Towards the Improvement of Ornamental Attributes in Chrysanthemum: Recent Progress in Biotechnological Advances
Abstract
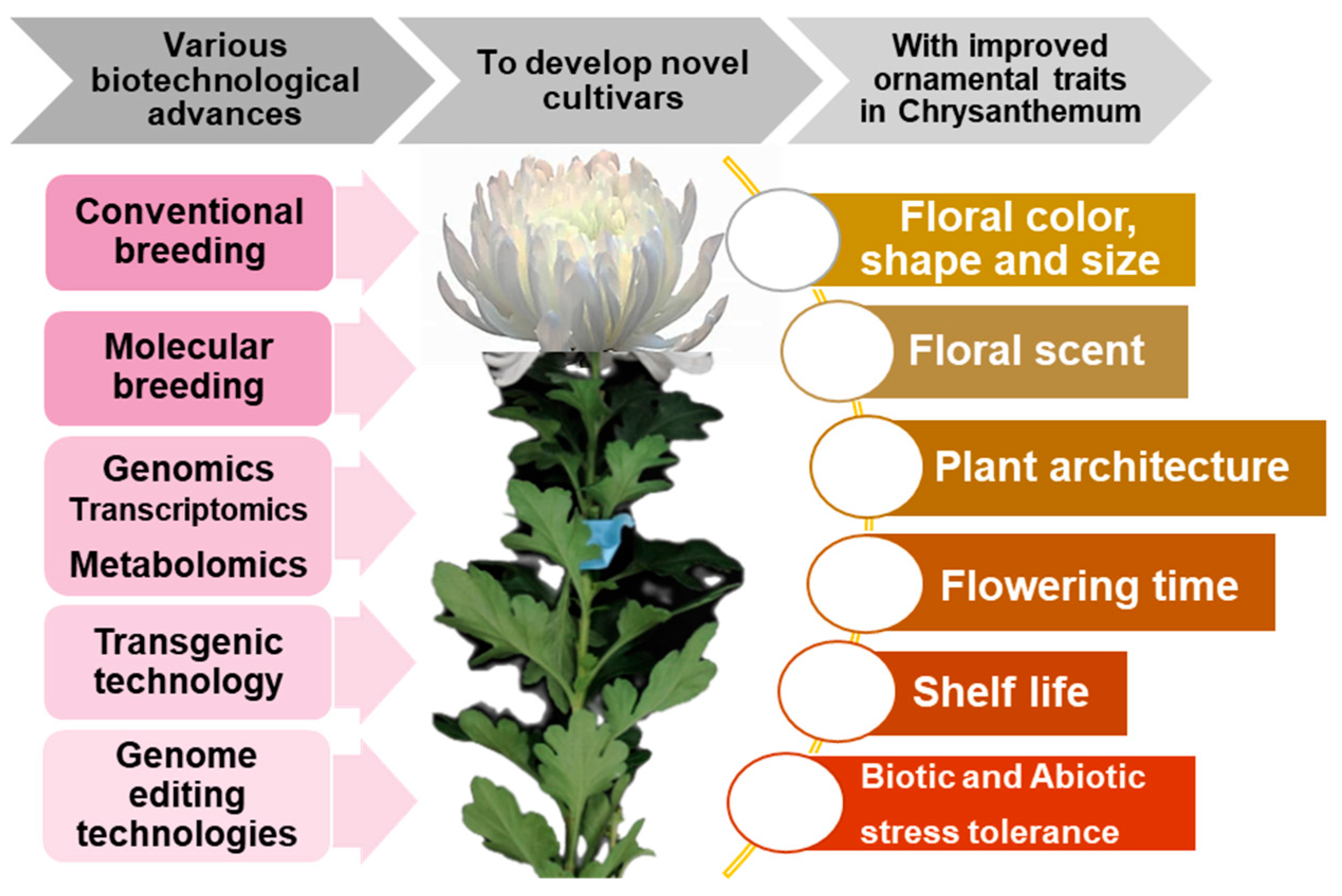
1. Introduction
2. Importance of Floral Attributes
3. Plant Architecture
4. Floral Color
5. Floral Scent
6. Vase Life
7. Flowering Time and Development
8. Floral Anatomy and Development
9. Biotic Stress Resistance
10. Abiotic Stress Tolerance
11. Advances in Genome Editing for Chrysanthemum
12. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dobres, M. Prospects for the Commercialization of Transgenic Ornamentals. In Transgenic HorTiculTural Crops; CRC Press: Boca Raton, FL, USA, 2011; p. 305. [Google Scholar]
- International Statistics Flowers and Plants 2021, 69th ed.; Union Fleurs, International Flower Trade Association: Brussels, Belgium, 2022; p. 16.
- Teixeira da Silva, J.A.; Shinoyama, H.; Aida, R.; Matsushita, Y.; Raj, S.K.; Chen, F. Chrysanthemum biotechnology: Quo vadis? Crit. Rev. Plant Sci. 2013, 32, 21–52. [Google Scholar] [CrossRef]
- Lin, L.-Z.; Harnly, J.M. Identification of the phenolic components of chrysanthemum flower (Chrysanthemum morifolium Ramat). Food Chem. Food Chem. 2010, 120, 319–326. [Google Scholar] [CrossRef]
- Anderson, N. Chrysanthemum. Dendranthema x grandiflora. In Flower Breeding and Genetics; Springer: Berlin/Heidelberg, Germany, 2006; pp. 389–438. [Google Scholar]
- Dowrick, G.J.H. The chromosomes of Chrysanthemum. Heredity 1953, 7, 59–72. [Google Scholar] [CrossRef]
- Liu, P.-L.; Wan, Q.; Guo, Y.-P.; Yang, J.; Rao, G.-Y. Phylogeny of the genus Chrysanthemum L.: Evidence from single-copy nuclear gene and chloroplast DNA sequences. PLoS ONE 2012, 7, e48970. [Google Scholar] [CrossRef]
- Dowrick, G.; El-Bayoumi, A. The origin of new forms of the garden Chrysanthemum. Euphytica 1966, 15, 32–38. [Google Scholar] [CrossRef]
- Ohmiya, A. Molecular mechanisms underlying the diverse array of petal colors in chrysanthemum flowers. Breed. Sci. 2018, 68, 119–127. [Google Scholar] [CrossRef]
- Su, J.; Jiang, J.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.; Chen, F. Current achievements and future prospects in the genetic breeding of chrysanthemum: A review. Hortic. Res. 2019, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Van Huylenbroeck, J. Ornamental Crops; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Anderson, N.O. Chrysanthemum. In Flower Breeding and Genetics; Springer: Dordrecht, The Netherlands, 2007; pp. 389–437. [Google Scholar]
- Azadi, P.; Bagheri, H.; Nalousi, A.M.; Nazari, F.; Chandler, S.F. Current status and biotechnological advances in genetic engineering of ornamental plants. Biotechnol. Adv. 2016, 34, 1073–1090. [Google Scholar] [CrossRef]
- Chandler, S.F.; Sanchez, C. Genetic modification; the development of transgenic ornamental plant varieties. Plant Biotechnol. J. 2012, 10, 891–903. [Google Scholar] [CrossRef]
- Parisi, C.; Tillie, P.; Rodríguez-Cerezo, E. The global pipeline of GM crops out to 2020. Nat. Biotechnol. 2016, 34, 31–36. [Google Scholar] [CrossRef]
- Mekapogu, M.; Kwon, O.K.; Hyun, D.Y.; Lee, K.J.; Ahn, M.S.; Park, J.T.; Jung, J. Environment,; Biotechnology. Identification of standard type cultivars in Chrysanthemum (Dendranthema grandiflorum) using SSR markers. Hortic. Environ. Biotechnol. 2020, 61, 153–161. [Google Scholar] [CrossRef]
- Debener, T. Current strategies and future prospects of resistance breeding in ornamentals. In Proceedings of the XXIII International Eucarpia Symposium, Section Ornamentals: Colourful Breeding and Genetics 836, Leiden, The Netherlands, 31 August–4 September 2009; pp. 125–130. [Google Scholar]
- Ibitoye, D.; Akin-Idowu, P.E. Marker-assisted-selection (MAS): A fast track to increase genetic gain in horticultural crop breeding. J. Afr. J. Biotechnol. 2011, 10, 11333–11339. [Google Scholar]
- Das, G.; Patra, J.K.; Baek, K.-H. Insight into MAS: A molecular tool for development of stress resistant and quality of rice through gene stacking. Front. Plant Sci. 2017, 8, 985. [Google Scholar] [CrossRef] [PubMed]
- Sumitomo, K.; Shirasawa, K.; Isobe, S.; Hirakawa, H.; Hisamatsu, T.; Nakano, Y.; Yagi, M.; Ohmiya, A. Genome-wide association study overcomes the genome complexity in autohexaploid chrysanthemum and tags SNP markers onto the flower color genes. Sci. Rep. 2019, 9, 13947. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Aqeel, M.; Deng, J.; Khalid, N.; Sanaullah, T.; Shuilin, H. Biotechnological advancements for improving floral attributes in ornamental plants. Front. Plant Sci. 2017, 8, 530. [Google Scholar] [CrossRef] [PubMed]
- Kishi-Kaboshi, M.; Aida, R.; Sasaki, K.J.P.; Physiology, C. Generation of gene-edited Chrysanthemum morifolium using multicopy transgenes as targets and markers. Plant Cell Physiol. 2017, 58, 216–226. [Google Scholar] [PubMed]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Gianfagna, T. Natural and synthetic growth regulators and their use in horticultural and agronomic crops. In Plant Hormones; Springer: Dordrecht, The Netherlands, 1995; pp. 751–773. [Google Scholar]
- Dole, J.M.; Wilkins, H.F. Floriculture: Principles and Species; Prentice-Hall Inc.: Upper. Saddle River, NJ, USA, 1999. [Google Scholar]
- Zheng, Z.-L.; Yang, Z.; Jang, J.-C.; Metzger, J.D. Modification of plant architecture in chrysanthemum by ectopic expression of the tobacco phytochrome B1 gene. J. Am. Soc. Hortic. Sci. 2001, 126, 19–26. [Google Scholar] [CrossRef][Green Version]
- Petty, L.M.; Harberd, N.P.; Carré, I.A.; Thomas, B.; Jackson, S. Expression of the Arabidopsis gai gene under its own promoter causes a reduction in plant height in chrysanthemum by attenuation of the gibberellin response. Plant Sci. 2003, 164, 175–182. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, G.; Liu, Q.; Zhu, Z.; Hu, Z. Dual silencing of DmCPD and DmGA20ox genes generates a novel miniature and delayed-flowering Dendranthema morifolium variety. Mol. Breed. 2015, 35, 67. [Google Scholar] [CrossRef]
- Evers, J.B.; van der Krol, A.R.; Vos, J.; Struik, P.C. Understanding shoot branching by modelling form and function. Trends Plant Sci. 2011, 16, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Kim, J.; Choi, S.; Woo, S.J. Effects of temperature and light on axillary bud initiation in branchless chrysanthemum. J. -Korean Soc. Hortic. Sci. 1999, 40, 368–370. [Google Scholar]
- Han, B.H.; Suh, E.J.; Lee, S.Y.; Shin, H.K.; Lim, Y.P. Selection of non-branching lines induced by introducing Ls-like cDNA into Chrysanthemum (Dendranthema × grandiflorum (Ramat.) Kitamura) “Shuho-no-chikara”. Sci. Hortic. 2007, 115, 70–75. [Google Scholar] [CrossRef]
