miRNA Expression May Have Implications for Immunotherapy in PDGFRA Mutant GISTs
Abstract
1. Introduction
2. Results
2.1. Differential miRNA Expression between PDGFRA D842V Mutant versus PDGFRA Non-D842V Mutant GIST
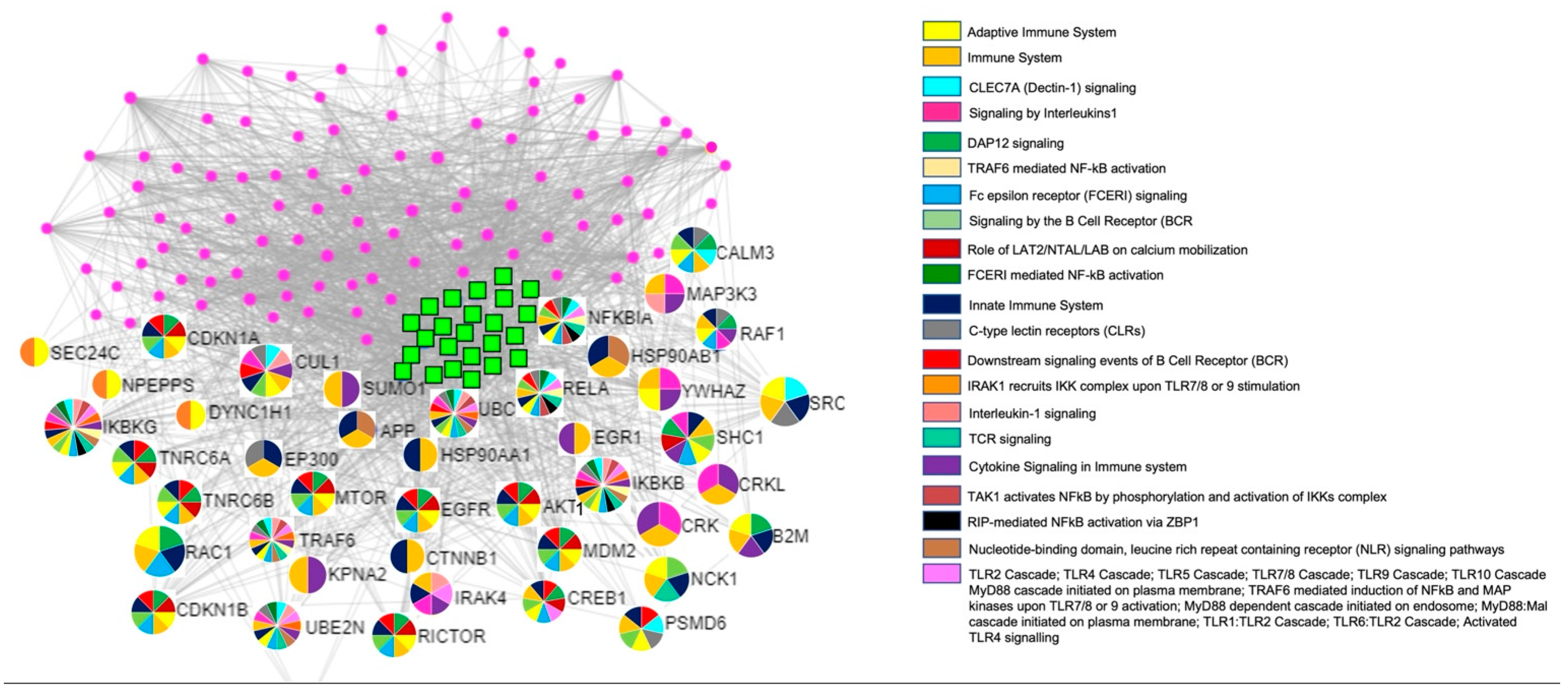
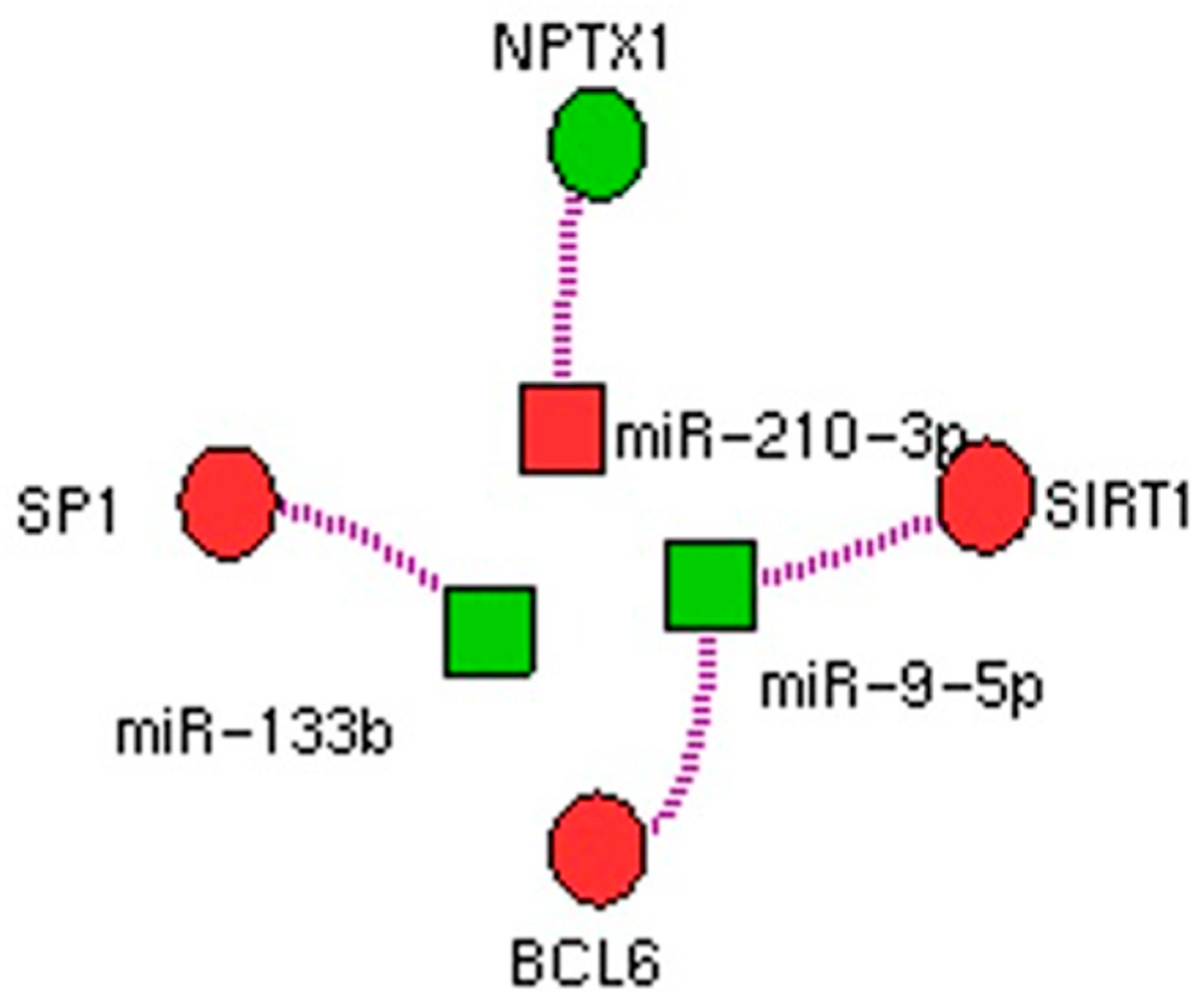
2.2. miRNA and mRNA Arrays Network
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. miRNA Expression Profiling
4.3. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corless, C.L.; Schroeder, A.; Griffith, D.; Town, A.; McGreevey, L.; Harrell, P.; Shiraga, S.; Bainbridge, T.; Morich, J.; Heinrich, M.C. PDGFRA Mutations in Gastrointestinal Stromal Tumors: Frequency, Spectrum and In Vitro Sensitivity to Imatinib. J. Clin. Oncol. 2005, 23, 5357–5364. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Ma, S.; Che, K.; Pobbati, A.V.; Rubin, B.P. Inhibition of PI3K and MAPK pathways along with KIT inhibitors as a strategy to overcome drug resistance in gastrointestinal stromal tumors. PLoS ONE 2021, 16, e0252689. [Google Scholar] [CrossRef]
- Angelini, S.; Ravegnini, G.; Fletcher, J.A.J.A.; Maffei, F.; Hrelia, P. Clinical relevance of pharmacogenetics in gastrointestinal stromal tumor treatment in the era of personalized therapy. Pharmacogenomics 2013, 14, 941–956. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Gasparotto, D.; Miceli, R.; Toffolatti, L.; Gallina, G.; Scaramel, E.; Marzotto, A.; Boscato, E.; Messerini, L.; Bearzi, I.; et al. KIT, PDGFRA, and BRAF mutational spectrum impacts on the natural history of imatinib-naive localized GIST. Am. J. Surg. Pathol. 2015, 39, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Farag, S.; Somaiah, N.; Choi, H.; Heeres, B.; Wang, W.L.; van Boven, H.; Nederlof, P.; Benjamin, R.; van der Graaf, W.; Grunhagen, D.; et al. Clinical characteristics and treatment outcome in a large multicentre observational cohort of PDGFRA exon 18 mutated gastrointestinal stromal tumour patients. Eur. J. Cancer 2017, 76, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Pantaleo, M.A.; Astolfi, A.; Indio, V.; Nannini, M. The Identity of PDGFRA D842V-Mutant Gastrointestinal Stromal Tumors (GIST). Cancers 2021, 13, 705. [Google Scholar] [CrossRef]
- Hirota, S.; Ohashi, A.; Nishida, T.; Isozaki, K.; Kinoshita, K.; Shinomura, Y.; Kitamura, Y. Gain-of-function mutations of platelet-derived growth factor receptor alpha gene in gastrointestinal stromal tumors. Gastroenterology 2003, 125, 660–667. [Google Scholar] [CrossRef]
