Chronic Arsenic Exposure Upregulates the Expression of Basal Transcriptional Factors and Increases Invasiveness of the Non-Muscle Invasive Papillary Bladder Cancer Line RT4
Abstract
1. Introduction
2. Results
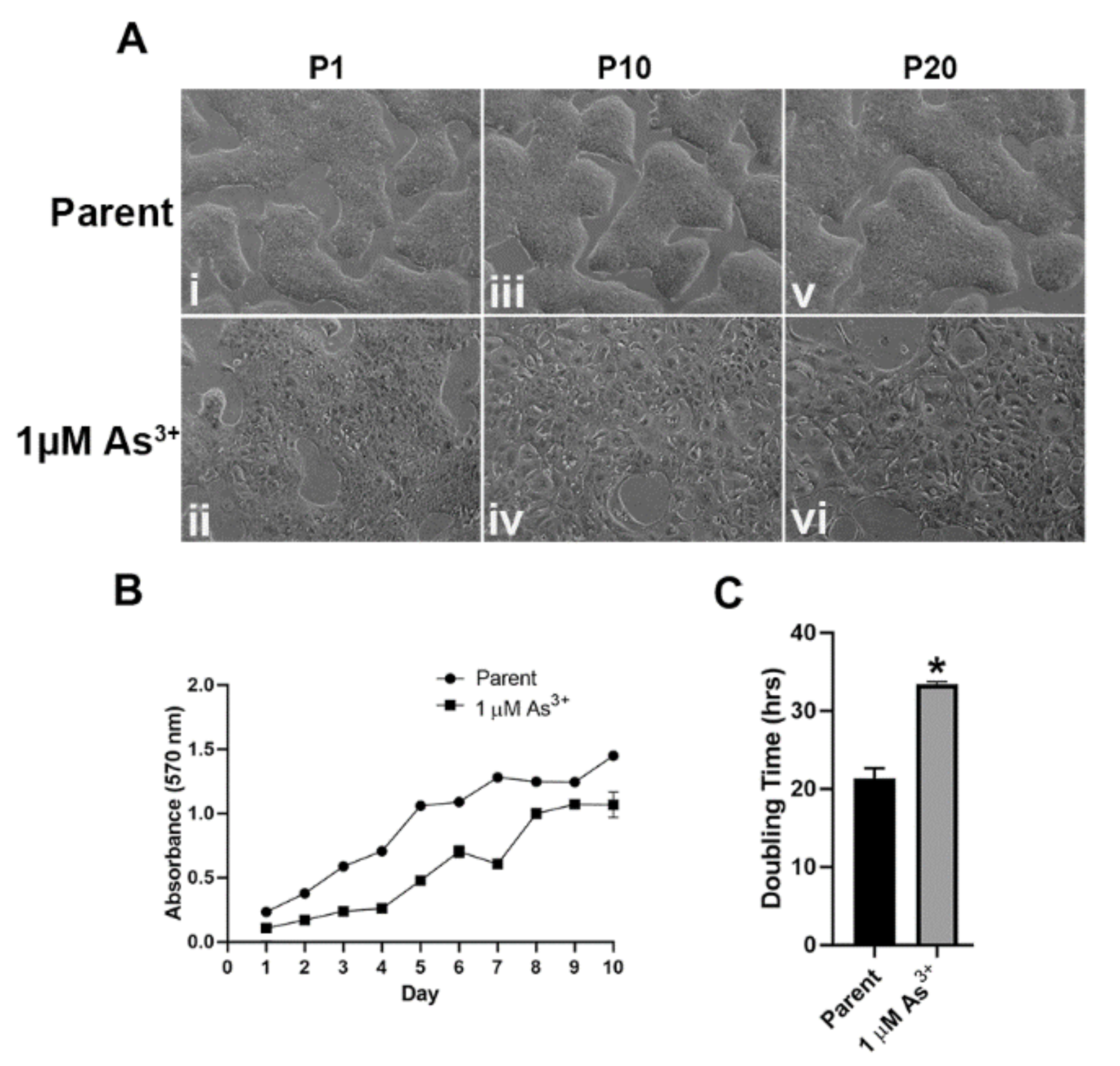
2.1. Morphology and Growth Rates of As3+-Exposed RT4 Cells
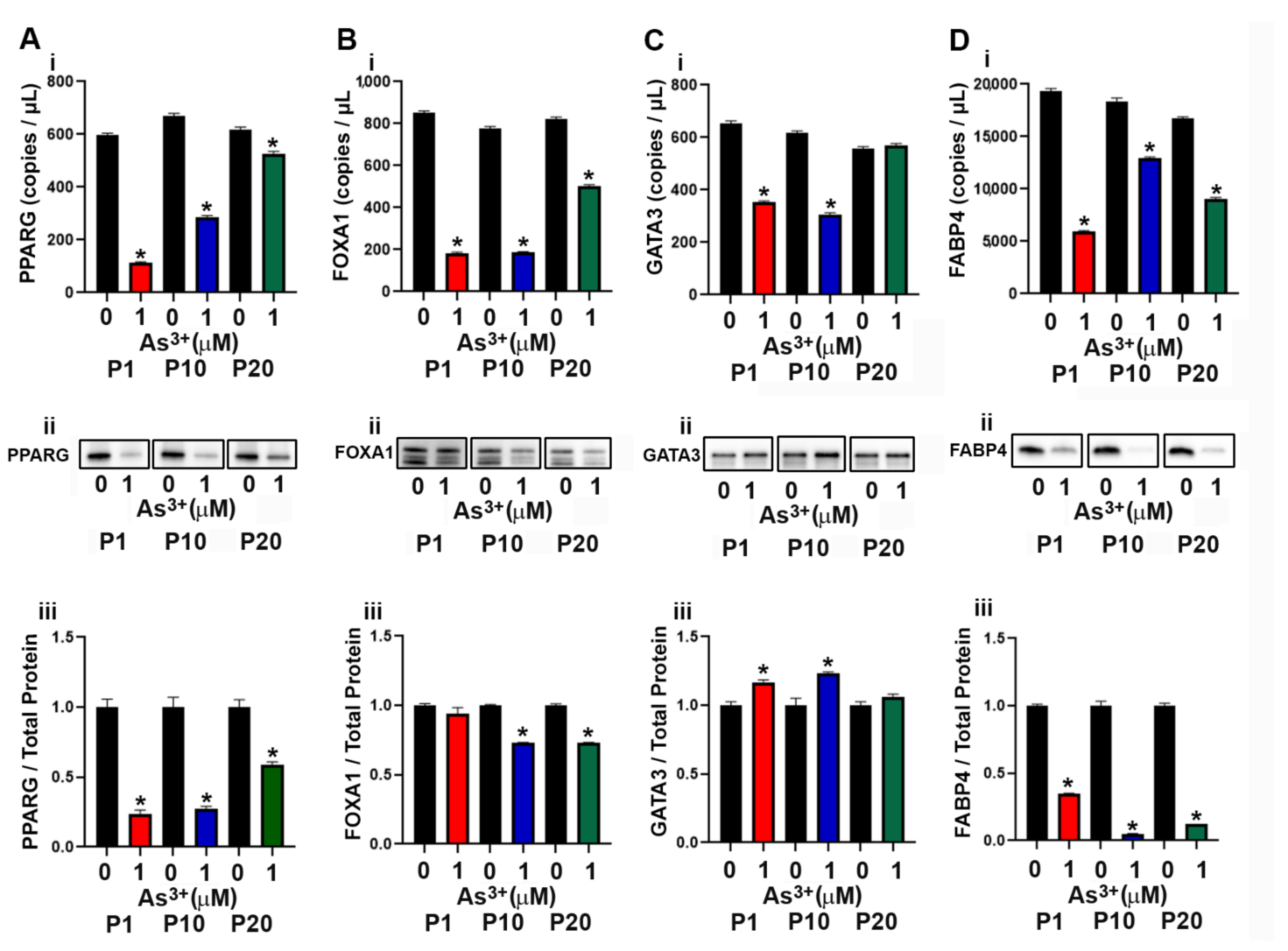
2.2. Expression of Luminal Genes and Proteins in As3+ Exposed RT4 Cells
2.3. Expression of Basal Genes and Proteins in As3+ Exposed RT4 Cells
2.4. Effect of As3+ Exposure on the Expression of Epithelial to Mesenchymal Transition (EMT) Genes and Proteins in RT4 Cells
2.5. Histology of the As3+-Exposed Tumor Heterotransplants
2.6. Immunohistochemical Analysis of As3+-Exposed Tumor Heterotransplants
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Cell Culture
4.3. Cell growth
4.4. RNA Isolation and Droplet Digital PCR Analysis
4.5. Western Blot Analysis
4.6. Immunohistochemical Staining
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Cancer Society. Key Statistics for Bladder Cancer. 2020. Available online: https://www.cancer.org/cancer/bladder-cancer/about/key-statistics.html (accessed on 30 April 2020).
