One Step Forward with Dry Surface Biofilm (DSB) of Staphylococcus aureus: TMT-Based Quantitative Proteomic Analysis Reveals Proteomic Shifts between DSB and Hydrated Biofilm
Abstract
1. Introduction
2. Results and Discussion
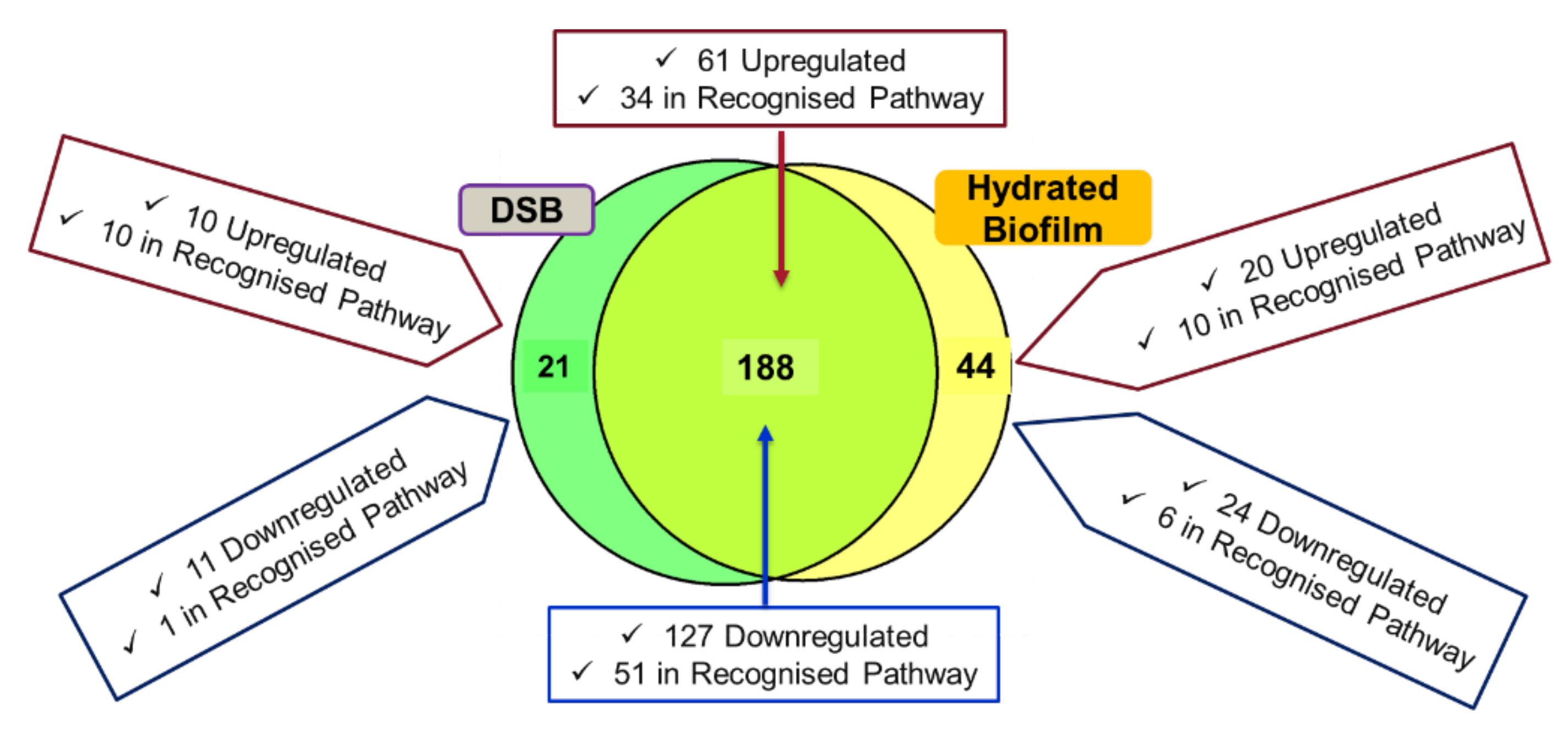
2.1. TMT Identification of Differentially Regulated Proteins in the Hydrated Biofilm and DSB
2.2. Energy Metabolism-Associated Proteins Are Significantly Upregulated in the Hydrated Biofilm
2.3. Sugar Transporter and Cell-Wall Synthesis Proteins Are Highly Abundant in the DSB
2.4. Fatty Acid Pathway Could Be a Potential Target for DSB
2.5. Ribosomal and ABC Transporter Proteins Revealed Decreased Abundance in the Hydrated Biofilm
2.6. Significant Differentially Expressed Stress Response Proteins in the Hydrated Biofilm and DSB
2.7. Scanning Electron Microscopy Observation of Hydrated Biofilm and DSB
2.8. Validation of TMT Data with qPCR Results
3. Materials and Methods
3.1. Microorganism and Culture Conditions
3.2. Protein Extraction and Fractionation
3.3. Protein Reduction, Alkylation, and Digestion
3.4. TMT Labelling and High pH Fractionation
3.5. Nanoflow LC-ESI-MS/MS
3.5.1. Nanoflow LC-ESI-MS/MS Using Orbitrap Elite
3.5.2. Nanoflow LC-ESI-MS/MS Using Q Exactive
3.6. Database Search, Statistical Analysis, and Bioinformatics
3.7. Validation of TMT Data with qPCR Results
3.8. Scanning Electron Microscopy Observation of Biofilm
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pajkos, A.; Vickery, K.; Cossart, Y. Is biofilm accumulation on endoscope tubing a contributor to the failure of cleaning and decontamination? J. Hosp. Infect. 2004, 58, 224–229. [Google Scholar] [CrossRef] [PubMed]
- de Melo Costa, D.; de Oliveira Lopes, L.K.; Tipple, A.F.V.; Johani, K.; Hu, H.; Deva, A.K.; Watanabe, E.; Vickery, K. Evaluation of stainless steel surgical instruments subjected to multiple use/processing. Infect. Dis. Health 2018, 23, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.M. Environmental contamination makes an important contribution to hospital infection. J. Hosp. Infect. 2007, 65, 50–54. [Google Scholar] [CrossRef]
- Chowdhury, D.; Tahir, S.; Legge, M.; Hu, H.; Prvan, T.; Johani, K.; Whiteley, G.S.; Glasbey, T.; Deva, A.K.; Vickery, K. Transfer of dry surface biofilm in the healthcare environment: The role of healthcare workers’ hands as vehicles. J. Hosp. Infect. 2018, 100, e85–e90. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.; Smith, J.; Watson, V.; Robertson, C.; Dancer, S. Examining the association between surface bioburden and frequently touched sites in intensive care. J. Hosp. Infect. 2017, 95, 76–80. [Google Scholar] [CrossRef]
- Dancer, S.J.; White, L.F.; Lamb, J.; Girvan, E.K.; Robertson, C. Measuring the effect of enhanced cleaning in a UK hospital: A prospective cross-over study. BMC Med. 2009, 7, 28. [Google Scholar] [CrossRef]
