A Perspective on the (Rise and Fall of) Protein β-Turns
Abstract
:1. Forewords
2. The Alpha and the Omega of the β
2.1. Superimposition of the β-Turns with Other Regular Structures
2.2. Superimposition of the Assignment with Other β-Turns
2.3. With or without Hydrogen Bonds
2.4. The Mess for Types (and Some Types Are for Nothing)
2.5. Only One Ancient Tool to Assign
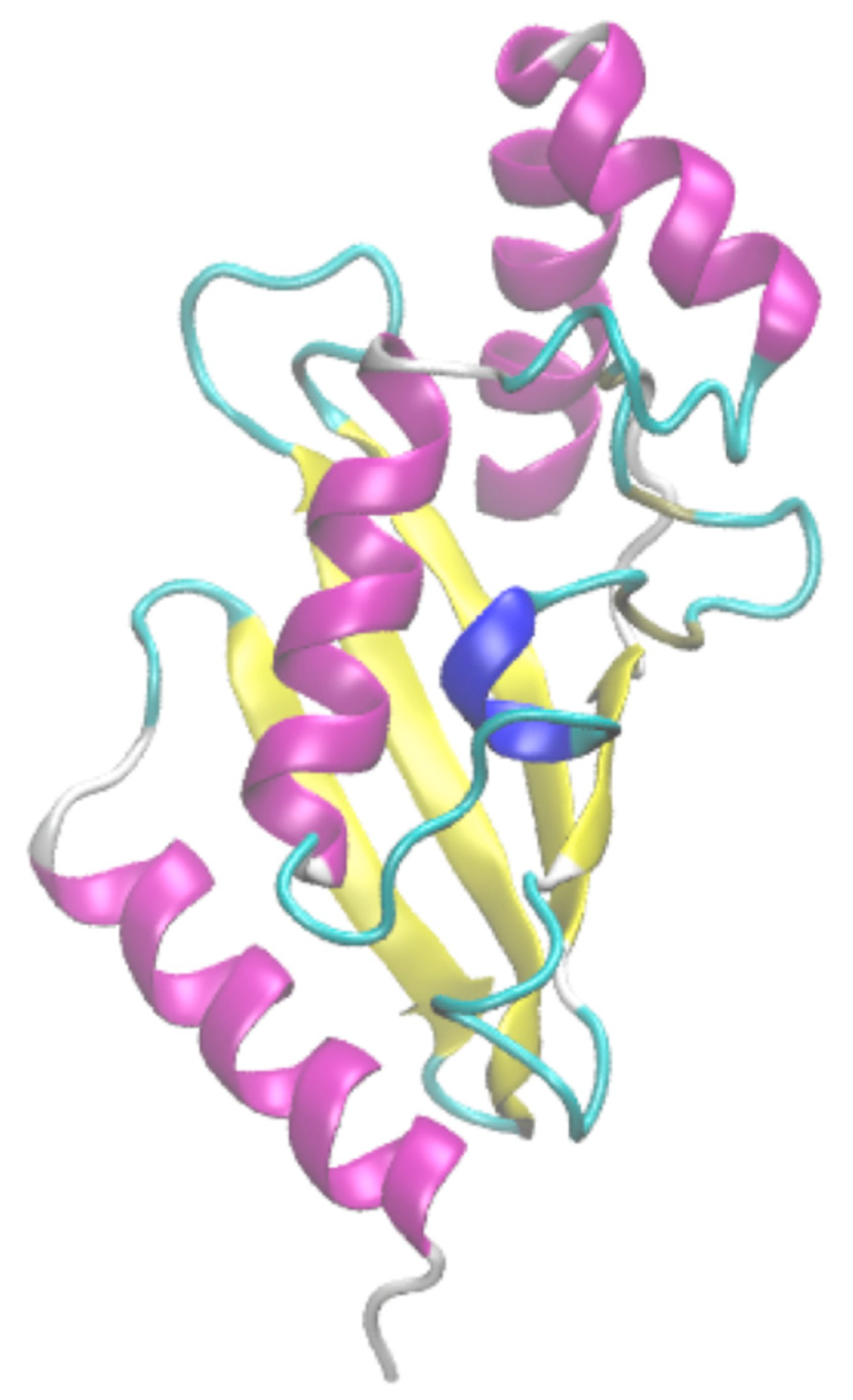
2.6. No (Easy) Specific Visualisation
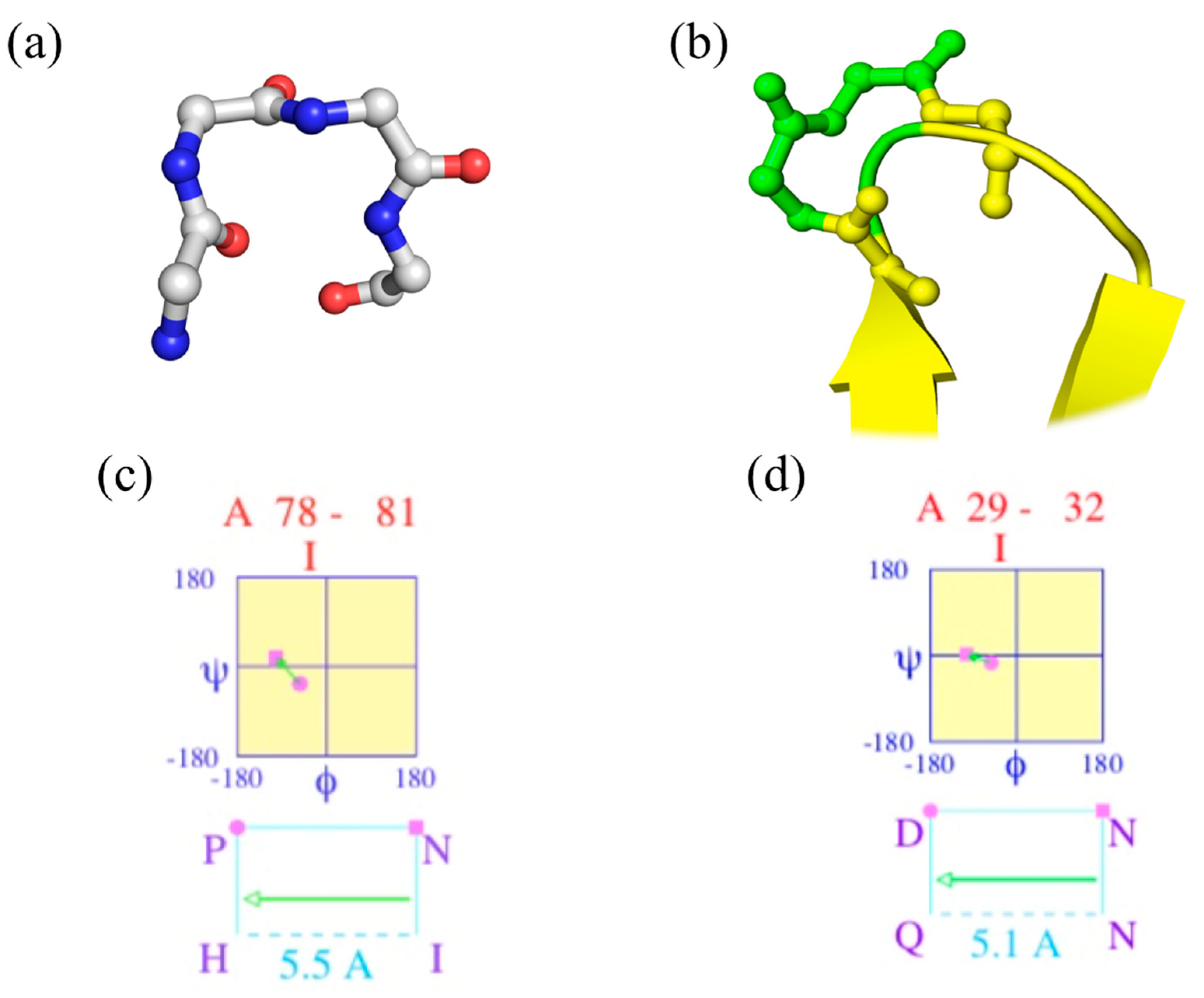
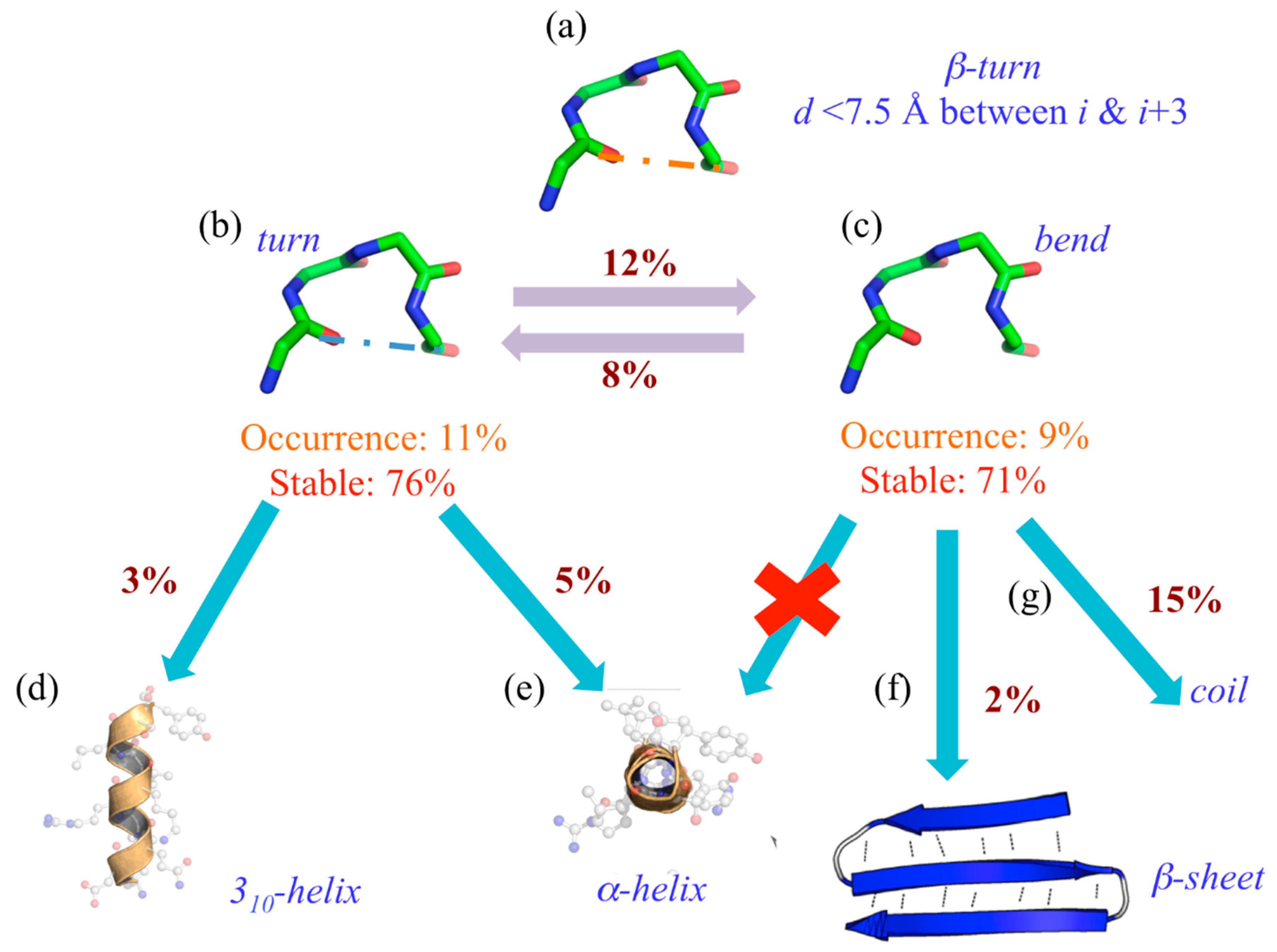
2.7. Confusion with Other Local Protein Structures
2.8. New Classifications
3. Conclusions and At Least Some Perspectives
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, G.; Jia, R.; Liang, D.; Yu, J.; Wu, Z.; Chen, C. Effects of music therapy on anxiety: A meta-analysis of randomized controlled trials. Psychiatry Res. 2021, 304, 114137. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; He, Z.; Shen, Z.; Huang, F. Potential benefits of music therapy on stroke rehabilitation. Oxidative Med. Cell. Longev. 2022, 2022, 9386095. [Google Scholar] [CrossRef] [PubMed]