- Kuper, H.; Boffetta, P.; Adami, H.-O. Tobacco use and cancer causation: Association by tumour type. J. Intern. Med. 2002, 252, 206–224. [Google Scholar] [CrossRef] [PubMed]
- Zeegers, M.P.A.; Kellen, E.; Buntinx, F.; van den Brandt, P.A. The association between smoking, beverage consumption, diet and bladder cancer: A systematic literature review. World J. Urol. 2003, 21, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Shariat, S.F.; Milowsky, M.; Droller, M.J. Bladder cancer in the elderly. Urol. Oncol. 2009, 27, 653–667. [Google Scholar] [CrossRef]
- Volanis, D.; Kadiyska, T.; Galanis, A.; Delakas, D.; Logotheti, S.; Zoumpourlis, V. Environmental factors and genetic susceptibility promote urinary bladder cancer. Toxicol. Lett. 2010, 193, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Bryan, R.T. Cell adhesion and urothelial bladder cancer: The role of cadherin switching and related phenomena. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140042. [Google Scholar] [CrossRef] [PubMed]
- Balci, M.G.; Tayfur, M. Loss of E-cadherin expression in recurrent non-invasive urothelial carcinoma of the bladder. Int. J. Clin. Exp. Pathol. 2018, 11, 4163–4168. [Google Scholar] [PubMed]
- Guo, C.C.; Czerniak, B. Bladder Cancer in the Genomic Era. Arch. Pathol. Lab. Med. 2019, 143, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.-L.; et al. Identification of Distinct Basal and Luminal Subtypes of Muscle-Invasive Bladder Cancer with Different Sensitivities to Frontline Chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Satyal, U.; Sikder, R.K.; McConkey, D.; Plimack, E.R.; Abbosh, P.H. Clinical implications of molecular subtyping in bladder cancer. Curr. Opin. Urol. 2019, 29, 350–356. [Google Scholar] [CrossRef]
- Cantor, K.P.; Lubin, J.H. Arsenic, internal cancers, and issues in inference from studies of low-level exposures in human popula-tions. Toxicol. Appl. Pharmacol. 2007, 222, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Akbani, R.; Broom, B.M.; Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar]
- Stewart, S.L.; Cardinez, C.J.; Richardson, L.C.; Norman, L.; Kaufmann, R.; Pechacek, T.F.; Thompson, T.D.; Weir, H.K.; A Sabatino, S. Surveillance for cancers associated with tobacco use—United States, 1999–2004. MMWR. Surveill. Summ. 2008, 57, 1–42. [Google Scholar] [PubMed]
- Iscovich, J.; Castelletto, R.; Estè, J.; Muñoz, N.; Colanzi, R.; Coronel, A.; Deamezola, I.; Tassi, V.; Arslan, A. Tobacco smoking, occupational exposure and bladder cancer in Argentina. Int. J. Cancer 1987, 40, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.C.; Skipper, P.L.; Tannenbaum, S.R.; Chan, K.K.; Ross, R.K. Arylamine exposures and bladder cancer risk. Mutat Res. 2002, 506, 21–28. [Google Scholar] [CrossRef]
- Snyderwine, E.G.; Sinha, R.; Felton, J.S.; Ferguson, L.R. Highlights of the Eighth International Conference on Carcinogenic/Mutagenic N-Substituted Aryl Compounds. Mutat. Res. Mol. Mech. Mutagen. 2002, 506, 1–8. [Google Scholar] [CrossRef]