- Jiang, B.; Miao, H.; Chen, S.; Zhang, S.; Chen, F.; Fang, W. The lateral suppressor-like gene, DgLsL, alternated the axillary branching in transgenic chrysanthemum (Chrysanthemum × morifolium) by modulating IAA and GA content. Plant Mol. Biol. Report. 2010, 28, 144–151. [Google Scholar] [CrossRef]
- Huh, Y.J.; Han, B.H.; Park, S.K.; Lee, S.Y.; Kil, M.J.; Pak, C.H.; Horticulture, Environment, and Biotechnology. Inhibition of chrysanthemum axillary buds via transformation with the antisense tomato lateral suppressor gene is season dependent. Hortic. Environ. Biotechnol. 2013, 54, 280–287. [Google Scholar] [CrossRef]
- Wen, C.; Zhao, Q.; Nie, J.; Liu, G.; Shen, L.; Cheng, C.; Xi, L.; Ma, N.; Zhao, L. Physiological controls of chrysanthemum DgD27 gene expression in regulation of shoot branching. Plant Cell Rep. 2016, 35, 1053–1070. [Google Scholar] [CrossRef]
- Dierck, R.; De Keyser, E.; De Riek, J.; Dhooghe, E.; Van Huylenbroeck, J.; Prinsen, E.; Van Der Straeten, D. Change in auxin and cytokinin levels coincides with altered expression of branching genes during axillary bud outgrowth in Chrysanthemum. PLoS ONE 2016, 11, e0161732. [Google Scholar] [CrossRef]
- Dierck, R.; Leus, L.; Dhooghe, E.; Van Huylenbroeck, J.; De Riek, J.; Van Der Straeten, D.; De Keyser, E. Branching gene expression during chrysanthemum axillary bud outgrowth regulated by strigolactone and auxin transport. Plant Growth Regul. 2018, 86, 23–36. [Google Scholar] [CrossRef]
- Ishak, A.; Dong, L.; Rong, H.; Zhang, S.; Zhao, L. Isolation and functional analysis of the regulation of branching by isopentenyl transferase gene CmIPT1 in Chrysanthemum morifolium cv.‘Jinba’. Am. J. Mol. Biol. 2018, 8, 92. [Google Scholar] [CrossRef]
- Klie, M.; Menz, I.; Linde, M.; Debener, T. Strigolactone pathway genes and plant architecture: Association analysis and QTL detection for horticultural traits in chrysanthemum. Mol. Genet. Genom. 2016, 291, 957–969. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, F.; Jiang, J.; Chen, S.; Fang, W.; Guan, Z.; Chen, F. Identification of quantitative trait loci for branching traits of spray cut chrysanthemum. Euphytica 2015, 202, 385–392. [Google Scholar] [CrossRef]
- Sun, W.; Yang, X.; Su, J.; Guan, Z.; Jiang, J.; Chen, F.; Fang, W.; Zhang, F. The genetics of planting density-dependent branching in chrysanthemum. Sci. Hortic. 2019, 256, 108598. [Google Scholar] [CrossRef]
- Sun, D.; Zhang, L.; Su, J.; Yu, Q.; Zhang, J.; Fang, W.; Wang, H.; Guan, Z.; Chen, F.; Song, A. Genetic Diversity and Genome-Wide Association Study of Architectural Traits of Spray Cut Chrysanthemum Varieties. Horticulturae 2022, 8, 458. [Google Scholar] [CrossRef]
- Behe, B.; Nelson, R.; Barton, S.; Hall, C.; Safley, C.D.; Turner, S. Consumer preferences for geranium flower color, leaf variegation, and price. HortScience 1999, 34, 740–742. [Google Scholar] [CrossRef]
- Posadas, B.C.; Coker, C.H.; Knight, P.R.; Fain, G. Consumer survey of selected garden chrysanthemum cultivars in Mississippi. HortTechnology 2006, 16, 539–543. [Google Scholar] [CrossRef]
- Samanta, A.; Das, G.; Das, S. Roles of flavonoids in plants. Carbon 2011, 100, 12–35. [Google Scholar]
- Hong, Y.; Bai, X.; Sun, W.; Jia, F. The numerical classification of chrysanthemum flower color phenotype. Acta Hortic. Sin. 2012, 39, 1330–1340. [Google Scholar]
- Wijayani, A.; Muafi, M.; Sukwadi, R. Retail Management Research Market actor’s response towards flower colours in d etermining the economic value of Chrysanthemum flowers. J. Bus. Retail. Manag. Res. 2017, 12, 69–75. [Google Scholar] [CrossRef][Green Version]
- Mekapogu, M.; Vasamsetti, B.M.K.; Kwon, O.-K.; Ahn, M.-S.; Lim, S.-H.; Jung, J.-A. Anthocyanins in floral colors: Biosynthesis and regulation in chrysanthemum flowers. Int. J. Mol. Sci. 2020, 21, 6537. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef]
- Kong, J.-M.; Chia, L.-S.; Goh, N.-K.; Chia, T.-F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochem. Rev. 2003, 64, 923–933. [Google Scholar] [CrossRef]
- Anderson, N.O.; Gesick, E.; Fritz, V.; Rohwer, C.; Yao, S.; Johnson, P.; Poppe, S.; Liedl, B.E.; Klossner, L.; Eash, N. Mammoth™ series garden chrysanthemum ‘Lavender Daisy’. HortScience 2014, 49, 1600–1604. [Google Scholar] [CrossRef]
- Anderson, N.O.; Ascher, P.D.; Fritz, V.; Rohwer, C.; Poppe, S.; Yao, S.; Johnson, P.; Klossner, L.; Eash, N.S.; Liedl, B.E. Chrysanthemum× hybridum MN 98-89-7 Shrub Garden Chrysanthemum. HortScience 2017, 52, 306–309. [Google Scholar] [CrossRef]
- Lim, J.; Park, S.; Cho, H.; Rhee, H.; Kim, M.; Joung, H.; Shin, H. A new spray chrysanthemum cultivar, “Pink PangPang201D with resistant to white rust, pompon type, and light pink color & pink center for cut flower. Korean J. Breed. Sci. 2007, 39, 520–521. [Google Scholar]
- Jung, Y.K.; Kim, S.K.; Kim, H.D.; Lee, Y.S. Breeding of a New Spray Chrysanthemum Cultivar,‘Dream Round’ with Dark Pink Petals and Thick Stem of Anemone Type for Cut-flower. Hortic. Sci. 2013, 31, 517–521. [Google Scholar]
- Jung, J.; Shin, H.; Lim, J.; Park, S.; Kwon, Y.; Choi, S. Standard chrysanthemum cultivar ‘Woonbaek’with white petals for cut-flower. Flower Res. J. 2016, 24, 362–367. [Google Scholar] [CrossRef]
- Kim, W.H.; Park, P.H.; Jung, J.A.; Park, K.Y.; Suh, J.-N.; Kwon, O.K.; Yoo, B.S.; Lee, S.Y.; Park, P.M.; Choi, Y. Achievement of flower breeding in Korea and its prospects. Korean Soc. Breed. Sci. 2020, 52, 161–169. [Google Scholar] [CrossRef]
- Song, C.; Liu, Y.; Song, A.; Dong, G.; Zhao, H.; Sun, W.; Ramakrishnan, S.; Wang, Y.; Wang, S.; Li, T. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Mol. Plant 2018, 11, 1482–1491. [Google Scholar] [CrossRef]
- Hirakawa, H.; Sumitomo, K.; Hisamatsu, T.; Nagano, S.; Shirasawa, K.; Higuchi, Y.; Kusaba, M.; Koshioka, M.; Nakano, Y.; Yagi, M. De novo whole-genome assembly in Chrysanthemum seticuspe, a model species of Chrysanthemums, and its application to genetic and gene discovery analysis. DNA Res. 2019, 26, 195–203. [Google Scholar] [CrossRef]
- Nakano, M.; Hirakawa, H.; Fukai, E.; Toyoda, A.; Kajitani, R.; Minakuchi, Y.; Itoh, T.; Higuchi, Y.; Kozuka, T.; Bono, H.; et al. A chromosome-level genome sequence of Chrysanthemum seticuspe, a model species for hexaploid cultivated chrysanthemum. Commun. Biol. 2021, 4, 1167. [Google Scholar] [CrossRef]
