Proteomic and miRNA Profiles of Exosomes Derived from Myometrial Tissue in Laboring Women
Abstract
1. Introduction
2. Results
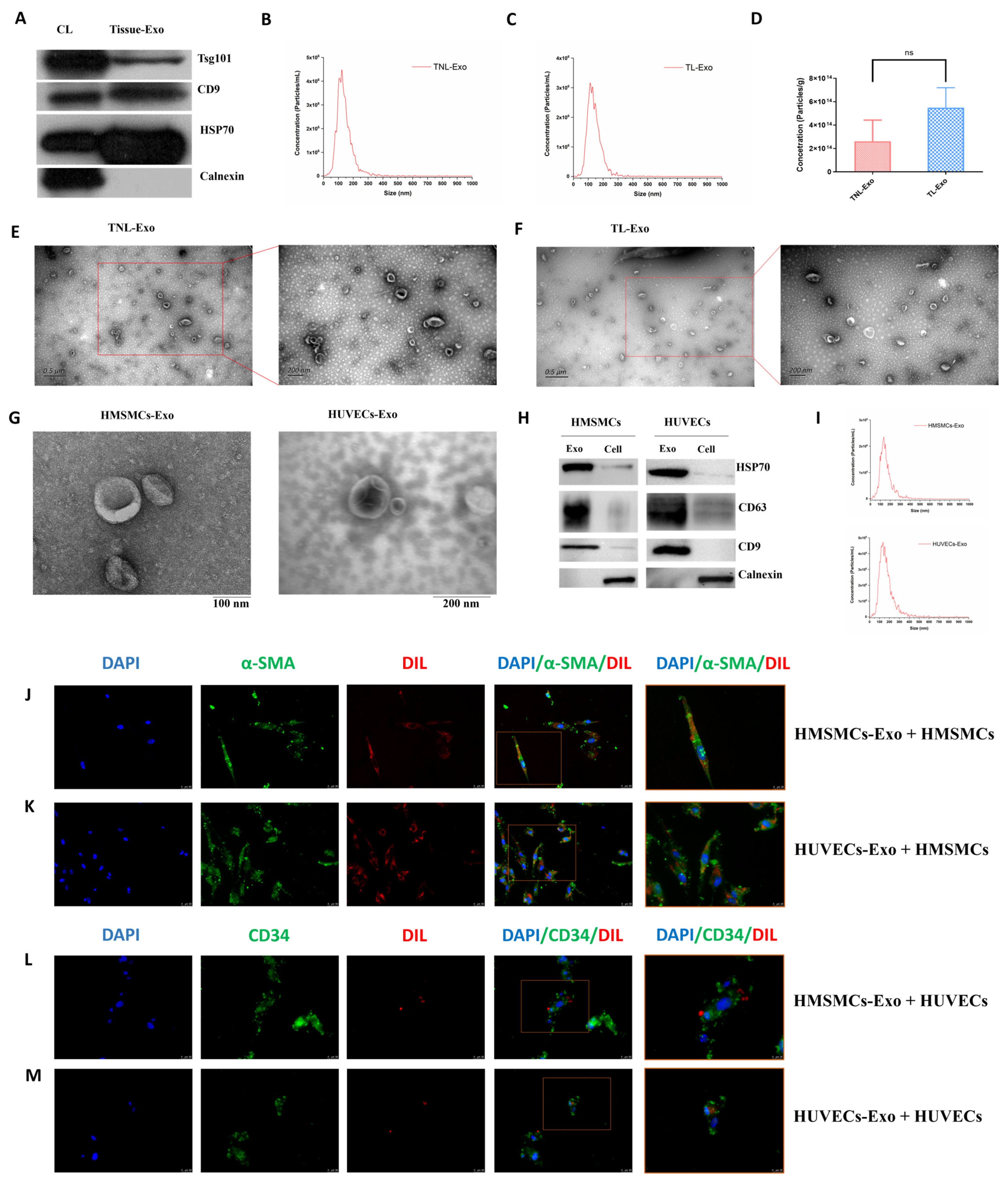
2.1. Characterization of Exosomes Isolated from Human Myometrial Tissue, HMSMCs, and HUVECs
2.2. Proteomic and miRNA Profiles of Exosomes in TNL and TL Myometrium
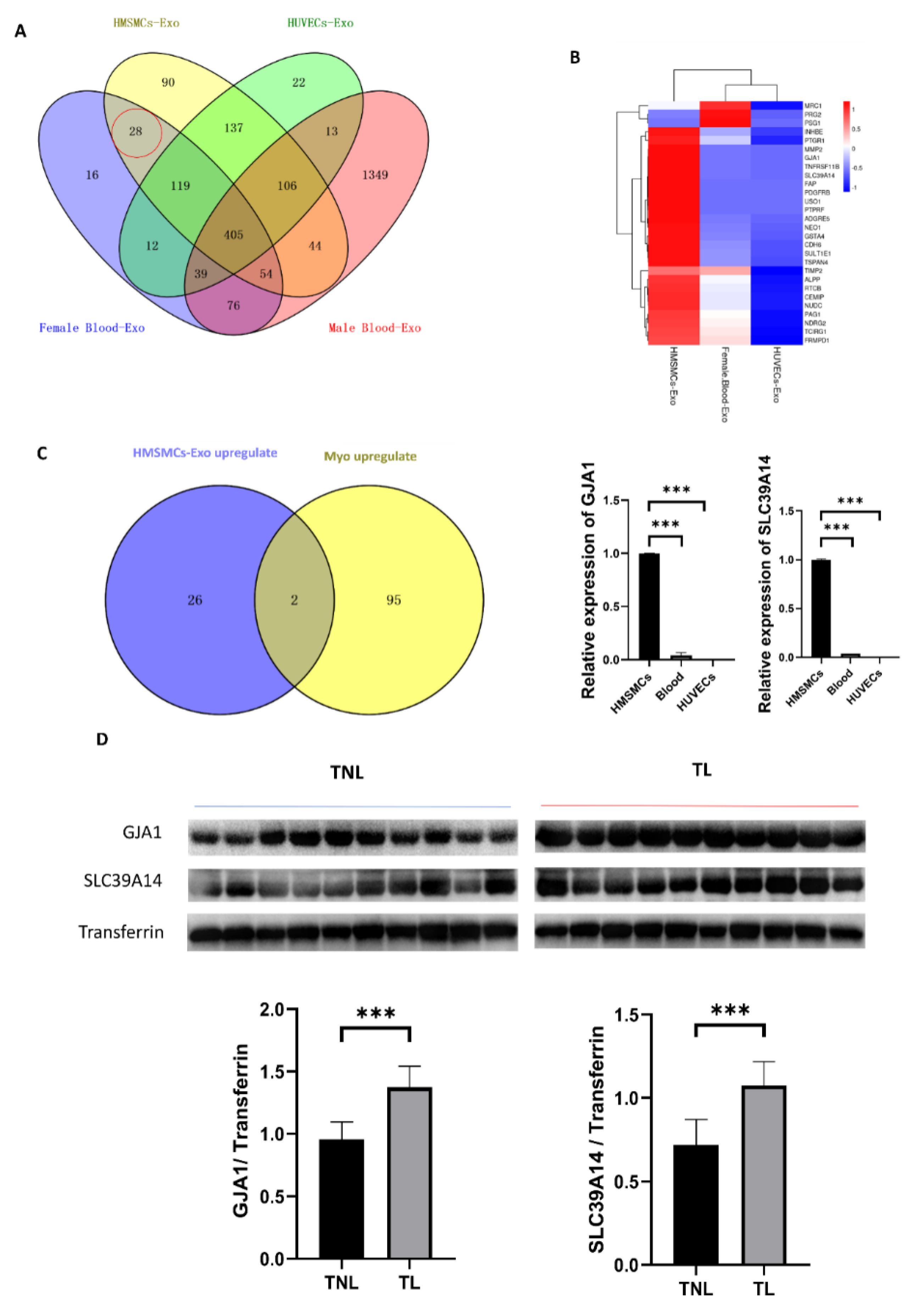
2.3. Cell-Specific Secreted miRNAs in the Tissue Exosomes
2.4. Myometrial Contraction-Specific Biomarkers among Exosomal Proteins
3. Discussion
4. Materials and Methods
4.1. Study Samples
4.2. Cell Culture
4.3. Exosome Isolation from Myometrial Tissue
4.4. Exosome Isolation from Plasma
4.5. Exosome Isolation from Cell Supernatant
4.6. NTA
4.7. Transmission Electron Microscopy
4.8. Immunofluorescence
4.9. Western Blotting
4.10. Label-free Quantitative Proteomics
4.10.1. Protein Extraction and Trypsin Treatment
4.10.2. LC-MS/MS Analysis
4.11. Exosomal MiRNA Analysis
4.11.1. miRNA Isolation and Sequencing
4.11.2. Bioinformatic Analysis of MiRNA
4.12. GO and KEGG Pathway Enrichment Analysis
4.13. RT-qPCR
4.14. Statistics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Barile, L.; Moccetti, T.; Marbán, E.; Vassalli, G. Roles of exosomes in cardioprotection. Eur. Heart J. 2017, 38, 1372–1379. [Google Scholar] [CrossRef]