- García-Pérez, J.; Pollán, M.; Boldo, E.; Pérez-Gómez, B.; Aragonés, N.; Lope, V.; Ramis, R.; Vidal, E.; López-Abente, G. Mortality due to lung, laryngeal and bladder cancer in towns lying in the vicinity of combustion installations. Sci. Total Environ. 2009, 407, 2593–2602. [Google Scholar] [CrossRef]
- Chen, H.I.; Liou, S.H.; Loh, C.H.; Uang, S.N.; Yu, Y.C.; Shih, T.S. Bladder cancer screening and monitoring of 4,4′-methylenebis(2-chloroaniline) exposure among workers in Taiwan. Urology 2005, 66, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Case, R.A.M.; Hosker, M.E. Tumour of the Urinary Bladder as an Occupational Disease in the Rubber Industry in England and Wales. J. Epidemiol. Community Health 1954, 8, 39–50. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Golka, K.; Wiese, A.; Assennato, G.; Bolt, H.M. Occupational exposure and urological cancer. World J. Urol. 2003, 21, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Gago-Dominguez, M.; Castelao, J.E.; Yuan, J.-M.; Yu, M.C.; Ross, R.K. Use of permanent hair dyes and bladder-cancer risk. Int. J. Cancer 2000, 91, 575–579. [Google Scholar] [CrossRef]
- Chung, J.-Y.; Yu, S.-D.; Hong, Y.-S. Environmental Source of Arsenic Exposure. J. Prev. Med. Public Health 2014, 47, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Kuo, T.L.; Wu, M.M. Arsenic and cancers. Lancet 1988, 1, 414–415. [Google Scholar] [CrossRef]
- Chen, C.J.; Chuang, Y.C.; Lin, T.M.; Wu, H.Y. Malignant neoplasms among residents of a blackfoot disease-endemic area in Taiwan: High-arsenic artesian well water and cancers. Cancer Res. 1985, 45, 5895–5899. [Google Scholar] [PubMed]
- Chen, C.-J.; Wu, M.-M.; Kuo, T.-L. Cancer potential in liver, lung, bladder and kidney due to ingested inorganic arsenic in drinking water. Br. J. Cancer 1992, 66, 888–892. [Google Scholar] [CrossRef]
- Smith, A.H.; Goycolea, M.; Haque, R.; Biggs, M.L. Marked Increase in Bladder and Lung Cancer Mortality in a Region of Northern Chile Due to Arsenic in Drinking Water. Am. J. Epidemiol. 1998, 147, 660–669. [Google Scholar] [CrossRef]
- Hopenhayn-Rich, C.; Biggs, M.L.; Fuchs, A.; Bergoglio, R.; Tello, E.E.; Nicolli, H.; Smith, A.H. Bladder Cancer Mortality Associated with Arsenic in Drinking Water in Argentina. Epidemiology 1996, 7, 117–124. [Google Scholar] [CrossRef]
- Hopenhayn-Rich, C.; Biggs, M.L.; Smith, A.H. Lung and kidney cancer mortality associated with arsenic in drinking water in Cordoba, Argentina. Int. J. Epidemiol. 1998, 27, 561–569. [Google Scholar] [CrossRef]
- Somji, S.; Zhou, X.D.; Garrett, S.H.; Sens, M.A.; Sens, D.A. Urothelial Cells Malignantly Transformed by Exposure to Cadmium (Cd+2) and Arsenite (As+3) Have Increased Resistance to Cd+2 and As+3-Induced Cell Death. Toxicol. Sci. 2006, 94, 293–301. [Google Scholar] [CrossRef]
- Cao, L.; Zhou, X.D.; Sens, M.A.; Garrett, S.H.; Zheng, Y.; Dunlevy, J.R.; Sens, D.A.; Somji, S. Keratin 6 expression correlates to areas of squamous dif-ferentiation in multiple independent isolates of As+3-induced bladder cancer. J. Appl. Toxicol. JAT 2010, 30, 416–430. [Google Scholar]