- Heinrich, M.C.; Corless, C.L.; Demetri, G.D.; Blanke, C.D.; von Mehren, M.; Joensuu, H.; McGreevey, L.S.; Chen, C.-J.; Van den Abbeele, A.D.; Druker, B.J.; et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 2003, 21, 4342–4349. [Google Scholar] [CrossRef]
- Debiec-Rychter, M.; Dumez, H.; Judson, I.; Wasag, B.; Verweij, J.; Brown, M.; Dimitrijevic, S.; Sciot, R.; Stul, M.; Vranck, H.; et al. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. Eur. J. Cancer 2004, 40, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Cassier, P.A.; Fumagalli, E.; Rutkowski, P.; Schöffski, P.; Van Glabbeke, M.; Debiec-Rychter, M.; Emile, J.-F.; Duffaud, F.; Martin-Broto, J.; Landi, B.; et al. Outcome of patients with platelet-derived growth factor receptor alpha-mutated gastrointestinal stromal tumors in the tyrosine kinase inhibitor era. Clin. Cancer Res. 2012, 18, 4458–4464. [Google Scholar] [CrossRef]
- Heinrich, M.C.; Maki, R.G.; Corless, C.L.; Antonescu, C.R.; Harlow, A.; Griffith, D.; Town, A.; McKinley, A.; Ou, W.-B.; Fletcher, J.A.; et al. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J. Clin. Oncol. 2008, 26, 5352–5359. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, P.; Demetri, G.D.; Gelderblom, H.; Rutkowski, P.; Im, S.-A.; Gupta, S.; Kang, Y.-K.; Schöffski, P.; Schuette, J.; Soulières, D.; et al. Correlation of KIT and PDGFRA mutational status with clinical benefit in patients with gastrointestinal stromal tumor treated with sunitinib in a worldwide treatment-use trial. BMC Cancer 2016, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Trullas-Jimeno, A.; Delgado, J.; Garcia-Ochoa, B.; Wang, I.; Sancho-Lopez, A.; Payares-Herrera, C.; Dalhus, M.L.; Strøm, B.O.; Egeland, E.J.; Enzmann, H.; et al. The EMA assessment of avapritinib in the treatment of gastrointestinal stromal tumours harbouring the PDGFRA D842V mutation. ESMO Open 2021, 6, 100159. [Google Scholar] [CrossRef]
- Jones, R.L.; Serrano, C.; von Mehren, M.; George, S.; Heinrich, M.C.; Kang, Y.K.; Schöffski, P.; Cassier, P.A.; Mir, O.; Chawla, S.P.; et al. Avapritinib in unresectable or metastatic PDGFRA D842V-mutant gastrointestinal stromal tumours: Long-term efficacy and safety data from the NAVIGATOR phase I trial. Eur. J. Cancer 2021, 145, 132–142. [Google Scholar] [CrossRef] [PubMed]
