, H.; Han, Z.; Han, Z.C.; Li, Z. Proangiogenic Features of Mesenchymal Stem Cells and Their Therapeutic Applications. Stem Cells Int. 2016, 2016, 1314709. [Google Scholar] [CrossRef] [Green Version]
- Stavely, R.; Nurgali, K. The emerging antioxidant paradigm of mesenchymal stem cell therapy. Stem Cells Transl. Med. 2020, 9, 985–1006. [Google Scholar] [CrossRef]
- Arrigoni, C.; D’Arrigo, D.; Rossella, V.; Candrian, C.; Albertini, V.; Moretti, M. Umbilical Cord MSCs and Their Secretome in the Therapy of Arthritic Diseases: A Research and Industrial Perspective. Cells 2020, 9, 1343. [Google Scholar] [CrossRef] [PubMed]
- Silini, A.R.; Papait, A.; Cargnoni, A.; Vertua, E.; Romele, P.; Bonassi Signoroni, P.; Magatti, M.; De Munari, S.; Masserdotti, A.; Pasotti, A.; et al. CM from intact hAM: An easily obtained product with relevant implications for translation in regenerative medicine. Stem Cell Res. Ther. 2021, 12, 540. [Google Scholar] [CrossRef]
- Cesarz, Z.; Tamama, K. Spheroid Culture of Mesenchymal Stem Cells. Stem Cells Int. 2016, 2016, 9176357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.C.; Wang, H.W.; Huang, C.C. Modulation of Inherent Niches in 3D Multicellular MSC Spheroids Reconfigures Metabolism and Enhances Therapeutic Potential. Cells 2021, 10, 2747. [Google Scholar] [CrossRef] [PubMed]
- Jaukovic, A.; Abadjieva, D.; Trivanovic, D.; Stoyanova, E.; Kostadinova, M.; Pashova, S.; Kestendjieva, S.; Kukolj, T.; Jeseta, M.; Kistanova, E.; et al. Specificity of 3D MSC Spheroids Microenvironment: Impact on MSC Behavior and Properties. Stem Cell Rev. Rep. 2020, 16, 853–875. [Google Scholar] [CrossRef]
- Miceli, V.; Chinnici, C.M.; Bulati, M.; Pampalone, M.; Amico, G.; Schmelzer, E.; Gerlach, J.C.; Conaldi, P.G. Comparative study of the production of soluble factors in human placenta-derived mesenchymal stromal/stem cells grown in adherent conditions or as aggregates in a catheter-like device. Biochem. Biophys. Res. Commun. 2020, 522, 171–176. [Google Scholar] [CrossRef]
- Santos, J.M.; Camoes, S.P.; Filipe, E.; Cipriano, M.; Barcia, R.N.; Filipe, M.; Teixeira, M.; Simoes, S.; Gaspar, M.; Mosqueira, D.; et al. Three-dimensional spheroid cell culture of umbilical cord tissue-derived mesenchymal stromal cells leads to enhanced paracrine induction of wound healing. Stem Cell Res. Ther. 2015, 6, 90. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, J.A.; Hettiaratchi, M.H.; McDevitt, T.C. Enhanced Immunosuppression of T Cells by Sustained Presentation of Bioactive Interferon-gamma within Three-Dimensional Mesenchymal Stem Cell Constructs. Stem Cells Transl. Med. 2017, 6, 223–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivanathan, K.N.; Rojas-Canales, D.; Grey, S.T.; Gronthos, S.; Coates, P.T. Transcriptome Profiling of IL-17A Preactivated Mesenchymal Stem Cells: A Comparative Study to Unmodified and IFN-gamma Modified Mesenchymal Stem Cells. Stem Cells Int. 2017, 2017, 1025820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zielniok, K.; Burdzinska, A.; Murcia Pienkowski, V.; Koppolu, A.; Rydzanicz, M.; Zagozdzon, R.; Paczek, L. Gene Expression Profile of Human Mesenchymal Stromal Cells Exposed to Hypoxic and Pseudohypoxic Preconditioning-An Analysis by RNA Sequencing. Int. J. Mol. Sci. 2021, 22, 8160. [Google Scholar] [CrossRef]
- Roson-Burgo, B.; Sanchez-Guijo, F.; Del Canizo, C.; De Las Rivas, J. Transcriptomic portrait of human Mesenchymal Stromal/Stem Cells isolated from bone marrow and placenta. BMC Genom. 2014, 15, 910. [Google Scholar] [CrossRef] [Green Version]
- Ulloa-Montoya, F.; Kidder, B.L.; Pauwelyn, K.A.; Chase, L.G.; Luttun, A.; Crabbe, A.; Geraerts, M.; Sharov, A.A.; Piao, Y.; Ko, M.S.; et al. Comparative transcriptome analysis of embryonic and adult stem cells with extended and limited differentiation capacity. Genome Biol. 2007, 8, R163. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Li, X.; Zhang, Y.; Han, Y.; Chang, F.; Ding, J. Mesenchymal Stem Cells for Regenerative Medicine. Cells 2019, 8, 886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittenger, M.F.; Discher, D.E.; Peault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizoso, F.J.; Eiro, N.; Cid, S.; Schneider, J.; Perez-Fernandez, R. Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. Int. J. Mol. Sci. 2017, 18, 1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLeod, C.M.; Mauck, R.L. On the origin and impact of mesenchymal stem cell heterogeneity: New insights and emerging tools for single cell analysis. Eur. Cells Mater. 2017, 34, 217–231. [Google Scholar] [CrossRef]
- Sun, C.; Wang, L.; Wang, H.; Huang, T.; Yao, W.; Li, J.; Zhang, X. Single-cell RNA-seq highlights heterogeneity in human primary Wharton’s jelly mesenchymal stem/stromal cells cultured in vitro. Stem Cell Res. Ther. 2020, 11, 149. [Google Scholar] [CrossRef] [Green Version]
- Wegmeyer, H.; Broske, A.M.; Leddin, M.; Kuentzer, K.; Nisslbeck, A.K.; Hupfeld, J.; Wiechmann, K.; Kuhlen, J.; von Schwerin, C.; Stein, C.; et al. Mesenchymal stromal cell characteristics vary depending on their origin. Stem Cells Dev. 2013, 22, 2606–2618. [Google Scholar] [CrossRef] [Green Version]
- Redondo-Castro, E.; Cunningham, C.; Miller, J.; Martuscelli, L.; Aoulad-Ali, S.; Rothwell, N.J.; Kielty, C.M.; Allan, S.M.; Pinteaux, E. Interleukin-1 primes human mesenchymal stem cells towards an anti-inflammatory and pro-trophic phenotype in vitro. Stem Cell Res. Ther. 2017, 8, 79. [Google Scholar] [CrossRef]