- Eisenhaber, F.; Verma, C.; Blundell, T. In memoriam of narayanaswamy srinivasan (1962–2021). Proteins 2022, 90, 909–911. [Google Scholar] [CrossRef]
- Gromiha, M.M.; Orengo, C.; Sowdhamini, R.; Thornton, J. Srinivasan (1962–2021) in bioinformatics and beyond. Bioinformatics 2022, 38, 2377–2379. [Google Scholar] [CrossRef] [PubMed]
- Sowdhamini, R. Biography of a scientist with strength, substance, sincerity and service: Late n. Srinivasan (1962–2021). Bioinformation 2022, 18, 600–604. [Google Scholar] [CrossRef]
- Varadarajan, R.N. Srinivasan (1962–2021). Curr. Sci. 2021, 121, 1252–1253. [Google Scholar]
- Jagger, M.; Richards, K. Paint it black. In Rolling Stones—Aftermath. 1966. [Google Scholar]
- Venkatachalam, C.M. Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units. Biopolymers 1968, 6, 1425–1436. [Google Scholar] [CrossRef] [Green Version]
- Lewis, P.N.; Momany, F.A.; Scheraga, H.A. Chain reversals in proteins. Biochim. Biophys. Acta 1973, 303, 211–229. [Google Scholar] [CrossRef]
- Rose, G.D.; Seltzer, J.P. A new algorithm for finding the peptide chain turns in a globular protein. J. Mol. Biol. 1977, 113, 153–164. [Google Scholar] [CrossRef]