- von Mehren, M.; Heinrich, M.C.; Shi, H.; Iannazzo, S.; Mankoski, R.; Dimitrijević, S.; Hoehn, G.; Chiroli, S.; George, S. Clinical efficacy comparison of avapritinib with other tyrosine kinase inhibitors in gastrointestinal stromal tumors with PDGFRA D842V mutation: A retrospective analysis of clinical trial and real-world data. BMC Cancer 2021, 21, 291. [Google Scholar] [CrossRef] [PubMed]
- Indio, V.; Astolfi, A.; Tarantino, G.; Urbini, M.; Patterson, J.; Nannini, M.; Saponara, M.; Gatto, L.; Santini, D.; do Valle, I.F.; et al. Integrated Molecular Characterization of Gastrointestinal Stromal Tumors (GIST) Harboring the Rare D842V Mutation in PDGFRA Gene. Int. J. Mol. Sci. 2018, 19, 732. [Google Scholar] [CrossRef]
- Berindan-Neagoe, I.; Monroig, P.D.C.; Pasculli, B.; Calin, G.A. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014, 64, 311–336. [Google Scholar] [CrossRef]
- Fernandez-Serra, A.; Moura, D.S.; Sanchez-Izquierdo, M.D.; Calabuig-Fariñas, S.; Lopez-Alvarez, M.; Martínez-Martínez, A.; Carrasco-Garcia, I.; Ramírez-Calvo, M.; Blanco-Alcaina, E.; López-Reig, R.; et al. Prognostic Impact of let-7e MicroRNA and Its Target Genes in Localized High-Risk Intestinal GIST: A Spanish Group for Research on Sarcoma (GEIS) Study. Cancers 2020, 12, 2979. [Google Scholar] [CrossRef] [PubMed]
- Nannini, M.; Ravegnini, G.; Angelini, S.; Astolfi, A.; Biasco, G.; Pantaleo, M.A. miRNA profiling in gastrointestinal stromal tumors: Implication as diagnostic and prognostic markers. Epigenomics 2015, 7, 1033–1049. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, M.A.; Ravegnini, G.; Astolfi, A.; Simeon, V.; Nannini, M.; Saponara, M.; Urbini, M.; Gatto, L.; Indio, V.; Sammarini, G.; et al. Integrating miRNA and gene expression profiling analysis revealed regulatory networks in gastrointestinal stromal tumors. Epigenomics 2016, 8, 1347–1366. [Google Scholar] [CrossRef] [PubMed]
- Indio, V.; Ravegnini, G.; Astolfi, A.; Urbini, M.; Saponara, M.; De Leo, A.; Gruppioni, E.; Tarantino, G.; Angelini, S.; Pession, A.; et al. Gene Expression Profiling of PDGFRA Mutant GIST Reveals Immune Signatures as a Specific Fingerprint of D842V Exon 18 Mutation. Front. Immunol. 2020, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Gill, E.E.; Hancock, R.E.W. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, G.A.; Bowler, T.G.; Liu, M.; Medina, B.D.; Zhang, J.Q.; Param, N.J.; Loo, J.K.; Goldfeder, R.L.; Chibon, F.; Rossi, F.; et al. Differential immune profiles distinguish the mutational subtypes of gastrointestinal stromal tumor. J. Clin. Investig. 2019, 129, 1863–1877. [Google Scholar] [CrossRef]