- Van Geest, G.; Bourke, P.M.; Voorrips, R.E.; Marasek-Ciolakowska, A.; Liao, Y.; Post, A.; van Meeteren, U.; Visser, R.G.; Maliepaard, C.; Arens, P.J.T.; et al. An ultra-dense integrated linkage map for hexaploid chrysanthemum enables multi-allelic QTL analysis. Theor. Appl. Genet. 2017, 130, 2527–2541. [Google Scholar] [CrossRef] [PubMed]
- Chong, X.; Zhang, F.; Wu, Y.; Yang, X.; Zhao, N.; Wang, H.; Guan, Z.; Fang, W.; Chen, F. A SNP-enabled assessment of genetic diversity, evolutionary relationships and the identification of candidate genes in chrysanthemum. Genome Biol. 2016, 8, 3661–3671. [Google Scholar] [CrossRef] [PubMed]
- Kee, E.S.; Naing, A.H.; Lim, S.H.; Han, J.S.; Kim, C. MYB transcription factor isolated from Raphanus sativus enhances anthocyanin accumulation in chrysanthemum cultivars. 3 Biotech 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y. Flower colour and cytochromes P450. Phytochem. Rev. 2006, 5, 283–291. [Google Scholar] [CrossRef]
- He, H.; Ke, H.; Keting, H.; Qiaoyan, X.; Silan, D. Flower colour modification of chrysanthemum by suppression of F3′H and overexpression of the exogenous Senecio cruentus F3′5′H gene. PLoS ONE 2013, 8, e74395. [Google Scholar] [CrossRef]
- Brugliera, F.; Tao, G.-Q.; Tems, U.; Kalc, G.; Mouradova, E.; Price, K.; Stevenson, K.; Nakamura, N.; Stacey, I.; Katsumoto, Y.; et al. Violet/blue chrysanthemums—Metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal colors. Plant Cell Physiol. 2013, 54, 1696–1710. [Google Scholar] [CrossRef]
- Noda, N.; Aida, R.; Kishimoto, S.; Ishiguro, K.; Fukuchi-Mizutani, M.; Tanaka, Y.; Ohmiya, A. Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant Cell Physiol. 2013, 54, 1684–1695. [Google Scholar] [CrossRef]
- Noda, N.; Yoshioka, S.; Kishimoto, S.; Nakayama, M.; Douzono, M.; Tanaka, Y.; Aida, R. Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci. Adv. 2017, 3, e1602785. [Google Scholar] [CrossRef]
- Hong, Y.; Tang, X.; Huang, H.; Zhang, Y.; Dai, S. Transcriptomic analyses reveal species-specific light-induced anthocyanin biosynthesis in chrysanthemum. BMC Genom. 2015, 16, 202. [Google Scholar] [CrossRef]
- Xiang, L.-l.; Liu, X.-f.; Li, X.; Yin, X.-r.; Grierson, D.; Li, F.; Chen, K.-S. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in Chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS ONE 2015, 10, e0143892. [Google Scholar] [CrossRef]
- Dong, W.; Li, M.; Li, Z.; Li, S.; Zhu, Y.; Wang, Z. Transcriptome analysis of the molecular mechanism of Chrysanthemum flower color change under short-day photoperiods. Plant Physiol. Biochem. 2020, 146, 315–328. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.-J.; Wang, Y.; Geng, Z.; Liu, S.; Chen, C.; Chen, S.; Jiang, J.; Chen, F. CmMYB9a activates floral coloration by positively regulating anthocyanin biosynthesis in chrysanthemum. Plant Mol. Biol. 2022, 108, 51–63. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.-J.; Wang, Y.; Geng, Z.; Ding, B.; Jiang, J.; Chen, S.; Chen, F. An R2R3-MYB transcription factor CmMYB21 represses anthocyanin biosynthesis in color fading petals of chrysanthemum. Sci. Hortic. 2022, 293, 110674. [Google Scholar] [CrossRef]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Piechulla, B.; Pott, M.B. Plant scents—Mediators of inter-and intraorganismic communication. Planta 2003, 217, 687–689. [Google Scholar] [CrossRef]
- Raguso, R.A. Start making scents: The challenge of integrating chemistry into pollination ecology. Entomol. Exp. Appl. 2008, 128, 196–207. [Google Scholar] [CrossRef]
- Zhou, F.; Pichersky, E. The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 2020, 226, 1341–1360. [Google Scholar] [CrossRef]
- Junker, R.R.; Blüthgen, N.J. Floral scents repel facultative flower visitors, but attract obligate ones. Ann. Bot. 2010, 105, 777–782. [Google Scholar] [CrossRef]
- Borghi, M.; Fernie, A.R.; Schiestl, F.P.; Bouwmeester, H.J. The sexual advantage of looking, smelling, and tasting good: The metabolic network that produces signals for pollinators. Sci. Agric. Sin. 2017, 22, 338–350. [Google Scholar] [CrossRef]
- Fenster, C.B.; Armbruster, W.S.; Wilson, P.; Dudash, M.R.; Thomson, J.D. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 375–403. [Google Scholar] [CrossRef]
- Dicke, M. Chemical ecology–from genes to communities. Chem. Ecol. Gene Ecosyst. 2006, 16, 175–189. [Google Scholar]
- Whitehead, M.R.; Peakall, R. Integrating floral scent, pollination ecology and population genetics. Funct. Ecol. 2009, 23, 863–874. [Google Scholar] [CrossRef]
- Amrad, A.; Moser, M.; Mandel, T.; de Vries, M.; Schuurink, R.C.; Freitas, L.; Kuhlemeier, C. Gain and loss of floral scent production through changes in structural genes during pollinator-mediated speciation. Curr. Biol. 2016, 26, 3303–3312. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhang, T.; Fan, Q.; Qi, X.; Zhang, F.; Fang, W.; Jiang, J.; Chen, F.; Chen, S. Identification of floral scent in chrysanthemum cultivars and wild relatives by gas chromatography-mass spectrometry. Molecules 2015, 20, 5346–5359. [Google Scholar] [CrossRef] [PubMed]
- Woo, K.-S.; Yu, J.-S.; Hwang, I.-G.; Lee, Y.-R.; Lee, C.-H.; Yoon, H.-S.; Lee, J.-S.; Jeong, H.-S. Antioxidative activity of volatile compounds in flower of Chrysanthemum indicum, C. morifolium, and C. zawadskii. J. Korean Soc. Food Sci. Nutr. 2008, 37, 805–809. [Google Scholar] [CrossRef]
- Turlings, T.C.; Ton, J. Exploiting scents of distress: The prospect of manipulating herbivore-induced plant odours to enhance the control of agricultural pests. Curr. Opin. Plant Biol. 2006, 9, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Pichersky, E. Metabolic engineering of plant volatiles. Curr. Opin. Biotechnol. 2008, 19, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Kamatou, G.P.; Viljoen, A.M. Linalool–A review of a biologically active compound of commercial importance. Nat. Prod. Commun. 2008, 3, 1183–1192. [Google Scholar] [CrossRef]
- Yang, T.; Stoopen, G.; Thoen, M.; Wiegers, G.; Jongsma, M.A. Chrysanthemum expressing a linalool synthase gene ‘smells good’, but ‘tastes bad’to western flower thrips. Plant Biotechnol. J. 2013, 11, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Mitsuda, N.; Nashima, K.; Kishimoto, K.; Katayose, Y.; Kanamori, H.; Ohmiya, A. Generation of expressed sequence tags for discovery of genes responsible for floral traits of Chrysanthemum morifolium by next-generation sequencing technology. BMC Genomics 2017, 18, 68. [Google Scholar]