- Minato, A.; Fujimoto, N.; Kubo, T. Squamous Differentiation Predicts Poor Response to Cisplatin-Based Chemotherapy and Un-favorable Prognosis in Urothelial Carcinoma of the Urinary Bladder. Clin. Genitourin. Cancer 2017, 15, e1063–e1067. [Google Scholar] [CrossRef]
- Li, G.; Yu, J.; Song, H.; Zhu, S.; Sun, L.; Shang, Z.; Niu, Y. Squamous differentiation in patients with superficial bladder urothelial car-cinoma is associated with high risk of recurrence and poor survival. BMC Cancer 2017, 17, 530. [Google Scholar] [CrossRef] [PubMed]
- Hoggarth, Z.E.; Osowski, D.B.; Freeberg, B.A.; Garrett, S.H.; Sens, D.A.; Sens, M.A.; Zhou, X.D.; Zhang, K.K.; Somji, S. The urothelial cell line UROtsa transformed by arsenite and cadmium display basal characteristics associated with muscle invasive urothelial cancers. PLoS ONE 2018, 13, e0207877. [Google Scholar] [CrossRef] [PubMed]
- Danes, J.M.; Abreu, A.L.P.; Kerketta, R.; Huang, Y.; Palma, F.R.; Gantner, B.N.; Mathison, A.J.; Urrutia, R.A.; Bonini, M.G. Inorganic arsenic promotes luminal to basal transition and metastasis of breast cancer. FASEB J. 2020, 34, 16034–16048. [Google Scholar] [CrossRef] [PubMed]
- Zuiverloon, T.C.; de Jong, F.C.; Costello, J.C.; Theodorescu, D. Systematic Review: Characteristics and Preclinical Uses of Bladder Cancer Cell Lines. Bl. Cancer 2018, 4, 169–183. [Google Scholar] [CrossRef]
- Crallan, R.; Georgopoulos, N.; Southgate, J. Experimental models of human bladder carcinogenesis. Carcinogenesis 2005, 27, 374–381. [Google Scholar] [CrossRef]
- Warrick, J.I.; Walter, V.; Yamashita, H.; Shuman, L.; Amponsa, V.O.; Zheng, Z.; Chan, W.; Whitcomb, T.L.; Yue, F.; Iyyanki, T.; et al. FOXA1, GATA3 and PPARɣ Cooperate to Drive Luminal Subtype in Bladder Cancer: A Molecular Analysis of Established Human Cell Lines. Sci. Rep. 2016, 6, 38531. [Google Scholar] [CrossRef]
- Fishwick, C.; Higgins, J.; Percival-Alwyn, L.; Hustler, A.; Pearson, J.; Bastkowski, S.; Moxon, S.; Swarbreck, D.; Greenman, C.D.; Southgate, J. Heterarchy of transcription factors driving basal and luminal cell phenotypes in human urothelium. Cell Death Differ. 2017, 24, 809–818. [Google Scholar] [CrossRef]
- Yadav, S.; Anbalagan, M.; Shi, Y.; Wang, F.; Wang, H. Arsenic inhibits the adipogenic differentiation of mesenchymal stem cells by down-regulating peroxisome proliferator-activated receptor gamma and CCAAT enhancer-binding proteins. Toxicol. In Vitro 2013, 27, 211–219. [Google Scholar] [CrossRef]
- Boiteux, G.; Lascombe, I.; Roche, E.; Plissonnier, M.-L.; Clairotte, A.; Bittard, H.; Fauconnet, S. A-FABP, a candidate progression marker of human transitional cell carcinoma of the bladder, is differentially regulated by PPAR in urothelial cancer cells. Int. J. Cancer 2009, 124, 1820–1828. [Google Scholar] [CrossRef]
- Varley, C.L.; Stahlschmidt, J.; Smith, B.; Stower, M.; Southgate, J. Activation of peroxisome proliferator-activated receptor-gamma reverses squamous metaplasia and induces transitional differentiation in normal human urothelial cells. Am. J. Pathol. 2004, 164, 1789–1798. [Google Scholar] [CrossRef]