- Thomas, R.E.; Thomas, B.C.; Conly, J.; Lorenzetti, D. Cleaning and disinfecting surfaces in hospitals and long-term care facilities for reducing hospital and facility-acquired bacterial and viral infections: A systematic review. J. Hosp. Infect. 2022, 122, 9–26. [Google Scholar] [CrossRef]
- Hu, H.; Johani, K.; Gosbell, I.B.; Jacombs, A.S.; Almatroudi, A.; Whiteley, G.S.; Deva, A.K.; Jensen, S.; Vickery, K. Intensive care unit environmental surfaces are contaminated by multidrug-resistant bacteria in biofilms: Combined results of conventional culture, pyrosequencing, scanning electron microscopy, and confocal laser microscopy. J. Hosp. Infect. 2015, 91, 35–44. [Google Scholar] [CrossRef]
- Marion-Ferey, K.; Pasmore, M.; Stoodley, P.; Wilson, S.; Husson, G.; Costerton, J.W. Biofilm removal from silicone tubing: An assessment of the efficacy of dialysis machine decontamination procedures using an in vitro model. J. Hosp. Infect. 2003, 53, 64–71. [Google Scholar] [CrossRef]
- Almatroudi, A.; Gosbell, I.B.; Hu, H.; Jensen, S.O.; Espedido, B.; Tahir, S.; Glasbey, T.; Legge, P.; Whiteley, G.; Deva, A. Staphylococcus aureus dry-surface biofilms are not killed by sodium hypochlorite: Implications for infection control. J. Hosp. Infect. 2016, 93, 263–270. [Google Scholar] [CrossRef]
- Vickery, K.; Ngo, Q.-D.; Zou, J.; Cossart, Y.E. The effect of multiple cycles of contamination, detergent washing, and disinfection on the development of biofilm in endoscope tubing. Am. J. Infect. Control. 2009, 37, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, D.; Rahman, A.; Hu, H.; Jensen, S.O.; Deva, A.K.; Vickery, K. Effect of disinfectant formulation and organic soil on the efficacy of oxidizing disinfectants against biofilms. J. Hosp. Infect. 2018, 103, e33–e41. [Google Scholar] [CrossRef] [PubMed]
- Otter, J.A.; Vickery, K.; Walker, J.T.; Pulcini, E.D.; Stoodley, P.; Goldenberg, S.D.; Salkeld, J.A.G.; Chewins, J.; Yezli, S.; Edgeworth, J.D. Surface-attached cells, biofilms and biocide susceptibility: Implications for hospital cleaning and disinfection. J. Hosp. Infect. 2015, 89, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Stoodley, P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005, 13, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Feazel, L.M.; Baumgartner, L.K.; Peterson, K.L.; Frank, D.N.; Harris, J.K.; Pace, N.R. Opportunistic pathogens enriched in showerhead biofilms. Proc. Natl. Acad. Sci. USA 2009, 106, 16393–16399. [Google Scholar] [CrossRef] [PubMed]
- Vickery, K.; Hu, H.; Jacombs, A.S.; Bradshaw, D.A.; Deva, A.K. A review of bacterial biofilms and their role in device-associated infection. Healthc. Infect. 2013, 18, 61–66. [Google Scholar] [CrossRef]
- Ledwoch, K.; Dancer, S.; Otter, J.; Kerr, K.; Roposte, D.; Rushton, L.; Weiser, R.; Mahenthiralingam, E.; Muir, D.; Maillard, J.-Y. Beware biofilm! Dry biofilms containing bacterial pathogens on multiple healthcare surfaces; a multi-centre study. J. Hosp. Infect. 2018, 100, e47–e56. [Google Scholar] [CrossRef]