- Cho, K.A.; Park, M.; Kim, Y.H.; Woo, S.Y.; Ryu, K.H. RNA sequencing reveals a transcriptomic portrait of human mesenchymal stem cells from bone marrow, adipose tissue, and palatine tonsils. Sci. Rep. 2017, 7, 17114. [Google Scholar] [CrossRef] [Green Version]
- Jansen, B.J.; Gilissen, C.; Roelofs, H.; Schaap-Oziemlak, A.; Veltman, J.A.; Raymakers, R.A.; Jansen, J.H.; Kogler, G.; Figdor, C.G.; Torensma, R.; et al. Functional differences between mesenchymal stem cell populations are reflected by their transcriptome. Stem Cells Dev. 2010, 19, 481–490. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, K.; Yue, J.; Meng, S.; Zhang, X. Deconstructing transcriptional variations and their effects on immunomodulatory function among human mesenchymal stromal cells. Stem Cell Res. Ther. 2021, 12, 53. [Google Scholar] [CrossRef]
- Kwon, A.; Kim, Y.; Kim, M.; Kim, J.; Choi, H.; Jekarl, D.W.; Lee, S.; Kim, J.M.; Shin, J.C.; Park, I.Y. Tissue-specific Differentiation Potency of Mesenchymal Stromal Cells from Perinatal Tissues. Sci. Rep. 2016, 6, 23544. [Google Scholar] [CrossRef]
- Reinisch, A.; Etchart, N.; Thomas, D.; Hofmann, N.A.; Fruehwirth, M.; Sinha, S.; Chan, C.K.; Senarath-Yapa, K.; Seo, E.Y.; Wearda, T.; et al. Epigenetic and in vivo comparison of diverse MSC sources reveals an endochondral signature for human hematopoietic niche formation. Blood 2015, 125, 249–260. [Google Scholar] [CrossRef]
- Sacchetti, B.; Funari, A.; Remoli, C.; Giannicola, G.; Kogler, G.; Liedtke, S.; Cossu, G.; Serafini, M.; Sampaolesi, M.; Tagliafico, E.; et al. No Identical “Mesenchymal Stem Cells” at Different Times and Sites: Human Committed Progenitors of Distinct Origin and Differentiation Potential Are Incorporated as Adventitial Cells in Microvessels. Stem Cell Rep. 2016, 6, 897–913. [Google Scholar] [CrossRef] [Green Version]
- Greenberg, D.A. Cerebral angiogenesis: A realistic therapy for ischemic disease? Methods Mol. Biol. 2014, 1135, 21–24. [Google Scholar]
- Kwon, H.M.; Hur, S.M.; Park, K.Y.; Kim, C.K.; Kim, Y.M.; Kim, H.S.; Shin, H.C.; Won, M.H.; Ha, K.S.; Kwon, Y.G.; et al. Multiple paracrine factors secreted by mesenchymal stem cells contribute to angiogenesis. Vasc. Pharmacol. 2014, 63, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jin, K.; Xie, L.; Childs, J.; Mao, X.O.; Logvinova, A.; Greenberg, D.A. VEGF-induced neuroprotection, neurogenesis, and angiogenesis after focal cerebral ischemia. J. Clin. Investig. 2003, 111, 1843–1851. [Google Scholar] [CrossRef] [PubMed]