- Aros, D.; Garrido, N.; Rivas, C.; Medel, M.; Müller, C.; Rogers, H.; Úbeda, C. Floral scent evaluation of three cut flowers through sensorial and gas chromatography analysis. Agron. J. 2020, 10, 131. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, Y.; Chen, S.; Chen, F.; Chen, F. Concentration-dependent emission of floral scent terpenoids from diverse cultivars of Chrysanthemum morifolium and their wild relatives. J. Plant Sci. 2021, 309, 110959. [Google Scholar] [CrossRef] [PubMed]
- Roh, Y.S.; Kim, I.K.; Yoo, Y.K. Vase life and quality of cut flower by NaOCl and sucrose treatment as wet harvesting solution in standard chrysanthemum ‘Baekma’. J. People Plants Environ. 2017, 20, 521–530. [Google Scholar] [CrossRef]
- Netam, N. Improving ornamental’s vase life through molecular approaches: A review. Pharm. Phytochem. 2018, 7, 1687–1691. [Google Scholar]
- Bowyer, M.; Wills, R.; Badiyan, D.; Ku, V. Extending the postharvest life of carnations with nitric oxide—Comparison of fumigation and in vivo delivery. Postharvest Biol. Technol. 2003, 30, 281–286. [Google Scholar] [CrossRef]
- Fanourakis, D.; Papadakis, V.M.; Psyllakis, E.; Tzanakakis, V.A.; Nektarios, P.A. The role of water relations and oxidative stress in the vase life response to prolonged storage: A case study in chrysanthemum. Agric. Hanbook 2022, 12, 185. [Google Scholar] [CrossRef]
- Doi, M.; Nakagawa, Y.; Watabe, S.; Aoe, K.; Inamoto, K.; Imanishi, H. Ethylene-induced leaf yellowing in cut chrysanthemums (Dendranthema grandiflora Kitamura). J. Jpn. Soc. Hortic. Sci. 2003, 72, 533–535. [Google Scholar] [CrossRef][Green Version]
- Narumi, T.; Aida, R.; Ohmiya, A.; Satoh, S. Transformation of chrysanthemum with mutated ethylene receptor genes: mDG-ERS1 transgenes conferring reduced ethylene sensitivity and characterization of the transformants. Postharvest Biol. Technol. 2005, 37, 101–110. [Google Scholar] [CrossRef]
- Satoh, S.; Watanabe, M.; Chisaka, K.; Narumi, T. Suppressed leaf senescence in chrysanthemum transformed with a mutated ethylene receptor genemDG-ERS1 (etr1-4). Plant Biol. 2008, 51, 424–427. [Google Scholar] [CrossRef]
- Liu, R.; Zuo, X.; Chen, Y.; Qian, Z.; Xu, C.; Wang, L.; Chen, S. Transcriptional Regulation in Leaves of Cut Chrysanthemum (Chrysanthemum morifolium)‘FenDante’in Response to Post-Harvest Ethylene Treatment. Horticulturae 2022, 8, 573. [Google Scholar] [CrossRef]
- Boss, P.K.; Bastow, R.M.; Mylne, J.S.; Dean, C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 2004, 16, S18–S31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Huang, H.; Wang, Q.; Dai, S. Cross breeding new cultivars of early-flowering multiflora chrysanthemum based on mathematical analysis. BMC Genom. 2018, 53, 421–426. [Google Scholar] [CrossRef]
- UPOV, 2010. UPOV (the International Union for the Protection of New Varieties of Plants) Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability (Chrysanthemum); UPOV: Geneva, Switzerland, 2010. [Google Scholar]
- Cockshull, K.E. Chrysanthemum morifolium. In CRC Handbook of Flowering; CRC Press: Boca Raton, FL, USA, 2019; pp. 238–257. [Google Scholar]
- Ochiai, M.; Liao, Y.; Shimazu, T.; Takai, Y.; Suzuki, K.; Yano, S.; Fukui, H. Varietal differences in flowering and plant growth under night-break treatment with LEDs in 12 chrysanthemum cultivars. Environ. Control Biol. 2015, 53, 17–22. [Google Scholar] [CrossRef]
- Litt, A.; Irish, V. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genet. Mol. Biol. 2003, 165, 821–833. [Google Scholar] [CrossRef]
- Shulga, O.A.; Mitiouchkina, T.Y.; Shchennikova, A.V.; Skryabin, K.G.; Dolgov, S.V.; Biology-Plant, D. Overexpression of AP1-like genes from Asteraceae induces early-flowering in transgenic Chrysanthemum plants. In Vitro Cell. Dev. Biol. Plant 2011, 47, 553–560. [Google Scholar] [CrossRef]
- Oda, A.; Narumi, T.; Li, T.; Kando, T.; Higuchi, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J. Exp. Bot. 2012, 63, 1461–1477. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, H.; Ren, L.; Chen, S.; Chen, F.; Jiang, J. CmFTL2 is involved in the photoperiod-and sucrose-mediated control of flowering time in chrysanthemum. Hortic. Res. 2017, 4, 17001. [Google Scholar] [CrossRef]
- Mao, Y.; Sun, J.; Cao, P.; Zhang, R.; Fu, Q.; Chen, S.; Chen, F.; Jiang, J. Functional analysis of alternative splicing of the FLOWERING LOCUS T orthologous gene in Chrysanthemum morifolium. Hortic. Res. 2016, 3, 16058. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. J. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Ma, C.; Xu, Y.; Wang, T.; Chen, Y.; Lü, J.; Zhang, L.; Jiang, C.-Z.; Hong, B.; Gao, J. Control of chrysanthemum flowering through integration with an aging pathway. Nat. Commun. 2017, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Jiang, J.; Huang, Y.; Zhang, Z.; Song, A.; Ding, L.; Wang, H.; Yao, J.; Chen, S.; Chen, F.; et al. The constitutive expression of a chrysanthemum ERF transcription factor influences flowering time in Arabidopsis thaliana. Mol. Biotechnol. 2019, 61, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Gao, Y.; Wu, Z.; Bu, X.; Fan, M.; Zhang, Q. Characterization of TEMINAL FLOWER1 homologs CmTFL1c gene from Chrysanthemum morifolium. Plant Mol. Biol. 2019, 99, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, M.; Li, Z.; Zhu, Y.; Ding, H.; Fan, X.; Li, F.; Wang, Z. Effects of the silencing of CmMET1 by RNA interference in chrysanthemum (Chrysanthemum morifolium). Plant Biotechnol. Rep. 2019, 13, 63–72. [Google Scholar] [CrossRef]
- Haider, S.; Gao, Y.; Gao, Y. Standardized Genetic Transformation Protocol for Chrysanthemum cv. ‘Jinba’ with TERMINAL FLOWER 1 Homolog CmTFL1a. Genes Dev. 2020, 11, 860. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Takase, T.; Sumitomo, K.; Suzuki, S.; Tsuda-Kawamura, K.; Hisamatsu, T. Delay of flowering at high temperature in chrysanthemum: Duration of darkness and transitions in lighting determine daily peak heat sensitivity. Hortic. J. 2020, 90, 255. [Google Scholar] [CrossRef]
- Nakano, Y.; Higuchi, Y.; Sumitomo, K.; Oda, A.; Hisamatsu, T.; Science, N. Delay of flowering by high temperature in chrysanthemum: Heat-sensitive time-of-day and heat effects on CsFTL3 and CsAFT gene expression. J. Hortic. Sci. Biotechnol. 2015, 90, 143–149. [Google Scholar] [CrossRef]
- Cho, A.R.; Kim, Y.J. Night temperature determines flowering time and quality of Chrysanthemum morifolium during a high day temperature. J. Hortic. Sci. Biotechnol. 2021, 96, 239–248. [Google Scholar] [CrossRef]
- Park, Y.G.; Jeong, B.R. How supplementary or night-interrupting low-intensity blue light affects the flower induction in chrysanthemum, a qualitative short-day plant. Plants 2020, 9, 1694. [Google Scholar] [CrossRef]