- Almatroudi, A.; Hu, H.; Deva, A.; Gosbell, I.B.; Jacombs, A.; Jensen, S.O.; Whiteley, G.; Glasbey, T.; Vickery, K. A new dry-surface biofilm model: An essential tool for efficacy testing of hospital surface decontamination procedures. J. Microbiol. Methods 2015, 117, 171–176. [Google Scholar] [CrossRef]
- Kranjec, C.; Morales Angeles, D.; Torrissen Mårli, M.; Fernández, L.; García, P.; Kjos, M.; Diep, D.B. Staphylococcal biofilms: Challenges and novel therapeutic perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef]
- Köcher, T.; Pichler, P.; Schutzbier, M.; Stingl, C.; Kaul, A.; Teucher, N.; Hasenfuss, G.; Penninger, J.M.; Mechtler, K. High precision quantitative proteomics using iTRAQ on an LTQ Orbitrap: A new mass spectrometric method combining the benefits of all. J. Proteome Res. 2009, 8, 4743–4752. [Google Scholar] [CrossRef]
- Pichler, P.; Köcher, T.; Holzmann, J.; Möhring, T.; Ammerer, G.; Mechtler, K. Improved precision of iTRAQ and TMT quantification by an axial extraction field in an Orbitrap HCD cell. Anal. Chem. 2011, 83, 1469–1474. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, W.M.; Bei, J.; Amigo, N.; Valacco, P.; Amadio, A.F.; Zhang, Q.; Wu, X.; Larzábal, M.; Chen, Z.; Cataldi, A. Quantification of Enterohemorrhagic Escherichia coli O157: H7 proteome using TMT-Based Analysis. PLoS ONE 2018, 13, e0208520. [Google Scholar] [CrossRef] [PubMed]
- Paulo, J.A.; O’Connell, J.D.; Everley, R.A.; O’Brien, J.; Gygi, M.A.; Gygi, S.P. Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J. Proteom. 2016, 148, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Emery, S.J.; Mirzaei, M.; Vuong, D.; Pascovici, D.; Chick, J.M.; Lacey, E.; Haynes, P.A. Induction of virulence factors in Giardia duodenalis independent of host attachment. Sci. Rep. 2016, 6, 20765. [Google Scholar] [CrossRef]
- Li, Z.; Adams, R.M.; Chourey, K.; Hurst, G.B.; Hettich, R.L.; Pan, C. Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos. J. Proteome Res. 2012, 11, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Paulo, J.A.; Jedrychowski, M.P.; Chouchani, E.T.; Kazak, L.; Gygi, S.P. Multiplexed isobaric tag-based profiling of seven murine tissues following in vivo nicotine treatment using a minimalistic proteomics strategy. Proteomics 2018, 18, 1700326. [Google Scholar] [CrossRef]
- Soares, N.C.; Bou, G.; Blackburn, J.M. Editorial: Proteomics of Microbial Human Pathogens. Front. Microbiol. 2016, 7, 1742. [Google Scholar] [CrossRef]
- Moche, M.; Schlüter, R.; Bernhardt, J.; Plate, K.; Riedel, K.; Hecker, M.; Becher, D. Time-resolved analysis of cytosolic and surface-associated proteins of Staphylococcus aureus HG001 under planktonic and biofilm conditions. J. Proteome Res. 2015, 14, 3804–3822. [Google Scholar] [CrossRef]
- Graf, A.C.; Leonard, A.; Schäuble, M.; Rieckmann, L.M.; Hoyer, J.; Maaß, S.; Lalk, M.; Becher, D.; Pané-Farré, J.; Riedel, K. Virulence factors produced by Staphylococcus aureus biofilms have a moonlighting function contributing to biofilm integrity. Mol. Cell. Proteom. 2019, 18, 1036–1053. [Google Scholar] [CrossRef]