- Bornot, A.; de Brevern, A.G. Protein beta-turn assignments. Bioinformation 2006, 1, 153–155. [Google Scholar] [CrossRef] [Green Version]
- Matthews, B.W. The gamma-turn. Evidence for a new folded conformation in proteins. Macromolecules 1972, 5, 818–819. [Google Scholar] [CrossRef]
- Pavone, V.; Gaeta, G.; Lombardi, A.; Nastri, F.; Maglio, O.; Isernia, C.; Saviano, M. Discovering protein secondary structures: Classification and description of isolated alpha-turns. Biopolymers 1996, 38, 705–721. [Google Scholar] [CrossRef]
- Rose, G.D.; Gierasch, L.M.; Smith, J.A. Turns in peptides and proteins. Adv. Protein Chem. 1985, 37, 1–109. [Google Scholar] [PubMed]
- Rajashankar, K.R.; Ramakumar, S. Pi-turns in proteins and peptides: Classification, conformation, occurrence, hydration and sequence. Protein Sci. A Publ. Protein Soc. 1996, 5, 932–946. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, B.; Chakrabarti, P. Pi-turns: Types, systematics and the context of their occurrence in protein structures. BMC Struct. Biol. 2008, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, P.F.; Alix, A.J. High accuracy prediction of beta-turns and their types using propensities and multiple alignments. Proteins 2005, 59, 828–839. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.S. The anatomy and taxonomy of protein structure. Adv Protein Chem 1981, 34, 167–339. [Google Scholar]
- de Brevern, A.G. Extension of the classical classification of β-turns. Sci. Rep. 2016, 6, 33191. [Google Scholar] [CrossRef] [Green Version]
- Cook, W.J.; Jeffrey, L.C.; Sullivan, M.L.; Vierstra, R.D. Three-dimensional structure of a ubiquitin-conjugating enzyme (e2). J. Biol. Chem. 1992, 267, 15116–15121. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Hutchinson, E.G.; Thornton, J.M. Promotif--a program to identify and analyze structural motifs in proteins. Protein Sci. 1996, 5, 212–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summer, B.; Hook, P.; Morris, S.; Curtis, I. Atmosphere. In Joy Division—Atmosphere. 1980. [Google Scholar]
- Delano, W.L. The Pymol Molecular Graphics System on World Wide Web. 2013. Available online: http://www.Pymol.Org (accessed on 16 September 2022).