- Karni-Schmidt, O.; Castillo-Martin, M.; Shen, T.H.; Gladoun, N.; Domingo-Domenech, J.; Sanchez-Carbayo, M.; Li, Y.; Lowe, S.; Prives, C.; Cordon-Cardo, C. Distinct ex-pression profiles of p63 variants during urothelial development and bladder cancer progression. Am. J. Pathol. 2011, 178, 1350–1360. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Kawasawa, Y.I.; Shuman, L.; Zheng, Z.; Tran, T.; Walter, V.; Warrick, J.I.; Chen, G.; Al-Ahmadie, H.; Kaag, M.; et al. Repression of transcription factor AP-2 alpha by PPARγ reveals a novel transcriptional circuit in basal-squamous bladder cancer. Oncogenesis 2019, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Palmbos, P.L.; Wang, Y.; Iii, A.B.; Kelleher, A.J.; Wang, L.; Yang, H.; Ahmet, M.L.; Gumkowski, E.R.; Welling, S.D.; Magnuson, B.; et al. ATDC mediates a TP63-regulated basal cancer invasive program. Oncogene 2019, 38, 3340–3354. [Google Scholar] [CrossRef]
- Rigby, C.C.; Franks, L.M. A Human Tissue Culture Cell Line from a Transitional Cell Tumour of the Urinary Bladder: Growth, Chromosome Pattern and Ultrastructure. Br. J. Cancer 1970, 24, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Czerniak, B.; Dinney, C.; McConkey, D. Origins of Bladder Cancer. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 149–174. [Google Scholar] [CrossRef]
- Bryan, R.T.; Tselepis, C. Cadherin Switching and Bladder Cancer. J. Urol. 2010, 184, 423–431. [Google Scholar] [CrossRef]
- Bryan, R.; Atherfold, P.; Yeo, Y.; Jones, L.; Harrison, R.; Wallace, D.; Jankowski, J. Cadherin switching dictates the biology of transitional cell carcinoma of the bladder: Ex vivo and in vitro studies. J. Pathol. 2008, 215, 184–194. [Google Scholar] [CrossRef]
- Schulte, J.; Weidig, M.; Balzer, P.; Richter, P.; Franz, M.; Junker, K.; Gajda, M.; Friedrich, K.; Wunderlich, H.; Östman, A.; et al. Expression of the E-cadherin repressors Snail, Slug and Zeb1 in urothelial carcinoma of the urinary bladder: Relation to stromal fibroblast activation and invasive behaviour of carcinoma cells. Histochem. Cell Biol. 2012, 138, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, N.A.; Neilson, E.G.; Moses, H.L. Stromal fibroblasts in cancer initiation and progression. Nature 2004, 432, 332–337. [Google Scholar] [CrossRef]
- Brun, R.P.; Tontonoz, P.; Forman, B.M.; Ellis, R.; Chen, J.; Evans, R.M.; Spiegelman, B.M. Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 1996, 10, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zhang, Y.; Davis, L.; Breyer, M.D. Expression of peroxisome proliferator-activated receptors in urinary tract of rabbits and humans. Am. J. Physiol. Physiol. 1997, 273, F1013–F1022. [Google Scholar] [CrossRef]
- Cheng, S.; Wang, G.; Wang, Y.; Cai, L.; Qian, K.; Ju, L.; Liu, X.; Xiao, Y.; Wang, X. Fatty acid oxidation inhibitor etomoxir suppresses tumor progression and induces cell cycle arrest via PPARγ-mediated pathway in bladder cancer. Clin. Sci. 2019, 133, 1745–1758. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Qian, K.; Wang, Y.; Wang, G.; Liu, X.; Xiao, Y.; Wang, X. PPARγ inhibition regulates the cell cycle, proliferation and motility of bladder cancer cells. J. Cell. Mol. Med. 2019, 23, 3724–3736. [Google Scholar] [CrossRef]