- Xu, C.-G.; Yang, Y.-B.; Zhou, Y.-H.; Hao, M.-Q.; Ren, Y.-Z.; Wang, X.-T.; Chen, J.-Q.; Muhammad, I.; Wang, S.; Liu, D. Comparative proteomic analysis provides insight into the key proteins as possible targets involved in aspirin inhibiting biofilm formation of Staphylococcus xylosus. Front. Pharmacol. 2017, 8, 543. [Google Scholar] [CrossRef]
- Zhu, Y.; Weiss, E.C.; Otto, M.; Fey, P.D.; Smeltzer, M.S.; Somerville, G.A. Staphylococcus aureus Biofilm Metabolism and the Influence of Arginine on Polysaccharide Intercellular Adhesin Synthesis, Biofilm Formation, and Pathogenesis. Infect. Immun. 2007, 75, 4219–4226. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-H.; Yu, H.-L.; Ba, Z.-Y.; Chen, J.-Y.; Sun, H.-G.; Han, B.-Z. Sampling methods for NMR-based metabolomics of Staphylococcus aureus. Biotechnol. J. 2010, 5, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Ammons, M.C.B.; Tripet, B.P.; Carlson, R.P.; Kirker, K.R.; Gross, M.A.; Stanisich, J.J.; Copié, V. Quantitative NMR metabolite profiling of methicillin-resistant and methicillin-susceptible Staphylococcus aureus discriminates between biofilm and planktonic phenotypes. J. Proteome Res. 2014, 13, 2973–2985. [Google Scholar] [CrossRef] [PubMed]
- Suo, B.; Yang, H.; Wang, Y.; Lv, H.; Li, Z.; Xu, C.; Ai, Z. Comparative proteomic and morphological change analyses of Staphylococcus aureus during resuscitation from prolonged freezing. Front. Microbiol. 2018, 9, 866. [Google Scholar] [CrossRef] [PubMed]
- Allan, R.N.; Skipp, P.; Jefferies, J.; Clarke, S.C.; Faust, S.N.; Hall-Stoodley, L.; Webb, J. Pronounced metabolic changes in adaptation to biofilm growth by Streptococcus pneumoniae. PLoS ONE 2014, 9, e107015. [Google Scholar] [CrossRef]
- Bøhle, L.A.; Færgestad, E.M.; Veiseth-Kent, E.; Steinmoen, H.; Nes, I.F.; Eijsink, V.G.; Mathiesen, G. Identification of proteins related to the stress response in Enterococcus faecalis V583 caused by bovine bile. Proteome Sci. 2010, 8, 37. [Google Scholar] [CrossRef]
- Navarre, W.W.; Schneewind, O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef]
- Lovering, A.L.; Safadi, S.S.; Strynadka, N.C. Structural perspective of peptidoglycan biosynthesis and assembly. Annu. Rev. Biochem. 2012, 81, 451–478. [Google Scholar] [CrossRef]
- Almatroudi, A.; Tahir, S.; Hu, H.; Chowdhury, D.; Gosbell, I.B.; Jensen, S.O.; Whiteley, G.S.; Deva, A.K.; Glasbey, T.; Vickery, K. Staphylococcus aureus dry-surface biofilms are more resistant to heat treatment than traditional hydrated biofilms. J. Hosp. Infect. 2018, 98, 161–167. [Google Scholar] [CrossRef]