- Usui, T.; Naruo, A.; Okada, M.; Hayabe, Y.; Yamawaki, H. Brain-derived neurotrophic factor promotes angiogenic tube formation through generation of oxidative stress in human vascular endothelial cells. Acta Physiol. 2014, 211, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.S.; Lee, J.H.; Yoon, Y.M.; Yun, C.W.; Noh, H.; Lee, S.H. Hypoxia-induced expression of cellular prion protein improves the therapeutic potential of mesenchymal stem cells. Cell Death Dis. 2016, 7, e2395. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Kumar, S.; Heinzel, A.; Gao, M.; Guo, J.; Alvarado, G.F.; Reindl-Schwaighofer, R.; Krautzberger, A.M.; Cippa, P.E.; McMahon, J.; et al. Renoprotective and Immunomodulatory Effects of GDF15 following AKI Invoked by Ischemia-Reperfusion Injury. J. Am. Soc. Nephrol. 2020, 31, 701–715. [Google Scholar] [CrossRef]
- Najar, M.; Raicevic, G.; Fayyad-Kazan, H.; Bron, D.; Toungouz, M.; Lagneaux, L. Mesenchymal stromal cells and immunomodulation: A gathering of regulatory immune cells. Cytotherapy 2016, 18, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Giri, J.; Das, R.; Nylen, E.; Chinnadurai, R.; Galipeau, J. CCL2 and CXCL12 Derived from Mesenchymal Stromal Cells Cooperatively Polarize IL-10+ Tissue Macrophages to Mitigate Gut Injury. Cell Rep. 2020, 30, 1923–1934.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, F.; Zhou, Y.; Wang, J.; Liu, C.; Xiao, Y. The Immunomodulatory Role of BMP-2 on Macrophages to Accelerate Osteogenesis. Tissue Eng. Part A 2018, 24, 584–594. [Google Scholar] [CrossRef]
- Curti, A.; Ratta, M.; Corinti, S.; Girolomoni, G.; Ricci, F.; Tazzari, P.; Siena, M.; Grande, A.; Fogli, M.; Tura, S.; et al. Interleukin-11 induces Th2 polarization of human CD4 (+) T cells. Blood 2001, 97, 2758–2763. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Yin, S.; Zhang, W.; Gao, F.; Liu, Y.; Chen, Z.; Zhang, M.; He, J.; Zheng, S. Hypoxia preconditioned bone marrow mesenchymal stem cells promote liver regeneration in a rat massive hepatectomy model. Stem Cell Res. Ther. 2013, 4, 83. [Google Scholar] [CrossRef] [Green Version]
- Gothelf, Y.; Abramov, N.; Harel, A.; Offen, D. Safety of repeated transplantations of neurotrophic factors-secreting human mesenchymal stromal stem cells. Clin. Transl. Med. 2014, 3, 21. [Google Scholar] [CrossRef]
- Bavaro, T.; Tengattini, S.; Rezwan, R.; Chiesa, E.; Temporini, C.; Dorati, R.; Massolini, G.; Conti, B.; Ubiali, D.; Terreni, M. Design of epidermal growth factor immobilization on 3D biocompatible scaffolds to promote tissue repair and regeneration. Sci. Rep. 2021, 11, 2629. [Google Scholar] [CrossRef]
- Xu, X.; Zheng, L.; Yuan, Q.; Zhen, G.; Crane, J.L.; Zhou, X.; Cao, X. Transforming growth factor-beta in stem cells and tissue homeostasis. Bone Res. 2018, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Xue, B.; Liu, D.; Song, M.; Zhao, G.; Cao, Y.; Yan, G.; Dai, J.; Hu, Y. Leukemia inhibitory factor promotes the regeneration of rat uterine horns with full-thickness injury. Wound Repair Regen. 2019, 27, 477–487. [Google Scholar] [CrossRef]