- Gasparotto, D.; Sbaraglia, M.; Rossi, S.; Baldazzi, D.; Brenca, M.; Mondello, A.; Nardi, F.; Racanelli, D.; Cacciatore, M.; Dei Tos, A.P.; et al. Tumor genotype, location, and malignant potential shape the immunogenicity of primary untreated gastrointestinal stromal tumors. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Chantharasamee, J.; Adashek, J.J.; Wong, K.; Eckardt, M.A.; Chmielowski, B.; Dry, S.; Eilber, F.C.; Singh, A.S. Translating Knowledge About the Immune Microenvironment of Gastrointestinal Stromal Tumors into Effective Clinical Strategies. Curr. Treat. Options Oncol. 2021, 22, 9. [Google Scholar] [CrossRef] [PubMed]
- Roulleaux Dugage, M.; Jones, R.L.; Trent, J.; Champiat, S.; Dumont, S. Beyond the Driver Mutation: Immunotherapies in Gastrointestinal Stromal Tumors. Front. Immunol. 2021, 12, 3372. [Google Scholar] [CrossRef]
- Sun, Q.; Li, J.; Li, F.; Li, H.; Bei, S.; Zhang, X.; Feng, L. LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Prolif. 2019, 52, e12687. [Google Scholar] [CrossRef]
- Yin, W.; Xu, J.; Li, C.; Dai, X.; Wu, T.; Wen, J. Circular RNA circ_0007142 Facilitates Colorectal Cancer Progression by Modulating CDC25A Expression via miR-122-5p. Onco. Targets. Ther. 2020, 13, 3689–3701. [Google Scholar] [CrossRef]
- Xu, X.; Gao, F.; Wang, J.; Tao, L.; Ye, J.; Ding, L.; Ji, W.; Chen, X. MiR-122-5p inhibits cell migration and invasion in gastric cancer by down-regulating DUSP4. Cancer Biol. Ther. 2018, 19, 427–435. [Google Scholar] [CrossRef]
- Walker, S.R.; Liu, S.; Xiang, M.; Nicolais, M.; Hatzi, K.; Giannopoulou, E.; Elemento, O.; Cerchietti, L.; Melnick, A.; Frank, D.A. The transcriptional modulator BCL6 as a molecular target for breast cancer therapy. Oncogene 2014, 34, 1073–1082. [Google Scholar] [CrossRef]
- Walters, M.P.; McPhail, E.D.; Law, M.E.; Folpe, A.L. BCL-6 expression in mesenchymal tumours: An immunohistochemical and fluorescence in situ hybridisation study. J. Clin. Pathol. 2011, 64, 866–869. [Google Scholar] [CrossRef]
- Ma, M.C.; Chiu, T.J.; Lu, H.I.; Huang, W.T.; Lo, C.M.; Tien, W.Y.; Lan, Y.C.; Chen, Y.Y.; Chen, C.H.; Li, S.H. SIRT1 overexpression is an independent prognosticator for patients with esophageal squamous cell carcinoma. J. Cardiothorac. Surg. 2018, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Eun, J.C.; Sang, J.N.; Keun, S.K.; Chan, Y.K.; Park, B.H.; Ho, S.P.; Lee, H.; Myoung, J.C.; Myoung, J.K.; Dong, G.L.; et al. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin. Cancer Res. 2009, 15, 4453–4459. [Google Scholar] [CrossRef]