- Setlow, P. Spore Resistance Properties. Microbiol. Spectr. 2014, 2, 5. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Lin, J.-S.; Ma, J.-C.; Wang, H.-H. Functional characterization of triclosan-resistant enoyl-acyl-carrier protein reductase (FabV) in Pseudomonas aeruginosa. Front. Microbiol. 2016, 7, 1903. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; Li, J.; Roland, G.E.; Rock, C.O. Inhibition of the staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J. Biol. Chem. 2000, 275, 4654–4659. [Google Scholar] [CrossRef] [PubMed]
- Latimer, J.; Forbes, S.; McBain, A.J. Attenuated virulence and biofilm formation in Staphylococcus aureus following sublethal exposure to triclosan. Antimicrob. Agents Chemother. 2012, 56, 3092–3100. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, Y.; Sternberg, C.; Molin, S. Evaluation of enoyl-acyl carrier protein reductase inhibitors as Pseudomonas aeruginosa quorum-quenching reagents. Molecules 2010, 15, 780–792. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; McBain, A.; Sreenivasan, P. Common therapeutic approaches for the control of oral biofilms: Microbiological safety and efficacy. Clin. Microbiol. Infect. 2007, 13, 17–24. [Google Scholar] [CrossRef]
- McMurry, L.M.; Oethinger, M.; Levy, S.B. Triclosan targets lipid synthesis. Nature 1998, 394, 531. [Google Scholar] [CrossRef]
- Heath, R.J.; Yu, Y.-T.; Shapiro, M.A.; Olson, E.; Rock, C.O. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 1998, 273, 30316–30320. [Google Scholar] [CrossRef]
- Jungermann, E. Soap bacteriostats. J. Am. Oil Chem. Soc. 1968, 45, 345–350. [Google Scholar] [CrossRef]
- Hiron, A.; Posteraro, B.; Carrière, M.; Remy, L.; Delporte, C.; La Sorda, M.; Sanguinetti, M.; Juillard, V.; Borezée-Durant, E. A nickel ABC-transporter of Staphylococcus aureus is involved in urinary tract infection. Mol. Microbiol. 2010, 77, 1246–1260. [Google Scholar] [CrossRef]
- Kiran, M.D.; Akiyoshi, D.E.; Giacometti, A.; Cirioni, O.; Scalise, G.; Balaban, N. OpuC—An ABC transporter that is associated with Staphylococcus aureus pathogenesis. Int. J. Artif. Organs 2009, 32, 600–610. [Google Scholar] [CrossRef]
- Tanaka, K.J.; Song, S.; Mason, K.; Pinkett, H.W. Selective substrate uptake: The role of ATP-binding cassette (ABC) importers in pathogenesis. Biochim. Biophys. Acta Biomembr. 2018, 1860, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Otto, M.; Götz, F. ABC transporters of staphylococci. Res. Microbiol. 2001, 152, 351–356. [Google Scholar] [CrossRef]
- Jonsson, M.; Juuti, J.T.; François, P.; AlMajidi, R.; Pietiäinen, M.; Girard, M.; Lindholm, C.; Saller, M.J.; Driessen, A.J.; Kuusela, P. Inactivation of the Ecs ABC transporter of Staphylococcus aureus attenuates virulence by altering composition and function of bacterial wall. PLoS ONE 2010, 5, e14209. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.; Kim, Y.; Ross, J.M.; Marten, M.R. Proteomic analysis of Staphylococcus aureus biofilm cells grown under physiologically relevant fluid shear stress conditions. Proteome Sci. 2014, 12, 21. [Google Scholar] [CrossRef]