- Wang, L.; Sun, J.; Ren, L.; Zhou, M.; Han, X.; Ding, L.; Zhang, F.; Guan, Z.; Fang, W.; Chen, S.; et al. CmBBX8 accelerates flowering by targeting CmFTL1 directly in summer chrysanthemum. Plant Biotechnol. J. 2020, 18, 1562–1572. [Google Scholar] [CrossRef]
- Chen, H.; Huang, F.; Liu, Y.; Cheng, P.; Guan, Z.; Fang, W.; Chen, S.; Chen, F.; Jiang, J. Constitutive expression of chrysanthemum CmBBX29 delays flowering time in transgenic Arabidopsis. Can. J. Plant Sci. 2019, 100, 86–94. [Google Scholar] [CrossRef]
- Zhu, L.; Guan, Y.; Liu, Y.; Zhang, Z.; Jaffar, M.A.; Song, A.; Chen, S.; Jiang, J.; Chen, F. Regulation of flowering time in chrysanthemum by the R2R3 MYB transcription factor CmMYB2 is associated with changes in gibberellin metabolism. Hortic. Res. 2020, 7, 96. [Google Scholar] [CrossRef]
- Aida, R.; Komano, M.; Saito, M.; Nakase, K.; Murai, K. Chrysanthemum flower shape modification by suppression of chrysanthemum-AGAMOUS gene. Plant Biotechnol. 2008, 25, 55–59. [Google Scholar] [CrossRef]
- Broholm, S.K.; Tähtiharju, S.; Laitinen, R.A.; Albert, V.A.; Teeri, T.H.; Elomaa, P.J. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 2008, 105, 9117–9122. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Li, X.; Sun, M.; Zhang, T.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q. Identification and characterization of CYC-like genes in regulation of ray floret development in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 1633. [Google Scholar] [CrossRef]
- Liu, H.; Sun, M.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q. Two Cyc2CL transcripts (Cyc2CL-1 and Cyc2CL-2) may play key roles in the petal and stamen development of ray florets in chrysanthemum. BMC Plant Biol. 2021, 21, 105. [Google Scholar] [CrossRef]
- Shen, C.Z.; Chen, J.; Zhang, C.J.; Rao, G.Y.; Guo, Y.P. Dysfunction of CYC2g is responsible for the evolutionary shift from radiate to disciform flowerheads in the Chrysanthemum group (Asteraceae: Anthemideae). Plant J. 2021, 106, 1024–1038. [Google Scholar] [CrossRef]
- Wang, J.; Guan, Y.; Ding, L.; Li, P.; Zhao, W.; Jiang, J.; Chen, S.; Chen, F. The CmTCP20 gene regulates petal elongation growth in Chrysanthemum morifolium. Plant Sci. 2019, 280, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, M.; Du, D.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q.; Gao, Y. Whole-transcriptome analysis of differentially expressed genes in the ray florets and disc florets of Chrysanthemum morifolium. BMC Genom. 2016, 17, 398. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Zhao, K.; Zhang, X.; Song, A.; Su, J.; Hu, Y.; Zhao, W.; Jiang, J.; Chen, F. Comprehensive characterization of a floral mutant reveals the mechanism of hooked petal morphogenesis in Chrysanthemum morifolium. Plant Biotechnol. J. 2019, 17, 2325–2340. [Google Scholar] [CrossRef]
- Yang, X.; Fang, X.; Su, J.; Ding, L.; Guan, Z.; Jiang, J.; Chen, S.; Chen, F.; Fang, W.; Zhang, F. Genetic dissection of floral traits in anemone-type chrysanthemum by QTL mapping. J. Mol. Breed. 2019, 39, 136. [Google Scholar] [CrossRef]
- Song, X.; Zhao, X.; Fan, G.; Gao, K.; Dai, S.; Zhang, M.; Ma, C.; Wu, X. Genetic analysis of the corolla tube merged degree and the relative number of ray florets in chrysanthemum (Chrysanthemum × morifolium Ramat.). Sci. Hortic. 2018, 242, 214–224. [Google Scholar] [CrossRef]
- Song, X.; Xu, Y.; Gao, K.; Fan, G.; Zhang, F.; Deng, C.; Dai, S.; Huang, H.; Xin, H.; Li, Y. High-density genetic map construction and identification of loci controlling flower-type traits in Chrysanthemum (Chrysanthemum × morifolium Ramat.). Hortic. Res. 2020, 7, 108. [Google Scholar] [CrossRef]
- Chong, X.; Su, J.; Wang, F.; Wang, H.; Song, A.; Guan, Z.; Fang, W.; Jiang, J.; Chen, S.; Chen, F. Identification of favorable SNP alleles and candidate genes responsible for inflorescence-related traits via GWAS in chrysanthemum. Plant Mol. Biol. 2019, 99, 407–420. [Google Scholar] [CrossRef]
- Mekapogu, M.; Jung, J.-A.; Kwon, O.-K.; Ahn, M.-S.; Song, H.-Y.; Jang, S.J. Recent progress in enhancing fungal disease resistance in ornamental plants. Int. J. Mol. Sci. 2021, 22, 7956. [Google Scholar] [CrossRef]
- Yu, M.; Liu, Z.; Chen, S.; Chen, F. Expression of P. mume PGIP gene in transgenic Dendranthema morifolium increased tolerance to disease resistance. J. Acta Bot. Boreali-Occident. Sin. 2010, 30, 1111–1116. [Google Scholar]
- Xu, G.; Chen, S.; Chen, F. Transgenic chrysanthemum plants expressing a harpinXoo gene demonstrate induced resistance to alternaria leaf spot and accelerated development. Russ. J. Plant Physiol. 2010, 57, 548–553. [Google Scholar] [CrossRef]
- Sen, S.; Kumar, S.; Ghani, M.; Thakur, M. Agrobacterium mediated genetic transformation of chrysanthemum (Dendranthema grandiflora Tzvelev) with rice chitinase gene for improved resistance against Septoria obesa. Plant Pathol. J. 2013, 12, 1–10. [Google Scholar] [CrossRef]
- Zhao, X.; Song, L.; Jiang, L.; Zhu, Y.; Gao, Q.; Wang, D.; Xie, J.; Lv, M.; Liu, P.; Li, M. The integration of transcriptomic and transgenic analyses reveals the involvement of the SA response pathway in the defense of chrysanthemum against the necrotrophic fungus Alternaria sp. Hortic. Res. 2020, 7, 80. [Google Scholar] [CrossRef]
- Xin, J.; Liu, Y.; Li, H.; Chen, S.; Jiang, J.; Song, A.; Fang, W.; Chen, F. CmMLO17 and its partner CmKIC potentially support Alternaria alternata growth in Chrysanthemum morifolium. Hortic. Res. 2021, 8, 101. [Google Scholar] [CrossRef]
- Takatsu, Y.; Nishizawa, Y.; Hibi, T.; Akutsu, K. Transgenic chrysanthemum (Dendranthema grandiflorum (Ramat.) Kitamura) expressing a rice chitinase gene shows enhanced resistance to gray mold (Botrytis cinerea). Sci. Hortic. 1999, 82, 113–123. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Lim, S.; Yoda, H.; Choi, Y.-E.; Sano, H.J. Simultaneous activation of salicylate production and fungal resistance in transgenic chrysanthemum producing caffeine. Plant Signal. Behav. 2011, 6, 409–412. [Google Scholar] [CrossRef]
- Ichikawa, H.; Kato, K.; Mochizuki, A.; Shinoyama, H.; Mitsuhara, I. Transgenic chrysanthemums (Chrysanthemum morifolium Ramat.) carrying both insect and disease resistance. In Proceedings of the XXV International EUCARPIA Symposium Section Ornamentals: Crossing Borders 1087, Melle, Belgium, 28 June–2 July 2015; pp. 485–497. [Google Scholar]
- Bi, M.; Li, X.; Yan, X.; Liu, D.; Gao, G.; Zhu, P.; Mao, H. Chrysanthemum WRKY15-1 promotes resistance to Puccinia horiana Henn. via the salicylic acid signaling pathway. Hortic. Res. 2021, 8, 6. [Google Scholar] [CrossRef]