- Oka, M.; Kobayashi, N.; Matsumura, K.; Nishio, M.; Nakano, K.; Okamura, T.; Okochi, H.; Minamisawa, T.; Shiba, K.; Saeki, K. New Role for Growth/Differentiation Factor 15 in the Survival of Transplanted Brown Adipose Tissues in Cooperation with Interleukin-6. Cells 2020, 9, 1365. [Google Scholar] [CrossRef] [PubMed]
- Gawriluk, T.R.; Simkin, J.; Hacker, C.K.; Kimani, J.M.; Kiama, S.G.; Ezenwa, V.O.; Seifert, A.W. Complex Tissue Regeneration in Mammals Is Associated with Reduced Inflammatory Cytokines and an Influx of T Cells. Front. Immunol. 2020, 11, 1695. [Google Scholar] [CrossRef] [PubMed]
- Gray, A.; Schloss, R.S.; Yarmush, M. Donor variability among anti-inflammatory pre-activated mesenchymal stromal cells. Technology 2016, 4, 201–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
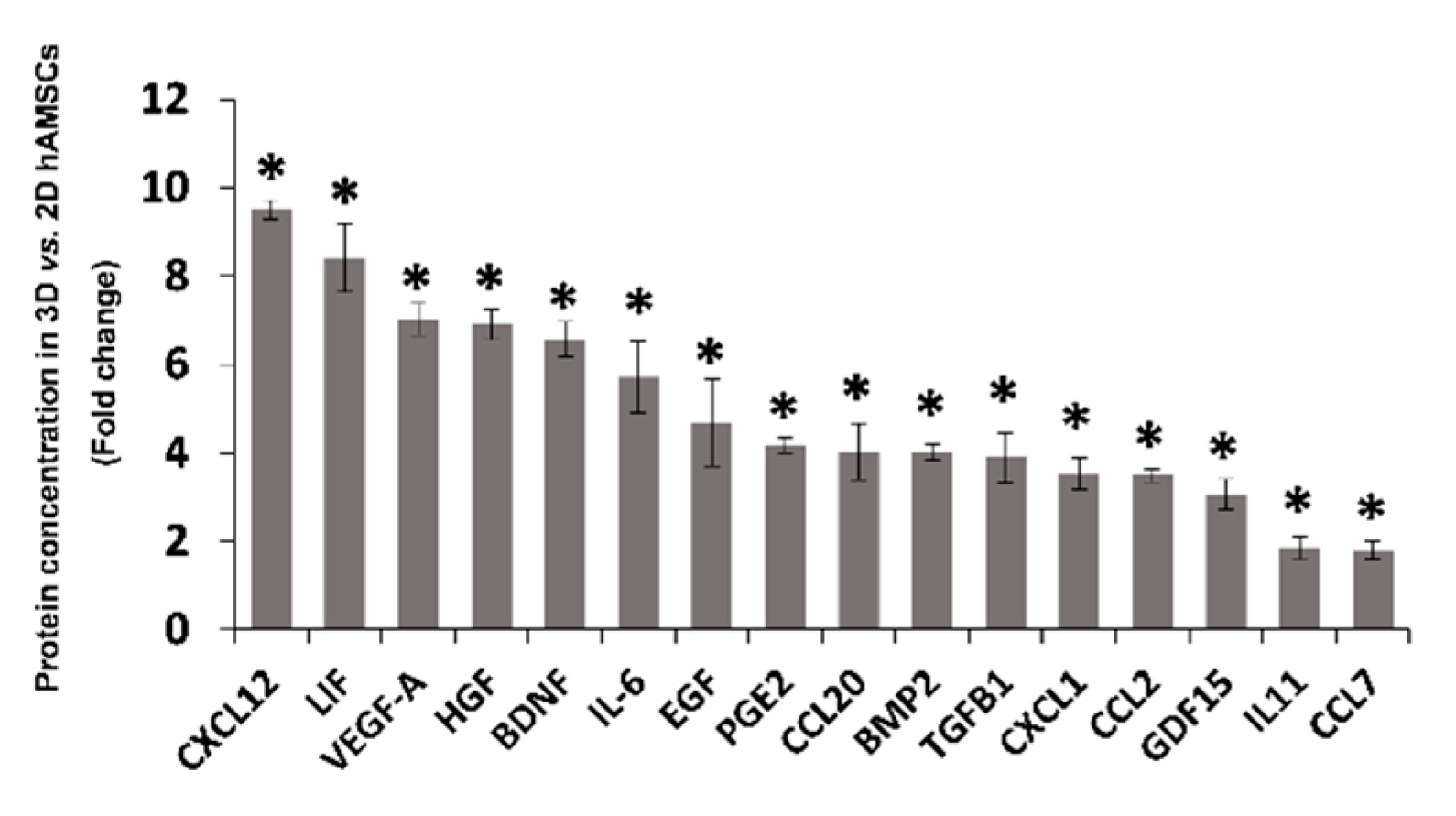
| Gene | Chromosome | Start | End | Description | 2D hAMSCs (%) | 3D hAMSCs (%) |
|---|---|---|---|---|---|---|
| AREG | 4 | 75480629 | 75490486 | Amphiregulin | 74.70 | 73.60 |
| BDNF | 11 | 27676440 | 27743605 | Brain-derived neurotrophic factor | 14.08 | 10.24 |
| BMP2 | 20 | 6748311 | 6760910 | Bone morphogenetic protein 2 | 7.32 | 4.45 |
| CCL2 | 17 | 32582313 | 32584222 | Chemokine (C-C motif) ligand 2 | 24.41 | 12.93 |
| CCL20 | 2 | 228678558 | 228682272 | Chemokine (C-C motif) ligand 20 | 84.90 | 77.06 |
| CCL3 | 17 | 34415602 | 34417515 | Chemokine (C-C motif) ligand 3 | 42.86 | 56.67 |
| CCL7 | 17 | 32597240 | 32599261 | Chemokine (C-C motif) ligand 7 | 45.19 | 39.34 |
| CHI3L1 | 1 | 203148059 | 203155877 | Chitinase 3-like 1 | 68.58 | 76.02 |
| CRLF1 | 19 | 18704037 | 18717660 | Cytokine receptor-like factor 1 | 17.79 | 17.66 |
| CXCL1 | 4 | 74735110 | 74736959 | Chemokine (C-X-C motif) ligand 1 | 18.06 | 11.30 |
| CXCL12 | 10 | 44793038 | 44881941 | Chemokine (C-X-C motif) ligand 12 | 30.98 | 26.83 |
| CXCR4 | 2 | 136871919 | 136875735 | Chemokine (C-X-C motif) receptor 4 | 7.47 | 6.95 |
| EGF | 4 | 110834040 | 110933422 | Epidermal growth factor | 60.13 | 56.20 |
| EREG | 4 | 75230860 | 75254468 | Epiregulin | 26.62 | 26.19 |
| GDF15 | 19 | 18496968 | 18499986 | Growth differentiation factor 15 | 20.99 | 16.99 |
| GDNF | 5 | 37812779 | 37839788 | Glial cell-derived neurotrophic factor | 16.70 | 16.05 |
| HGF | 7 | 81328322 | 81399754 | Hepatocyte growth factor | 50.33 | 41.92 |
| IL11 | 19 | 55875757 | 55881814 | Interleukin 11 | 26.80 | 21.64 |
| IL24 | 1 | 207070788 | 207077484 | Interleukin 24 | 71.90 | 70.12 |