- Yang, J.; He, Y.; Jiang, J.; Chen, W.; Gao, Q.; Pan, L.; Shi, C. Comparative proteomic analysis by iTRAQ-2DLC-MS/MS provides insight into the key proteins involved in Cronobacter sp. biofilm formation. Food Control. 2016, 63, 93–100. [Google Scholar] [CrossRef]
- Zhu, X.; Long, F.; Chen, Y.; Knøchel, S.; She, Q.; Shi, X. A putative ABC transporter is involved in negative regulation of biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 7675–7683. [Google Scholar] [CrossRef]
- Beenken, K.E.; Dunman, P.M.; McAleese, F.; Macapagal, D.; Murphy, E.; Projan, S.J.; Blevins, J.S.; Smeltzer, M.S. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 2004, 186, 4665–4684. [Google Scholar] [CrossRef]
- Ferreira, M.T.; Manso, A.S.; Gaspar, P.; Pinho, M.G.; Neves, A.R. Effect of oxygen on glucose metabolism: Utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS ONE 2013, 8, e58277. [Google Scholar] [CrossRef]
- Kolbeck, S.; Behr, J.; Vogel, R.F.; Ludwig, C.; Ehrmann, M.A. Acid stress response of Staphylococcus xylosus elicits changes in the proteome and cellular membrane. J. Appl. Microbiol. 2019, 126, 1480–1495. [Google Scholar] [CrossRef]
- Pagels, M.; Fuchs, S.; Pané-Farré, J.; Kohler, C.; Menschner, L.; Hecker, M.; McNamarra, P.J.; Bauer, M.C.; Von Wachenfeldt, C.; Liebeke, M. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 2010, 76, 1142–1161. [Google Scholar] [CrossRef]
- Chiu, K.-C.; Lin, C.-J.; Shaw, G.-C. Transcriptional regulation of the L-lactate permease gene lutP by the LutR repressor of Bacillus subtilis RO-NN-1. Microbiology 2014, 160, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Kolter, R.; Losick, R. A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J. Bacteriol. 2009, 191, 2423–2430. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.E.; He, Z.; Redding, A.M.; Joachimiak, M.P.; Keasling, J.D.; Zhou, J.Z.; Arkin, A.P.; Mukhopadhyay, A.; Fields, M.W. Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: Carbon and energy flow contribute to the distinct biofilm growth state. BMC Genom. 2012, 13, 138. [Google Scholar] [CrossRef] [PubMed]
- Morrison, M.D.; Nicholson, W.L. Meta-analysis of data from spaceflight transcriptome experiments does not support the idea of a common bacterial “spaceflight response”. Sci. Rep. 2018, 8, 14403. [Google Scholar] [CrossRef] [PubMed]
- Junka, A.F.; Deja, S.; Smutnicka, D.; Szymczyk, P.; Ziółkowski, G.; Bartoszewicz, M.; Młynarz, P. Differences in metabolic profiles of planktonic and biofilm cells in Staphylococcus aureus—(1)H Nuclear Magnetic Resonance search for candidate biomarkers. Acta Biochim. Pol. 2013, 60, 701–706. [Google Scholar] [CrossRef]
- Santi, L.; Beys-da-Silva, W.O.; Berger, M.; Calzolari, D.; Guimarães, J.A.; Moresco, J.J.; Yates, J.R., III. Proteomic profile of Cryptococcus neoformans biofilm reveals changes in metabolic processes. J. Proteome Res. 2014, 13, 1545–1559. [Google Scholar] [CrossRef]
- Borriello, G.; Werner, E.; Roe, F.; Kim, A.M.; Ehrlich, G.D.; Stewart, P.S. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2004, 48, 2659–2664. [Google Scholar] [CrossRef]