- Hutchinson, E.G.; Thornton, J.M. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 1994, 3, 2207–2216. [Google Scholar] [CrossRef] [PubMed]
- Fourrier, L.; Benros, C.; de Brevern, A.G. Use of a structural alphabet for analysis of short loops connecting repetitive structures. BMC Bioinform. 2004, 5, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, M.; Bornot, A.; Offmann, B.; de Brevern, A.G. Protein short loop prediction in terms of a structural alphabet. Comput. Biol. Chem. 2009, 33, 329–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, M.; Bornot, A.; Offmann, B.; de Brevern, A.G. Analysis of loop boundaries using different local structure assignment methods. Protein Sci. 2009, 18, 1869–1881. [Google Scholar] [CrossRef] [Green Version]
- Offmann, B.; Tyagi, M.; de Brevern, A.G. Local protein structures. Curr. Bioinform. 2007, 3, 165–202. [Google Scholar] [CrossRef] [Green Version]
- Burns, P.; Coy, S.; Hussey, W.; Lever, T.; Percy, M. You spin me round (like a record). In Dead or Alive—Youthquake. 1984. [Google Scholar]
- Guruprasad, K.; Prasad, M.S.; Kumar, G.R. Analysis of gammabeta, betagamma, gammagamma, betabeta multiple turns in proteins. J. Pept. Res. Off. J. Am. Pept. Soc. 2000, 56, 250–263. [Google Scholar] [CrossRef]
- Guruprasad, K.; Rao, M.J.; Adindla, S.; Guruprasad, L. Combinations of turns in proteins. J. Pept. Res. Off. J. Am. Pept. Soc. 2003, 62, 167–174. [Google Scholar] [CrossRef]
- Bono. With or without you. In U2—The Joshua Tree. 1987. [Google Scholar]
- Brinkjost, T.; Ehrt, C.; Koch, O.; Mutzel, P. Scot: Rethinking the classification of secondary structure elements. Bioinformatics 2020, 36, 2417–2428. [Google Scholar] [CrossRef] [PubMed]
- Richards, F.M.; Kundrot, C.E. Identification of structural motifs from protein coordinate data: Secondary structure and first-level supersecondary structure. Proteins 1988, 3, 71–84. [Google Scholar] [CrossRef]
- Cubellis, M.V.; Cailliez, F.; Lovell, S.C. Secondary structure assignment that accurately reflects physical and evolutionary characteristics. BMC Bioinform. 2005, 6, S8. [Google Scholar] [CrossRef] [Green Version]
- King, S.M.; Johnson, W.C. Assigning secondary structure from protein coordinate data. Proteins 1999, 35, 313–320. [Google Scholar] [CrossRef]
- Labesse, G.; Colloc’h, N.; Pothier, J.; Mornon, J.P. P-sea: A new efficient assignment of secondary structure from c alpha trace of proteins. Comput. Appl. Biosci. 1997, 13, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.; Letellier, G.; Marin, A.; Taly, J.F.; de Brevern, A.G.; Gibrat, J.F. Protein secondary structure assignment revisited: A detailed analysis of different assignment methods. BMC Struct. Biol. 2005, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Antony, J.V.; Madhu, P.; Balakrishnan, J.P.; Yadav, H. Assigning secondary structure in proteins using ai. J. Mol. Model. 2021, 27, 252. [Google Scholar] [CrossRef]