- Cao, R.; Wang, G.; Qian, K.; Chen, L.; Qian, G.; Xie, C.; Dan, H.C.; Jiang, W.; Wu, M.; Wu, C.L.; et al. Silencing of HJURP induces dysregulation of cell cycle and ROS me-tabolism in bladder cancer cells via PPARγ-SIRT1 feedback loop. J. Cancer 2017, 8, 2282–2295. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Wang, G.; Cheng, S.; Xiong, Y.; Cao, R.; Qian, K.; Ju, L.; Wang, X.; Xiao, Y. The role and function of PPARγ in bladder cancer. J. Cancer 2020, 11, 3965–3975. [Google Scholar] [CrossRef] [PubMed]
- Mylona, E.; Giannopoulou, I.; Diamantopoulou, K.; Bakarakos, P.; Nomikos, A.; Zervas, A.; Nakopoulou, L. Peroxisome proliferator-activated receptor gamma expression in urothelial carcinomas of the bladder: Association with differentiation, proliferation and clinical outcome. Eur. J. Surg. Oncol. EJSO 2009, 35, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Osei-Amponsa, V.; Buckwalter, J.M.; Shuman, L.; Zheng, Z.; Yamashita, H.; Walter, V.; Wildermuth, T.; Ellis-Mohl, J.; Liu, C.; Warrick, J.I.; et al. Hypermethylation of FOXA1 and allelic loss of PTEN drive squamous differentiation and promote heterogeneity in bladder cancer. Oncogene 2019, 39, 1302–1317. [Google Scholar] [CrossRef]
- DeGraff, D.J.; Clark, P.E.; Cates, J.M.; Yamashita, H.; Robinson, V.L.; Yu, X.; Smolkin, M.E.; Chang, S.S.; Cookson, M.S.; Herrick, M.K.; et al. Loss of the Urothelial Differentiation Marker FOXA1 Is Associated with High Grade, Late Stage Bladder Cancer and Increased Tumor Proliferation. PLoS ONE 2012, 7, e36669. [Google Scholar]
- Wang, C.C.; Tsai, Y.C.; Jeng, Y.M. Biological significance of GATA3, cytokeratin 20, cytokeratin 5/6 and p53 expression in mus-cle-invasive bladder cancer. PLoS ONE 2019, 14, e0221785. [Google Scholar]
- Wauson, E.M.; Langan, A.S.; Vorce, R.L. Sodium arsenite inhibits and reverses expression of adipogenic and fat cell-specific genes during in vitro adipogenesis. Toxicol. Sci. 2002, 65, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Volkmer, J.-P.; Sahoo, D.; Chin, R.K.; Ho, P.L.; Tang, C.; Kurtova, A.V.; Willingham, S.B.; Pazhanisamy, S.K.; Contreras-Trujillo, H.; Storm, T.A.; et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc. Natl. Acad. Sci. USA 2012, 109, 2078–2083. [Google Scholar] [CrossRef] [PubMed]
- Bellmunt, J.; Hussain, M.; Dinney, C. Novel approaches with targeted therapies in bladder cancer: Therapy of bladder cancer by blockade of the epidermal growth factor receptor family. Crit. Rev. Oncol. 2003, 46, 85–104. [Google Scholar] [CrossRef]
- Rebouissou, S.; Bernard-Pierrot, I.; de Reyniès, A.; Lepage, M.-L.; Krucker, C.; Chapeaublanc, E.; Hérault, A.; Kamoun, A.; Caillault, A.; Letouzé, E.; et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci. Transl. Med. 2014, 6, 244ra91. [Google Scholar] [CrossRef] [PubMed]