- Rahman, M.A.; Amirkhani, A.; Chowdhury, D.; Mempin, M.; Molloy, M.P.; Deva, A.K.; Vickery, K.; Hu, H. Proteome of Staphylococcus aureus Biofilm Changes Significantly with Aging. Int. J. Mol. Sci. 2022, 23, 6415. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Jacombs, A.; Tahir, S.; Hu, H.; Deva, A.K.; Almatroudi, A.; Wessels, W.L.F.; Bradshaw, D.A.; Vickery, K. In vitro and in vivo investigation of the influence of implant surface on the formation of bacterial biofilm in mammary implants. Plast. Reconstr. Surg. 2014, 133, 471e–480e. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Hewapathirana, S.; García-Seisdedos, D.; Kamatchinathan, S.; Kundu, D.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]




| Accession ID | Protein Name | Fold Change | Protein Pathways |
|---|---|---|---|
| AIO20800.1 | phosphoribosyl transferase, PyrR | 2.5 | Pyrimidine metabolism |
| AIO20657.1 | cytochrome C oxidase subunit III, QoxC | 2.3 | Oxidative phosphorylation |
| AIO21790.1 | glucosamine--fructose-6-phosphate aminotransferase, GlmS | 2.1 | Biosynthesis of antibiotics, Alanine, aspartate and glutamate metabolism, Amino sugar and nucleotide sugar metabolism |
| AIO21830.1 | galactose-6-phosphate isomerase, LacA | 2.1 | Galactose metabolism |
| AIO22037.1 | nitrite reductase, NasD | 2.1 | Microbial metabolism in diverse environments, Nitrogen metabolism |
| AIO21798.1 | Arginase, Arg | 2.01 | Biosynthesis of secondary metabolites, Biosynthesis of antibiotics, Biosynthesis of amino acids, Arginine biosynthesis, Arginine and proline metabolism |
| AIO21892.1 | Aminoacyltransferase, FemX | 2.01 | Peptidoglycan biosynthesis |
| OOC91313.1 | 5-(carboxyamino)imidazole ribonucleotide mutase, PurE | 2.01 | Biosynthesis of secondary metabolites, Biosynthesis of antibiotics, Purine metabolism |
| AIO20536.1 | argininosuccinate synthase, ArgG | 2.01 | Biosynthesis of secondary metabolites, Biosynthesis of antibiotics, Biosynthesis of amino acids, Arginine biosynthesis, Alanine, aspartate and glutamate metabolism |
| AIO20949.1 | threonine synthase, ThrC | 2.01 | Microbial metabolism in diverse environments, Glycine, serine and threonine metabolism, Vitamin B6 metabolism, Biosynthesis of amino acids |
| Accession ID | Protein Name | Fold Change | Protein Pathways |
|---|---|---|---|
| OOC91425.1 | serine acetyltransferase, CysE | 2.3 | Microbial metabolism in diverse environments, Cysteine and methionine metabolism, Sulfur metabolism, Biosynthesis of amino acids, Carbon metabolism |
| AIO21414.1 | PTS glucose transporter subunit IIBC, PtaA | 2.3 | Amino sugar and nucleotide sugar metabolism, Phosphotransferase system |
| AIO21824.1 | 6-phospho-beta-galactosidase, LacG | 2.1 | Galactose metabolism |
| AIO20705.1 | inositol monophosphatase, SACOL1116 | 2.1 | Streptomycin biosynthesis, Inositol phosphate metabolism |