- Sklenar, H.; Etchebest, C.; Lavery, R. Describing protein structure: A general algorithm yielding complete helicoidal parameters and a unique overall axis. Proteins 1989, 6, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Fodje, M.N.; Al-Karadaghi, S. Occurrence, conformational features and amino acid propensities for the pi-helix. Protein Eng. 2002, 15, 353–358. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins 1995, 23, 566–579. [Google Scholar] [CrossRef]
- Ball, J.B.; Alewood, P.F. Conformational constraints: Nonpeptide beta-turn mimics. J. Mol. Recognit. JMR 1990, 3, 55–64. [Google Scholar] [CrossRef]
- Chou, P.Y.; Fasman, G.D. Prediction of beta-turns. Biophys. J. 1979, 26, 367–383. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.; Shang, Y.; Xu, D. A deep dense inception network for protein beta-turn prediction. Proteins 2020, 88, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Narwani, T.J.; Craveur, P.; Shinada, N.K.; Floch, A.; Santuz, H.; Vattekatte, A.M.; Srinivasan, N.; Rebehmed, J.; Gelly, J.C.; Etchebest, C.; et al. Discrete analyses of protein dynamics. J. Biomol. Struct. Dyn. 2020, 38, 2988–3002. [Google Scholar] [CrossRef]
- Knopfler, M.; Sting. Money for nothing. In Dire Straits—Brothers in Arms. 1985. [Google Scholar]
- Pal, L.; Chakrabarti, P.; Basu, G. Sequence and structure patterns in proteins from an analysis of the shortest helices: Implications for helix nucleation. J. Mol. Biol. 2003, 326, 273–291. [Google Scholar] [CrossRef]
- Wilmot, C.M.; Thornton, J.M. Analysis and prediction of the different types of beta-turn in proteins. J. Mol. Biol. 1988, 203, 221–232. [Google Scholar] [CrossRef]
- Evans, R. In the year 2525 (exordium & terminus). In Zager and Evans—2525 (Exordium & Terminus). 1969. [Google Scholar]
- Roberto Di, C.; Stefano, Z. Software heritage: Why and how to preserve software source code. In Proceedings of the iPRES 2017-14th International Conference on Digital Preservation, Kyoto, Japan, 25–29 September 2017. [Google Scholar]
- Touw, W.G.; Baakman, C.; Black, J.; te Beek, T.A.; Krieger, E.; Joosten, R.P.; Vriend, G. A series of pdb-related databanks for everyday needs. Nucleic Acids Res. 2015, 43, D364–D368. [Google Scholar] [CrossRef] [PubMed]
- Bender’s Olde Fortran Malt Liquor 1.0. Available online: http://www.beginbrewing.com/2019/06/benders-olde-fortran-malt-liquor-10.html (accessed on 26 March 2022).