- Li, H.; Chen, S.; Song, A.; Wang, H.; Fang, W.; Guan, Z.; Jiang, J.; Chen, F. RNA-Seq derived identification of differential transcription in the chrysanthemum leaf following inoculation with Alternaria tenuissima. BMC Genom. 2014, 15, 9. [Google Scholar] [CrossRef]
- Lu, D.; Zhiqiang, H.; Di, L.; Pengfang, Z.; Shengjin, L.; Na, L.; Hongyu, M. Transcriptome analysis of chrysanthemum in responses to white rust. Sci. Hortic. 2018, 233, 421–430. [Google Scholar] [CrossRef]
- Shinoyama, H.; Mochizuki, A.; Komano, M.; Nomura, Y.; Nagai, T. Insect resistance in transgenic chrysanthemum [Dendranthema × grandiflorum (Ramat.) Kitamura] by the introduction of a modified δ-endotoxin gene of Bacillus thuringiensis. Breed. Sci. 2003, 53, 359–367. [Google Scholar] [CrossRef][Green Version]
- Leiss, K.A.; Maltese, F.; Choi, Y.H.; Verpoorte, R.; Klinkhamer, P.G. Identification of chlorogenic acid as a resistance factor for thrips in chrysanthemum. Plant Physiol. Biochem. 2009, 150, 1567–1575. [Google Scholar] [CrossRef]
- Xia, X.; Shao, Y.; Jiang, J.; Du, X.; Sheng, L.; Chen, F.; Fang, W.; Guan, Z.; Chen, S. MicroRNA expression profile during aphid feeding in chrysanthemum (Chrysanthemum morifolium). PLoS ONE 2015, 10, e0143720. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, T.; Li, P.; Tian, C.; Song, A.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F.; Chen, S. Chrysanthemum CmWRKY53 negatively regulates the resistance of chrysanthemum to the aphid Macrosiphoniella sanborni. Hortic. Res. 2020, 7, 109. [Google Scholar] [CrossRef]
- Sherman, J.M.; Moyer, J.W.; Daub, M.E.J.P.D. Tomato spotted wilt virus resistance in chrysanthemum expressing the viral nucleocapsid gene. Plant Dis. 1998, 82, 407–414. [Google Scholar] [CrossRef]
- Kumar, S.; Raj, S.; Sharma, A.; Varma, H. Genetic transformation and development of Cucumber mosaic virus resistant transgenic plants of Chrysanthemum morifolium cv. Kundan. Sci. Hortic. 2012, 134, 40–45. [Google Scholar] [CrossRef]
- Mitiouchkina, T.Y.; Firsov, A.P.; Titova, S.M.; Pushin, A.S.; Shulga, O.A.; Dolgov, S.V. Different approaches to produce transgenic virus B Resistant Chrysanthemum. Agron. J. 2018, 8, 28. [Google Scholar] [CrossRef]
- Choi, H.; Jo, Y.; Lian, S.; Jo, K.-M.; Chu, H.; Yoon, J.-Y.; Choi, S.-K.; Kim, K.-H.; Cho, W.K. Comparative analysis of chrysanthemum transcriptome in response to three RNA viruses: Cucumber mosaic virus, Tomato spotted wilt virus and Potato virus X. Plant Mol. Biol. 2015, 88, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Jo, K.-M.; Jo, Y.; Choi, H.; Chu, H.; Lian, S.; Yoon, J.-Y.; Choi, S.-K.; Kim, K.-H.; Cho, W.K. Development of genetically modified chrysanthemums resistant to Chrysanthemum stunt viroid using sense and antisense RNAs. Sci. Hortic. 2015, 195, 17–24. [Google Scholar] [CrossRef]
- Nabeshima, T.; Doi, M.; Hosokawa, M. Comparative analysis of Chrysanthemum stunt viroid accumulation and movement in two Chrysanthemum (Chrysanthemum morifolium) cultivars with differential susceptibility to the viroid infection. Front. Plant Sci. 2017, 8, 1940. [Google Scholar] [CrossRef]
- Takino, H.; Furuya, M.; Sakuma, A.; Yamamoto, S.; Hirano, S.; Tsuro, M.; Yanagimoto, T.; Tanaka, Y.; Mino, M. The siRNAs targeting the left or right terminal region of chrysanthemum stunt viroid (CSVd) sequence suppress the development of disease symptoms caused by CSVd infection of chrysanthemum, but do not suppress viroid propagation. J. Hortic. Sci. Biotechnol. 2018, 93, 491–499. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef]
- Hong, B.; Ma, C.; Yang, Y.; Wang, T.; Yamaguchi-Shinozaki, K.; Gao, J. Over-expression of AtDREB1A in chrysanthemum enhances tolerance to heat stress. Plant Mol. Biol. 2009, 70, 231–240. [Google Scholar] [CrossRef]
- Du, X.; Li, W.; Sheng, L.; Deng, Y.; Wang, Y.; Zhang, W.; Yu, K.; Jiang, J.; Fang, W.; Guan, Z. Over-expression of chrysanthemum CmDREB6 enhanced tolerance of chrysanthemum to heat stress. BMC Plant Biol. 2018, 18, 178. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, Y.; Zhang, Z.; Gao, J.; Guan, Z.; Fang, W.; Chen, S.; Chen, F.; Jiang, J. The over-expression of a chrysanthemum gene encoding an RNA polymerase II CTD phosphatase-like 1 enzyme enhances tolerance to heat stress. Hortic. Res. 2018, 5, 37. [Google Scholar] [CrossRef]
- Xing, X.; Ding, Y.; Jin, J.; Song, A.; Chen, S.; Chen, F.; Fang, W.; Jiang, J. Physiological and transcripts analyses reveal the mechanism by which melatonin alleviates heat stress in chrysanthemum seedlings. Front. Plant Sci. 2021, 12, 673236. [Google Scholar] [CrossRef]
- Wang, X.; Wu, J.; Wang, Y.; Jiang, Y.; Li, F.; Chen, Y.; Jiang, J.; Wang, L.; Guan, Z.; Chen, F. Chrysanthemum CmHSP90. 5 as a Tool to Regulate Heat and Salt Stress Tolerance. Horticulturae 2022, 8, 532. [Google Scholar] [CrossRef]
- Bo, H.; Zheng, T.; Qiuhua, L. Regeneration and transformation through somatic embryogenesis, and determination of cold stress tolerance in ground cover Chrysanthemum cv. Fall color. Sci. Agric. Sin. 2006, 39, 1443–1450. [Google Scholar]
- Huang, Q.; Liao, X.; Yang, X.; Luo, Y.; Lin, P.; Zeng, Q.; Bai, H.; Jiang, B.; Pan, Y.; Zhang, F. Lysine crotonylation of DgTIL1 at K72 modulates cold tolerance by enhancing DgnsLTP stability in chrysanthemum. Plant Biotechnol. J. 2021, 19, 1125–1140. [Google Scholar] [CrossRef]
- Yang, X.; Lin, P.; Luo, Y.; Bai, H.; Liao, X.; Li, X.; Tian, Y.; Jiang, B.; Pan, Y.; Zhang, F. Lysine decrotonylation of glutathione peroxidase at lysine 220 site increases glutathione peroxidase activity to resist cold stress in chrysanthemum. J. Ecotoxicol. Environ. Saf. 2022, 232, 113295. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Luo, Y.; Bai, H.; Li, X.; Tang, S.; Liao, X.; Zhang, L.; Liu, Q. DgMYB2 improves cold resistance in chrysanthemum by directly targeting DgGPX1. Hortic. Res. 2022, 9, uhab028. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Liao, X.; Li, X.; Wang, B.; Luo, Y.; Yang, X.; Tian, Y.; Zhang, L.; Zhang, F.; Pan, Y. DgbZIP3 interacts with DgbZIP2 to increase the expression of DgPOD for cold stress tolerance in chrysanthemum. Hortic. Res. 2022, 9, uhac105. [Google Scholar] [CrossRef]