| AIO21431.1 | UDP-N-acetylmuramate--alanine ligase, MurC | 2.1 | D-Glutamine and D-glutamate metabolism, Peptidoglycan biosynthesis |
| AIO20408.1 | UDP-N-acetylenolpyruvoylglucosamine reductase, MurB | 2.1 | Amino sugar and nucleotide sugar metabolism, Peptidoglycan biosynthesis |
| AIO22036.1 | nitrite reductase, NasE | 2.1 | Microbial metabolism in diverse environments, Nitrogen metabolism |
| AIO20589.1 | enoyl-ACP reductase, FabI | 2.1 | Biotin metabolism, Fatty acid biosynthesis and metabolism, |
| AIO20891.1 | Recombinase, RecA | 2.01 | Homologous recombination |
| AIO20673.1 | phosphoribosylamine-glycine ligase, PurD | 2.01 | Biosynthesis of secondary metabolites, Purine metabolism |
| Accession ID | Protein Name | Fold Change | Protein Pathways |
|---|---|---|---|
| AIO21853.1 | 50S ribosomal protein L17, RplQ | 0.4 | Ribosome |
| AIO21993.1 | hemin ABC transporter ATP-binding protein, HrtA | 0.4 | ABC transporters |
| AIO22260.1 | sulfite reductase [NADPH] flavoprotein alpha-component, SA2413 | 0.4 | Microbial metabolism in diverse environments, Sulfur metabolism |
| AIO20838.1 | cell division protein FtsY | 0.4 | Protein export, Bacterial secretion system, Quorum sensing |
| AIO20584.1 | GTP pyrophosphokinase, SA0864 | 0.5 | Purine metabolism |
| AIO22050.1 | amino acid ABC transporter substrate-binding protein, SACOL2412 | 0.5 | ABC transporters |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.A.; Amirkhani, A.; Parvin, F.; Chowdhury, D.; Molloy, M.P.; Deva, A.K.; Vickery, K.; Hu, H. One Step Forward with Dry Surface Biofilm (DSB) of Staphylococcus aureus: TMT-Based Quantitative Proteomic Analysis Reveals Proteomic Shifts between DSB and Hydrated Biofilm. Int. J. Mol. Sci. 2022, 23, 12238. https://doi.org/10.3390/ijms232012238
Rahman MA, Amirkhani A, Parvin F, Chowdhury D, Molloy MP, Deva AK, Vickery K, Hu H. One Step Forward with Dry Surface Biofilm (DSB) of Staphylococcus aureus: TMT-Based Quantitative Proteomic Analysis Reveals Proteomic Shifts between DSB and Hydrated Biofilm. International Journal of Molecular Sciences. 2022; 23(20):12238. https://doi.org/10.3390/ijms232012238
Chicago/Turabian StyleRahman, Md. Arifur, Ardeshir Amirkhani, Farhana Parvin, Durdana Chowdhury, Mark P. Molloy, Anand Kumar Deva, Karen Vickery, and Honghua Hu. 2022. "One Step Forward with Dry Surface Biofilm (DSB) of Staphylococcus aureus: TMT-Based Quantitative Proteomic Analysis Reveals Proteomic Shifts between DSB and Hydrated Biofilm" International Journal of Molecular Sciences 23, no. 20: 12238. https://doi.org/10.3390/ijms232012238
APA StyleRahman, M. A., Amirkhani, A., Parvin, F., Chowdhury, D., Molloy, M. P., Deva, A. K., Vickery, K., & Hu, H. (2022). One Step Forward with Dry Surface Biofilm (DSB) of Staphylococcus aureus: TMT-Based Quantitative Proteomic Analysis Reveals Proteomic Shifts between DSB and Hydrated Biofilm. International Journal of Molecular Sciences, 23(20), 12238. https://doi.org/10.3390/ijms232012238