- Starr, R. It don’t come easy. In Ringo Starr—Ringo. 1973. [Google Scholar]
- Martinez, X.; Chavent, M.; Baaden, M. Visualizing protein structures—Tools and trends. Biochem. Soc. Trans. 2020, 48, 499–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, X.; Krone, M.; Alharbi, N.; Rose, A.S.; Laramee, R.S.; O’Donoghue, S.; Baaden, M.; Chavent, M. Molecular graphics: Bridging structural biologists and computer scientists. Structure 2019, 27, 1617–1623. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. Ucsf chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Summer, B.; Hook, P.; Morris, S.; Curtis, I. She’s lost control. In Joy Division—Unknown Pleasures. 1979. [Google Scholar]
- Crawford, J.L.; Lipscomb, W.N.; Schellman, C.G. The reverse turn as a polypeptide conformation in globular proteins. Proc. Natl. Acad. Sci. USA 1973, 70, 538–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, R.M.; Waters, M.L. Model systems for beta-hairpins and beta-sheets. Curr. Opin. Struct. Biol. 2006, 16, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, A.L.; Daggett, V. The effect of context on the folding of β-hairpins. J. Struct. Biol. 2011, 176, 143–150. [Google Scholar] [CrossRef]
- Mahalakshmi, R. Aromatic interactions in β-hairpin scaffold stability: A historical perspective. Arch. Biochem. Biophys. 2019, 661, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Roche, D.B.; Viet, P.D.; Bakulina, A.; Hirsh, L.; Tosatto, S.C.E.; Kajava, A.V. Classification of β-hairpin repeat proteins. J. Struct. Biol. 2018, 201, 130–138. [Google Scholar] [CrossRef] [PubMed]
- DuPai, C.D.; Davies, B.W.; Wilke, C.O. A systematic analysis of the beta hairpin motif in the protein data bank. Protein Sci. A Publ. Protein Soc. 2021, 30, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, K.; Ramakrishnan, C.; Balaram, P. Beta-hairpins in proteins revisited: Lessons for de novo design. Protein Eng. 1997, 10, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Newley, A.; Bricusse, L. Feeling good. In Nina Simone—I Put a Spell on You. 1965. [Google Scholar]
- Wilmot, C.M.; Thornton, J.M. Beta-turns and their distortions: A proposed new nomenclature. Protein Eng. 1990, 3, 479–493. [Google Scholar] [CrossRef]
- Efimov, A.V. Structure of coiled beta-beta-hairpins and beta-beta-corners. FEBS Lett. 1991, 284, 288–292. [Google Scholar] [CrossRef] [Green Version]
- Rooman, M.J.; Rodriguez, J.; Wodak, S.J. Automatic definition of recurrent local structure motifs in proteins. J. Mol. Biol. 1990, 213, 327–336. [Google Scholar] [CrossRef]
- Kohonen, T. Self-organized formation of topologically correct feature maps. Biol. Cybern 1982, 43, 59–69. [Google Scholar] [CrossRef]
- Kohonen, T. Self-Organizing Maps, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2001; p. 501. [Google Scholar]
- Koch, O.; Klebe, G. Turns revisited: A uniform and comprehensive classification of normal, open, and reverse turn families minimizing unassigned random chain portions. Proteins 2009, 74, 353–367. [Google Scholar] [CrossRef]
- Meissner, M.; Koch, O.; Klebe, G.; Schneider, G. Prediction of turn types in protein structure by machine-learning classifiers. Proteins 2009, 74, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Koch, O.; Cole, J.; Block, P.; Klebe, G. Secbase: Database module to retrieve secondary structure elements with ligand binding motifs. J. Chem. Inf. Model. 2009, 49, 2388–2402. [Google Scholar] [CrossRef] [PubMed]
- Brinkjost, T. Eldorado, T.-D. Seconds First! A Thesis Dedicated to Secondary Structure Elements. Ph.D. Thesis, Universitätsbibliothek Dortmund, Dortmund, Germany, 2020. [Google Scholar]
- Shapovalov, M.; Vucetic, S.; Dunbrack, R.L., Jr. A new clustering and nomenclature for beta turns derived from high-resolution protein structures. PLoS Comput. Biol. 2019, 15, e1006844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining (KDD-96), Portland, OR, USA, 2–4 August 1996. [Google Scholar]