| IL33 | 9 | 6215805 | 6257983 | Interleukin 33 | 60.55 | 68.69 |
| IL6 | 7 | 22765503 | 22771621 | Interleukin 6 | 19.70 | 12.45 |
| LIF | 22 | 30636436 | 30642840 | Leukemia inhibitory factor | 48.59 | 41.73 |
| NRG1 | 8 | 31496902 | 32622548 | Neuregulin 1 | 37.85 | 36.44 |
| PTGS2 | 1 | 186640923 | 186649559 | Prostaglandin-endoperoxide synthase 2 | 32.49 | 27.00 |
| SPHK1 | 17 | 74372742 | 74383941 | Sphingosine kinase 1 | 14.60 | 13.79 |
| TGFB1 | 19 | 41836813 | 41859831 | Transforming growth factor, beta 1 | 20.05 | 16.55 |
| TGFB3 | 14 | 76424442 | 76449334 | Transforming growth factor, beta 3 | 20.21 | 19.32 |
| VEGF-A | 6 | 43737921 | 43754224 | Vascular endothelial growth factor A | 25.56 | 21.88 |
| WNT4 | 1 | 22446461 | 22470462 | Wingless-type MMTV integration site family, member 4 | 43.70 | 48.77 |
| WNT5A | 3 | 55499743 | 55523973 | Wingless-type MMTV integration site family, member 5A | 28.81 | 31.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallo, A.; Cuscino, N.; Contino, F.; Bulati, M.; Pampalone, M.; Amico, G.; Zito, G.; Carcione, C.; Centi, C.; Bertani, A.; et al. Changes in the Transcriptome Profiles of Human Amnion-Derived Mesenchymal Stromal/Stem Cells Induced by Three-Dimensional Culture: A Potential Priming Strategy to Improve Their Properties. Int. J. Mol. Sci. 2022, 23, 863. https://doi.org/10.3390/ijms23020863
Gallo A, Cuscino N, Contino F, Bulati M, Pampalone M, Amico G, Zito G, Carcione C, Centi C, Bertani A, et al. Changes in the Transcriptome Profiles of Human Amnion-Derived Mesenchymal Stromal/Stem Cells Induced by Three-Dimensional Culture: A Potential Priming Strategy to Improve Their Properties. International Journal of Molecular Sciences. 2022; 23(2):863. https://doi.org/10.3390/ijms23020863
Chicago/Turabian StyleGallo, Alessia, Nicola Cuscino, Flavia Contino, Matteo Bulati, Mariangela Pampalone, Giandomenico Amico, Giovanni Zito, Claudia Carcione, Claudio Centi, Alessandro Bertani, and et al. 2022. "Changes in the Transcriptome Profiles of Human Amnion-Derived Mesenchymal Stromal/Stem Cells Induced by Three-Dimensional Culture: A Potential Priming Strategy to Improve Their Properties" International Journal of Molecular Sciences 23, no. 2: 863. https://doi.org/10.3390/ijms23020863
APA StyleGallo, A., Cuscino, N., Contino, F., Bulati, M., Pampalone, M., Amico, G., Zito, G., Carcione, C., Centi, C., Bertani, A., Conaldi, P. G., & Miceli, V. (2022). Changes in the Transcriptome Profiles of Human Amnion-Derived Mesenchymal Stromal/Stem Cells Induced by Three-Dimensional Culture: A Potential Priming Strategy to Improve Their Properties. International Journal of Molecular Sciences, 23(2), 863. https://doi.org/10.3390/ijms23020863