- Tian, C.; Zhai, L.; Zhu, W.; Qi, X.; Yu, Z.; Wang, H.; Chen, F.; Wang, L.; Chen, S. Characterization of the TCP Gene Family in Chrysanthemum nankingense and the Role of CnTCP4 in Cold Tolerance. Plants 2022, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Song, A.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The over-expression of Chrysanthemum crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol. Biol. Rep. 2014, 41, 4155–4162. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.; Zhao, H.; Gao, T.; Song, A.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum Cm HSFA 4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 2018, 16, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wu, Y.-H.; Tian, X.-Q.; Bai, Z.-Y.; Liang, Q.-Y.; Liu, Q.-L.; Pan, Y.-Z.; Zhang, L.; Jiang, B.-B. Overexpression of DgWRKY4 enhances salt tolerance in chrysanthemum seedlings. Front. Plant Sci. 2017, 8, 1592. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.-Y.; Wu, Y.-H.; Wang, K.; Bai, Z.-Y.; Liu, Q.-L.; Pan, Y.-Z.; Zhang, L.; Jiang, B.-B. Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci. Rep. 2017, 7, 4799. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhong, M.; Wu, Y.-H.; Bai, Z.-Y.; Liang, Q.-Y.; Liu, Q.-L.; Pan, Y.-Z.; Zhang, L.; Jiang, B.-B.; Jia, Y.J. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017, 36, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xie, L.; Sun, T.; Ding, B.; Li, Y.; Zhang, Y. Chrysanthemum morifolium aquaporin genes CmPIP1 and CmPIP2 are involved in tolerance to salt stress. J. Sci. Hortic. 2019, 256, 108627. [Google Scholar] [CrossRef]
- Wang, H.; Li, T.; Li, W.; Wang, W.; Zhao, H. Identification and analysis of Chrysanthemum nankingense NAC transcription factors and an expression analysis of OsNAC7 subfamily members. PeerJ 2021, 9, e11505. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Y.; Xu, N.; Sun, Y.; Li, Q.; Yue, L.; Zhou, Y.; He, M. Chrysanthemum × grandiflora leaf and root transcript profiling in response to salinity stress. BMC Plant Biol. 2022, 22, 240. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Meng, R.; Liu, Y.; Chen, S.; Jiang, J.; Wang, L.; Zhao, S.; Wang, Z.; Fang, W.; Chen, F.; et al. Heterografted chrysanthemums enhance salt stress tolerance by integrating reactive oxygen species, soluble sugar, and proline. Hortic. Res. 2022, 9, uhac073. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.; Tong, Z.; Ma, N.; Kasuga, M.; Yamaguchi-Shinozaki, K.; Gao, J.-P. Expression of the Arabidopsis DREB1A gene in transgenic chrysanthemum enhances tolerance to low temperature. J. Hortic. Sci. Biotechnol. 2006, 81, 1002–1008. [Google Scholar] [CrossRef]
- Chen, S.; Cui, X.; Chen, Y.; Gu, C.; Miao, H.; Gao, H.; Chen, F.; Liu, Z.; Guan, Z.; Fang, W.; et al. CgDREBa transgenic chrysanthemum confers drought and salinity tolerance. Environ. Exp. Bot. 2011, 74, 255–260. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Y.; Jiang, J.; Chen, S.; Chen, F.; Guan, Z.; Fang, W. The constitutive expression of Chrysanthemum dichrum ICE1 in Chrysanthemum grandiflorum improves the level of low temperature, salinity and drought tolerance. Plant Cell Rep. 2012, 31, 1747–1758. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, S.; Yang, Y.; Huang, M.; Cheng, L.; Wei, Q.; Fei, Z.; Gao, J.; Hong, B. Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genom. 2013, 14, 662. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Su, J.; Guan, Z.; Fang, W.; Chen, F.; Zhang, F. Association analysis of drought tolerance in cut chrysanthemum (Chrysanthemum morifolium Ramat.) at seedling stage. 3 Biotech. 2018, 8, 226. [Google Scholar] [CrossRef]
- Fan, Q.; Song, A.; Jiang, J.; Zhang, T.; Sun, H.; Wang, Y.; Chen, S.; Chen, F. CmWRKY1 enhances the dehydration tolerance of chrysanthemum through the regulation of ABA-associated genes. PLoS ONE 2016, 11, e0150572. [Google Scholar] [CrossRef] [PubMed]
- Jaffar, M.A.; Song, A.; Faheem, M.; Chen, S.; Jiang, J.; Liu, C.; Fan, Q.; Chen, F. Involvement of CmWRKY10 in drought tolerance of chrysanthemum through the ABA-signaling pathway. Int. J. Mol. Sci. 2016, 17, 693. [Google Scholar] [CrossRef]
- Gao, W.; He, M.; Liu, J.; Ma, X.; Zhang, Y.; Dai, S.; Zhou, Y. Overexpression of Chrysanthemum lavandulifolium ClCBF1 in Chrysanthemum morifolium ‘White Snow’improves the level of salinity and drought tolerance. Plant Physiol. Biochem. 2018, 124, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Wen, C.; Xi, L.; Lv, S.; Zhao, Q.; Kou, Y.; Ma, N.; Zhao, L.; Zhou, X. The AP2/ERF transcription factor CmERF053 of chrysanthemum positively regulates shoot branching, lateral root, and drought tolerance. Plant Cell Rep. 2018, 37, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhong, M.; He, L.; Wang, B.; Liu, Q.-L.; Pan, Y.-Z.; Jiang, B.-B.; Zhang, L.; Culture, O. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves drought tolerance in chrysanthemum. J. Plant Cell Tissue 2018, 135, 119–132. [Google Scholar] [CrossRef]
- Liu, Y.; He, M.; Dong, F.; Cai, Y.; Gao, W.; Zhou, Y.; Huang, H.; Dai, S. The Chrysanthemum lavandulifolium ClNAC9 gene positively regulates saline, alkaline, and drought stress in transgenic Chrysanthemum grandiflora. J. Am. Soc. Hortic. Sci. 2019, 144, 280–288. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, X.; Aiwaili, P.; Mu, X.; Zhao, M.; Zhao, J.; Cheng, L.; Ma, C.; Gao, J.; Hong, B. A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 2020, 103, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wei, Q.; Wang, Z.; Liu, W.; Zhao, X.; Ma, C.; Gao, J.; Xu, Y.; Hong, B. CmNF-YB8 affects drought resistance in chrysanthemum by altering stomatal status and leaf cuticle thickness. J. Integr. Plant Biol. 2022, 64, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Zhu, X.; Yang, S.; Gu, C.; Liu, P.; Song, A.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F. Constitutive expression of a chrysanthemum phospholipase Dα gene in Chrysanthemum morifolium enhances drought tolerance. Ornam. Plant Res. 2021, 1, 8. [Google Scholar]
- Zhang, W.; Xu, H.; Duan, X.; Hu, J.; Li, J.; Zhao, L.; Ma, Y. Characterizing the leaf transcriptome of Chrysanthemum rhombifolium (Ling et C. Shih), a drought resistant, endemic plant from China. Front. Genet. 2021, 12, 625985. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Liu, C.; Yin, H.; Liu, H.; Luo, H.; He, M.; Zhou, Y. CgbZIP1: A bZIP Transcription Factor from Chrysanthemum Grandiflora Confers Plant Tolerance to Salinity and Drought Stress. Agron. J. 2022, 12, 556. [Google Scholar] [CrossRef]
- Shinoyama, H.; Ichikawa, H.; Nishizawa-Yokoi, A.; Skaptsov, M.; Toki, S. Simultaneous TALEN-mediated knockout of chrysanthemum DMC1 genes confers male and female sterility. Sci. Rep. 2020, 10, 16165. [Google Scholar] [CrossRef]
- Royal FloraHolland in Facts and Figures. 2021. Annual Report. Available online: www.floraholland.com (accessed on 10 August 2022).