- Zhang, R.; Stahr, M.C.; Kennedy, M.A. Introduction of a new scheme for classifying β-turns in protein structures. Proteins 2022, 90, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.; Manzarek, R.; Krieger, R.; Densmore, J. The end. In The Doors—The Doors. 1967. [Google Scholar]
- Fuchs, P.F.; Bonvin, A.M.; Bochicchio, B.; Pepe, A.; Alix, A.J.; Tamburro, A.M. Kinetics and thermodynamics of type viii beta-turn formation: A cd, nmr, and microsecond explicit molecular dynamics study of the gdnp tetrapeptide. Biophys. J. 2006, 90, 2745–2759. [Google Scholar] [CrossRef] [Green Version]
- de Brevern, A.G.; Etchebest, C.; Hazout, S. Bayesian probabilistic approach for predicting backbone structures in terms of protein blocks. Proteins 2000, 41, 271–287. [Google Scholar] [CrossRef]
- Joseph, A.P.; Agarwal, G.; Mahajan, S.; Gelly, J.C.; Swapna, L.S.; Offmann, B.; Cadet, F.; Bornot, A.; Tyagi, M.; Valadié, H. A short survey on protein blocks. Bio. Rev. 2010, 2, 137–145. [Google Scholar] [CrossRef]
- Mansiaux, Y.; Joseph, A.P.; Gelly, J.C.; de Brevern, A.G. Assignment of polyproline ii conformation and analysis of sequence--structure relationship. PLoS ONE 2011, 6, e18401. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Kennedy, M.A. Structural dynamics of pentapeptide repeat proteins. Proteins 2020, 88, 1493–1512. [Google Scholar] [CrossRef] [PubMed]
- Odolczyk, N.; Marzec, E.; Winiewska-Szajewska, M.; Poznański, J.; Zielenkiewicz, P. Native structure-based peptides as potential protein-protein interaction inhibitors of sars-cov-2 spike protein and human ace2 receptor. Molecules 2021, 26, 2157. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Bansal, M. Identification of local variations within secondary structures of proteins. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1077–1086. [Google Scholar] [CrossRef]
- Kneller, G.R.; Hinsen, K. Protein secondary-structure description with a coarse-grained model. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, J.; Knapp, E.W. Protein secondary structure classification revisited: Processing dssp information with pssc. J. Chem. Inf. Model. 2014, 54, 2166–2179. [Google Scholar] [CrossRef] [PubMed]
- Salawu, E.O. Rafosa: Random forests secondary structure assignment for coarse-grained and all-atom protein systems. Cogent Biol. 2016, 2, 1214061. [Google Scholar] [CrossRef]
- Cao, C.; Wang, G.; Liu, A.; Xu, S.; Wang, L.; Zou, S. A new secondary structure assignment algorithm using cα backbone fragments. Int. J. Mol. Sci. 2016, 17, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adasme-Carreño, F.; Caballero, J.; Ireta, J. Psique: Protein secondary structure identification on the basis of quaternions and electronic structure calculations. J. Chem. Inf. Model. 2021, 61, 1789–1800. [Google Scholar] [CrossRef] [PubMed]
- Kee, K.S.; Jois, S.D. Design of beta-turn based therapeutic agents. Curr. Pharm. Des. 2003, 9, 1209–1224. [Google Scholar]
- Mahmud, S.; Paul, G.K.; Biswas, S.; Afrose, S.; Mita, M.A.; Hasan, M.R.; Shimu, M.S.S.; Hossain, A.; Promi, M.M.; Ema, F.K.; et al. Prospective role of peptide-based antiviral therapy against the main protease of sars-cov-2. Front. Mol. Biosci. 2021, 8, 628585. [Google Scholar] [CrossRef]
- Vaquer-Alicea, J.; Diamond, M.I.; Joachimiak, L.A. Tau strains shape disease. Acta Neuropathol. 2021, 142, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with alphafold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Brevern, A.G. A Perspective on the (Rise and Fall of) Protein β-Turns. Int. J. Mol. Sci. 2022, 23, 12314. https://doi.org/10.3390/ijms232012314
de Brevern AG. A Perspective on the (Rise and Fall of) Protein β-Turns. International Journal of Molecular Sciences. 2022; 23(20):12314. https://doi.org/10.3390/ijms232012314
Chicago/Turabian Stylede Brevern, Alexandre G. 2022. "A Perspective on the (Rise and Fall of) Protein β-Turns" International Journal of Molecular Sciences 23, no. 20: 12314. https://doi.org/10.3390/ijms232012314
APA Stylede Brevern, A. G. (2022). A Perspective on the (Rise and Fall of) Protein β-Turns. International Journal of Molecular Sciences, 23(20), 12314. https://doi.org/10.3390/ijms232012314