- Stenzinger, A.; Endris, V.; Klauschen, F.; Sinn, B.; Lorenz, K.; Warth, A.; Goeppert, B.; Ehemann, V.; Muckenhuber, A.; Kamphues, C.; et al. High SIRT1 expression is a negative prognosticator in pancreatic ductal adenocarcinoma. BMC Cancer 2013, 13, 450. [Google Scholar] [CrossRef]
- Chevrier, S.; Kratina, T.; Emslie, D.; Tarlinton, D.M.; Corcoran, L.M. IL4 and IL21 cooperate to induce the high Bcl6 protein level required for germinal center formation. Immunol. Cell Biol. 2017, 95, 925–932. [Google Scholar] [CrossRef]
- Pelham, S.J.; Caldirola, M.S.; Avery, D.T.; Mackie, J.; Rao, G.; Gothe, F.; Peters, T.J.; Guerin, A.; Neumann, D.; Vokurkova, D.; et al. STAT5B restrains human B-cell differentiation to maintain humoral immune homeostasis. J. Allergy Clin. Immunol. 2022, 150, 931–946. [Google Scholar] [CrossRef]
- Subramanian, S.; Lui, W.O.; Lee, C.H.; Espinosa, I.; Nielsen, T.O.; Heinrich, M.C.; Corless, C.L.; Fire, A.Z.; Van De Rijn, M. MicroRNA expression signature of human sarcomas. Oncogene 2007, 27, 2015–2026. [Google Scholar] [CrossRef]
- Yamamoto, H.; Kohashi, K.; Fujita, A.; Oda, Y. Fascin-1 overexpression and miR-133b downregulation in the progression of gastrointestinal stromal tumor. Mod. Pathol. 2012, 26, 563–571. [Google Scholar] [CrossRef]
- Tong, H.X.; Zhou, Y.H.; Hou, Y.Y.; Zhang, Y.; Huang, Y.; Xie, B.; Wang, J.Y.; Jiang, Q.; He, J.Y.; Shao, Y.B.; et al. Expression profile of microRNAs in gastrointestinal stromal tumors revealed by high throughput quantitative RT-PCR microarray. World J. Gastroenterol. 2015, 21, 5843–5855. [Google Scholar] [CrossRef]
- Yang, X.; Shi, L.; Yi, C.; Yang, Y.; Chang, L.; Son, D. MiR-210-3p inhibits the tumor growth and metastasis of bladder cancer via targeting fibroblast growth factor receptor-like 1. Am. J. Cancer Res. 2017, 7, 1738–1753. [Google Scholar]
- Hong, L.; Han, Y.; Zhang, H.; Zhao, Q.; Qiao, Y. miR-210: A therapeutic target in cancer. Expert Opin. Ther. Targets 2013, 17, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Rankin, E.B.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Xia, J. miRNet—Functional Analysis and Visual Exploration of miRNA–Target Interactions in a Network Context; Humana Press: New York, NY, USA, 2018; pp. 215–233. [Google Scholar]