| Ser. No. | Gene | Source | Resulting Trait | Reference |
|---|---|---|---|---|
| 1 | Phytochrome b1 | Tobacco | shorter plant with larger branch angles | [26] |
| 2 | gai | Arabidopsis | shorter plant height | [27] |
| 3 | DmCPD and DmGA20ox (silencing) | Chrysanthemum | dwarf phenotype | [28] |
| 4 | DgLsL (silencing) | Chrysanthemum | suppressed axillary bud growth | [32] |
| 5 | DgD27 | Chrysanthemum | less number of tillers | [34] |
| 6 | CmDRM1, CmBRC1 and CmMAX1 | Chrysanthemum | axillary bud outgrowth | [35] |
| 7 | CmIPT1 | Chrysanthemum | regulation of lateral branching | [37] |
| 8 | PhyB, BRH1, CPC and bZIP16 | Chrysanthemum | regulation of plant architecture | [41] |
| 9 | F3′5′H | Senecio cruentus | anthocyanin biosynthesis | [63] |
| 10 | F3′5′H | Pansy | anthocyanin biosynthesis | [64] |
| 11 | F3′5′H | Campanula | anthocyanin biosynthesis | [65] |
| 12 | 3′,5′-O-glucosyltransferase and F3′5′H | butterfly pea and Canterbury bells | true-blue colored flower | [66] |
| 13 | CmMYB5-1, CmMYB6, CmMYB7-1 and CmbHLH24 | Chrysanthemum | light-induced anthocyanin accumulation | [67] |
| 14 | CmbHLH2+CmMYB6 | Chrysanthemum | regulation of anthocyanin accumulation | [68] |
| 15 | CmMYB9a | Chrysanthemum | regulation of anthocyanin accumulation | [70] |
| 16 | CmMYB21 | Chrysanthemum | repression of anthocyanin biosynthesis | [71] |
| 17 | FaNES1 | Strawberry | floral scent | [109] |
| 18 | mDG-ERS1 | Chrysanthemum | reduced ethylene sensitivity | [96] |
| 19 | mDG-ERS1 (etr1-4) | Chrysanthemum | leaf senescence suppression | [97] |
| 20 | AP1 | Asteraceae | early flowering | [105] |
| 21 | CsFTL3 | Chrysanthemum seticuspe | early flowering | [106] |
| 22 | CmFTL2 | Chrysanthemum | regulation of photoperiodic flowering under short day | [107] |
| 23 | CmBBX24 (silencing) | Chrysanthemum | early flowering | [109] |
| 24 | miR156 | Chrysanthemum | regulation of flowering | [110] |
| 25 | CmERF110 | Chrysanthemum | early flowering | [111] |
| 26 | CmTFL1a | Chrysanthemum | delayed flowering | [114] |
| 27 | FTL3 (silencing) | Chrysanthemum | heat-induced flowering delay | [116] |
| 28 | CmBBX8 | Chrysanthemum | regulation of early flowering | [119] |
| 29 | CmBBX29 | Chrysanthemum | regulation of delayed flowering | [120] |
| 30 | CmMYB2+CmBBX24 | Chrysanthemum | regulation of early flowering | [121] |
| 31 | AGAMOUS (silencing) | Chrysanthemum | alteration of floral shape | [122] |
| 31 | CmCYC2c | Chrysanthemum | enhanced ray floret length and number of flowers | [124] |
| 32 | Cyc2CL-1 and Cyc2CL-2 | Chrysanthemum | stamen and ray floret development | [125] |
| 33 | CmTCP20 | Chrysanthemum | petal elongation | [127] |
| 34 | CmYAB1 | Chrysanthemum | reduction in petal curvature for flat petals | [129] |
| 35 | PGIP | Prunus mumei | tolerance to Alternaria leaf spot | [135] |
| 36 | hpaGX00 | Xanthomonas oryzae pv.oryzae | tolerance to Alternaria tenuissima | [136] |
| 37 | chiII | Rice | tolerance to leaf spot caused by Septoria obesa | [137] |
| 38 | CmNPR1 | Chrysanthemum | tolerance to black spot | [138] |
| 39 | CmMLO17+CmKIC | Chrysanthemum | support the fungal growth of Alternata alternata | [139] |
| 40 | RCC2 | Rice | tolerance to gray mold caused Botrytis cinerea | [140] |
| 41 | CaXMT1, CaMXMT1 and CaDXMT1 | Coffea arabica | tolerance to B.cinerea | [141] |
| 42 | sarcotoxin IA+Cry1Ab | Sarcophaga peregrine and Bacillus thuringiensis | tolerance to white rust and Helicoverpa armigera | [142] |
| 43 | CmWRKY15-1 | Chrysanthemum | white rust resistance | [143] |
| 44 | δ-endotoxin | Bacillus thuringiensis | tolerance to Helicoverpa armigera | [146] |
| 45 | CmWRKY53 | Chrysanthemum | susceptibility to aphids | [138] |
| 46 | nucleocapsid (N) | Virus | resistance to TSWV | [150] |
| 47 | coat protein (CP) | Virus | resitance to CMV | [151] |
| 48 | coat protein (CP) | CVB virus | resistance to chrysanthemum virus B | [152] |
| 49 | CSVd | Stunt Viroid | tolerance to CSVd | [154] |
| 50 | AtDREB1A | Arabidopsis | enhanced heat tolerance | [158] |
| 51 | CmDREB6 | Chrysanthemum | enhanced heat tolerance | [159] |
| 52 | CmCPL1 | Chrysanthemum | enhanced heat tolerance | [160] |
| 53 | CmHSP90.5 | Chrysanthemum | sensitivity to heat stress | [162] |
| 54 | AtDREB1A | Arabidopsis | tolerance to low temperature | [179] |
| 55 | DgTIL1 | Chrysanthemum | enhanced tolerance to cold stress | [164] |
| 56 | DgGPX1 | Chrysanthemum | enhanced tolerance to cold stress | [166] |
| 57 | DgMYB2 | Chrysanthemum | improved tolerance to cold stress | [167] |
| 58 | CnTCP4 | Chrysanthemum nankingense | hypersensitivity to cold stress | [168] |
| 59 | CcSOS1 | Chrysanthemum crissum | tolerance to salt stress | [169] |
| 60 | CmHSF4 | Chrysanthemum | tolerance to salt stress | [183] |
| 61 | DgWRKY4 and DGWRKY5 | Chrysanthemum | tolerance to salt stress | [172,174] |
| 62 | CmWRKY17 | Chrysanthemum | negative regulation of salt stress | [173] |
| 63 | DgNAC1 | Chrysanthemum | improved tolerance to salt stress | [174] |
| 64 | Aquaporin | Chrysanthemum | tolerance to salt stress | [175] |
| 65 | DREB1A | Arabidopsis | tolerance to salt and water deficiency stress | [179] |
| 66 | CgDREB1A | Chrysanthemum | enhanced tolerance to drought | [180] |
| 67 | CdICE1 | Chrysanthemum dichrum | tolerance to drought, salinity and low temperature stress | [181] |
| 68 | CmWRKY1 and CmWRKY10 | Chrysanthemum | tolerance to drought | [184,185] |
| 69 | ClCBF1 | Chrysanthemum lavandulifolium | tolerance to drought and salt stress | [186] |
| 70 | CmERF053 | Chrysanthemum | tolerance to drought stress | [187] |
| 71 | DgNAC1 | Chrysanthemum | tolerance to drought stress | [188] |
| 72 | ClNAC9 | Chrysanthemum lavandulifolium | tolerance to drought, salinity, and alkaline stress | [189] |
| 73 | CmBBX19 (silencing) | Chrysanthemum | tolerance to drought stress | [190] |
| 74 | CmNF-YB8 | Chrysanthemum | negative influence of drought stress | [191] |
| 75 | CmPLDα | Chrysanthemum | tolerance to drought stress | [192] |
| 76 | CgbZIP1 | Chrysanthemum | enhanced tolerance to drought and salt stress | [194] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mekapogu, M.; Kwon, O.-K.; Song, H.-Y.; Jung, J.-A. Towards the Improvement of Ornamental Attributes in Chrysanthemum: Recent Progress in Biotechnological Advances. Int. J. Mol. Sci. 2022, 23, 12284. https://doi.org/10.3390/ijms232012284
Mekapogu M, Kwon O-K, Song H-Y, Jung J-A. Towards the Improvement of Ornamental Attributes in Chrysanthemum: Recent Progress in Biotechnological Advances. International Journal of Molecular Sciences. 2022; 23(20):12284. https://doi.org/10.3390/ijms232012284
Chicago/Turabian StyleMekapogu, Manjulatha, Oh-Keun Kwon, Hyun-Young Song, and Jae-A Jung. 2022. "Towards the Improvement of Ornamental Attributes in Chrysanthemum: Recent Progress in Biotechnological Advances" International Journal of Molecular Sciences 23, no. 20: 12284. https://doi.org/10.3390/ijms232012284
APA StyleMekapogu, M., Kwon, O.-K., Song, H.-Y., & Jung, J.-A. (2022). Towards the Improvement of Ornamental Attributes in Chrysanthemum: Recent Progress in Biotechnological Advances. International Journal of Molecular Sciences, 23(20), 12284. https://doi.org/10.3390/ijms232012284