- Somji, S.; Zhou, X.D.; Mehus, A.; Sens, M.A.; Garrett, S.H.; Lutz, K.L.; Dunlevy, J.R.; Zheng, Y.; Sens, D.A. Variation of Keratin 7 Expression and Other Phenotypic Characteristics of Independent Isolates of Cadmium Transformed Human Urothelial Cells (UROtsa). Chem. Res. Toxicol. 2009, 23, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Denizot, F.; Lang, R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]
- Garrett, S.H.; Somji, S.; Todd, J.H.; Sens, M.A.; Sens, D.A. Differential expression of human metallothionein isoform I mRNA in human proximal tubule cells exposed to metals. Environ. Health Perspect. 1998, 106, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Mehus, A.A.; Bergum, N.; Knutson, P.; Shrestha, S.; Zhou, X.D.; Garrett, S.H.; Sens, D.A.; Sens, M.A.; Somji, S. Activation of PPARγ and inhibition of cell prolif-eration reduces key proteins associated with the basal subtype of bladder cancer in As3+-transformed UROtsa cells. PLoS ONE 2020, 15, e0237976. [Google Scholar] [CrossRef] [PubMed]
- Janes, K.A. An analysis of critical factors for quantitative immunoblotting. Sci. Signal. 2015, 8, rs2. [Google Scholar] [CrossRef]





| Parent | 1 µM As3+ | |||
|---|---|---|---|---|
| Protein | INT | % | INT | % |
| PPARγ | 3+ | 60 | 1+ | 20 |
| FOXA1 | 1+ | 35 | 2+ | 60 |
| GATA3 | 1+ | 50 | 2+ | 60 |
| EGFR | 1+ | 10 | 3+ | 80 |
| TFAP2A | 0 | 0 | 1+ | 60 |
| P63 | 2+ | 30 | 3+ | 80 |
| E-Cadherin | 3+ | 90 | 3+ | 90 |
| N-Cadherin | 0 | 0 | 1+ | 10 |
| ACTA2 | 1+ | 10 | 1+ | 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehus, A.A.; Bergum, N.; Knutson, P.; Shrestha, S.; Kalonick, M.; Zhou, X.; Garrett, S.H.; Sens, D.A.; Sens, M.A.; Somji, S. Chronic Arsenic Exposure Upregulates the Expression of Basal Transcriptional Factors and Increases Invasiveness of the Non-Muscle Invasive Papillary Bladder Cancer Line RT4. Int. J. Mol. Sci. 2022, 23, 12313. https://doi.org/10.3390/ijms232012313
Mehus AA, Bergum N, Knutson P, Shrestha S, Kalonick M, Zhou X, Garrett SH, Sens DA, Sens MA, Somji S. Chronic Arsenic Exposure Upregulates the Expression of Basal Transcriptional Factors and Increases Invasiveness of the Non-Muscle Invasive Papillary Bladder Cancer Line RT4. International Journal of Molecular Sciences. 2022; 23(20):12313. https://doi.org/10.3390/ijms232012313
Chicago/Turabian StyleMehus, Aaron A., Nicholas Bergum, Peter Knutson, Swojani Shrestha, Matthew Kalonick, Xudong Zhou, Scott H. Garrett, Donald A. Sens, Mary Ann Sens, and Seema Somji. 2022. "Chronic Arsenic Exposure Upregulates the Expression of Basal Transcriptional Factors and Increases Invasiveness of the Non-Muscle Invasive Papillary Bladder Cancer Line RT4" International Journal of Molecular Sciences 23, no. 20: 12313. https://doi.org/10.3390/ijms232012313
APA StyleMehus, A. A., Bergum, N., Knutson, P., Shrestha, S., Kalonick, M., Zhou, X., Garrett, S. H., Sens, D. A., Sens, M. A., & Somji, S. (2022). Chronic Arsenic Exposure Upregulates the Expression of Basal Transcriptional Factors and Increases Invasiveness of the Non-Muscle Invasive Papillary Bladder Cancer Line RT4. International Journal of Molecular Sciences, 23(20), 12313. https://doi.org/10.3390/ijms232012313