| miRNA ID | Delta Ct | D842V vs. Non-D842V | p-Value | Predicted Targets among the Deregulated Gene in Our Cohort of Patients * |
|---|---|---|---|---|
| hsa-miR-1825 | −5.31 | ↑ | 0.027 | NLK(2) |
| hsa-miR-431-3p | −4.96 | ↑ | 0.009 | |
| hsa-miR-20b-3p | −3.87 | ↑ | 0.015 | |
| hsa-miR-149-3p | −3.40 | ↑ | 0.037 | SPRY4(2) |
| hsa-miR-9-5p | −2.98 | ↑ | 0.038 | GNPNAT1(2), SIRT1(3), CREB5(2), POU2F1(3), BCL6(3), PXDN(3), RNF169(2), FBN2(3), PTAR1(2), NIN(2) |
| hsa-miR-604 | −2.82 | ↑ | 0.044 | |
| hsa-miR-661 | −1.64 | ↑ | 0.049 | IL17RA(2) |
| hsa-miR-133b | −1.43 | ↑ | 0.032 | PPP2R2D(2), SP1(2), ZHX3(2), CREB5(2), |
| hsa-miR-133a-3p | −1.38 | ↑ | 0.044 | |
| hsa-miR-1233-3p | −1.07 | ↑ | 0.042 | |
| hsa-miR-545-3p | 1.32 | ↓ | 0.048 | TSPAN2(2) |
| hsa-miR-210-3p | 1.56 | ↓ | 0.046 | NPTX1(1) |
| hsa-miR-221-3p | 1.59 | ↓ | 0.039 | |
| hsa-miR-135b-5p | 2.10 | ↓ | 0.027 | |
| hsa-miR-33a-5p | 2.12 | ↓ | 0.043 | |
| hsa-miR-452-5p | 2.18 | ↓ | 0.044 | |
| hsa-miR-219a-5p | 2.43 | ↓ | 0.019 | |
| hsa-miR-499a-5p | 2.72 | ↓ | 0.017 | |
| hsa-miR-517c-3p | 2.79 | ↓ | 0.024 | |
| hsa-miR-873-5p | 3.07 | ↓ | 0.027 | |
| hsa-miR-512-3p | 3.60 | ↓ | 0.032 | |
| hsa-miR-708-5p | 3.92 | ↓ | 0.002 | |
| hsa-miR-122-5p | 4.45 | ↓ | 0.010 | CD320(2) |
| hsa-miR-15a-5p | 6.43 | ↓ | 0.025 | RSPO3(2) |
| Pathway | Adjusted p Value |
|---|---|
| Fc epsilon receptor (FCERI) signaling | 1.88 × 10−11 |
| Signaling by the B Cell Receptor (BCR) | 4.35 × 10−10 |
| Innate Immune System | 6.25 × 10−10 |
| Downstream signaling events of B Cell Receptor (BCR) | 7.55 × 10−9 |
| Adaptive Immune System | 1.82 × 10−7 |
| TAK1 activates NFkB by phosphorylation and activation of IKKs complex | 2.35 × 10−6 |
| Toll Like Receptor 10 (TLR10) Cascade | 2.51 × 10−6 |
| Toll Like Receptor 5 (TLR5) Cascade | 2.51 × 10−6 |
| MyD88 cascade initiated on plasma membrane | 2.51 × 10−6 |
| TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation | 2.74 × 10−6 |
| Toll Like Receptor 7/8 (TLR7/8) Cascade | 2.92 × 10−6 |
| MyD88 dependent cascade initiated on endosome | 2.92 × 10−6 |
| MyD88:Mal cascade initiated on plasma membrane | 4.24 × 10−6 |
| Toll Like Receptor TLR1:TLR2 Cascade | 4.24 × 10−6 |
| Toll Like Receptor TLR6:TLR2 Cascade | 4.24 × 10−6 |
| Toll Like Receptor 2 (TLR2) Cascade | 4.24 × 10−6 |
| IRAK1 recruits IKK complex | 4.26 × 10−6 |
| IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation | 4.26 × 10−6 |
| Cytokine Signaling in Immune system | 4.27 × 10−6 |
| TRAF6 mediated NF-kB activation | 5.53 × 10−6 |
| Toll Like Receptor 9 (TLR9) Cascade | 3.67 × 10−6 |
| CLEC7A (Dectin-1) signaling | 1.29 × 10−6 |
| Interleukin-1 signaling | 1.29 × 10−6 |
| Role of LAT2/NTAL/LAB on calcium mobilization | 1.81 × 10−6 |
| DAP12 signaling | 1.09 × 10−4 |
| Patient ID | Size (cm) | Mitotic Index (HPF) * | Last Follow Up § | PDGRA Molecular Analysis |
|---|---|---|---|---|
| GIST140 | 15 | 3/50 | AWOD | Exon 18 D842V |
| GIST165 | 12 | 2/50 | AWOD | Exon 18 D842V |
| GIST138 | 7 | 8/50 | AWOD | Exon 18 D842V |
| GIST142 | 3 | 5/50 | AWOD | Exon 18 D842V |
| GIST136 | 4.5 | 6/50 | DNFD | Exon 18 D842V |
| GIST05 | 7 | 4/50 | AWOD | Exon 12 del 16117-20 CCCG + ins 16124 TC + del 16124-30 GGACATG |
| GIST12 | NA | NA | NA | Exon 14 K646E |
| GIST15 | NA | NA | NA | Exon 18 DIMH842-845del |
| GIST26 | NA | NA | NA | Exon 12 V561D |
| GIST168 | 5.5 | 4/50 | AWOD | Exon 12 S566_E571 > R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravegnini, G.; Nannini, M.; Indio, V.; Serrano, C.; Gorini, F.; Astolfi, A.; Di Vito, A.; Morroni, F.; Pantaleo, M.A.; Hrelia, P.; et al. miRNA Expression May Have Implications for Immunotherapy in PDGFRA Mutant GISTs. Int. J. Mol. Sci. 2022, 23, 12248. https://doi.org/10.3390/ijms232012248
Ravegnini G, Nannini M, Indio V, Serrano C, Gorini F, Astolfi A, Di Vito A, Morroni F, Pantaleo MA, Hrelia P, et al. miRNA Expression May Have Implications for Immunotherapy in PDGFRA Mutant GISTs. International Journal of Molecular Sciences. 2022; 23(20):12248. https://doi.org/10.3390/ijms232012248
Chicago/Turabian StyleRavegnini, Gloria, Margherita Nannini, Valentina Indio, Cesar Serrano, Francesca Gorini, Annalisa Astolfi, Aldo Di Vito, Fabiana Morroni, Maria Abbondanza Pantaleo, Patrizia Hrelia, and et al. 2022. "miRNA Expression May Have Implications for Immunotherapy in PDGFRA Mutant GISTs" International Journal of Molecular Sciences 23, no. 20: 12248. https://doi.org/10.3390/ijms232012248
APA StyleRavegnini, G., Nannini, M., Indio, V., Serrano, C., Gorini, F., Astolfi, A., Di Vito, A., Morroni, F., Pantaleo, M. A., Hrelia, P., & Angelini, S. (2022). miRNA Expression May Have Implications for Immunotherapy in PDGFRA Mutant GISTs. International Journal of Molecular Sciences, 23(20), 12248. https://doi.org/10.3390/ijms232012248