- Jin, J.; Menon, R. Placental exosomes: A proxy to understand pregnancy complications. Am. J. Reprod. Immunol. 2018, 79, e12788. [Google Scholar] [CrossRef] [PubMed]
- Shahin, H.I.; Radnaa, E.; Tantengco, O.A.G.; Kechichian, T.; Kammala, A.K.; Sheller-Miller, S.; Taylor, B.D.; Menon, R. Microvesicles and Exosomes Released by Amnion Epithelial Cells Under Oxidative Stress Cause Inflammatory Changes in Uterine Cells. Biol. Reprod. 2021, 105, 464–480. [Google Scholar] [CrossRef] [PubMed]
- Li, S.R.; Man, Q.W.; Gao, X.; Lin, H.; Wang, J.; Su, F.C.; Wang, H.Q.; Bu, L.L.; Liu, B.; Chen, G. Tissue-derived extracellular vesicles in cancers and non-cancer diseases: Present and future. J. Extracell. Vesicles 2021, 10, e12175. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, B.; Yang, Y.; Jia, Q.; Qi, Z.; Zhang, A.; Lv, S.; Zhang, J. Exosomes in ischemic heart disease: Novel carriers for bioinformation. Biomed. Pharmacother. 2019, 120, 109451. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, X.; Hu, J.; Chen, F.; Qiao, S.; Sun, X.; Gao, L.; Xie, J.; Xu, B. Mesenchymal stromal cell-derived exosomes attenuate myocardial ischaemia-reperfusion injury through miR-182-regulated macrophage polarization. Cardiovasc. Res. 2019, 115, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Eldh, M.; Mints, M.; Hiltbrunner, S.; Ladjevardi, S.; Alamdari, F.; Johansson, M.; Jakubczyk, T.; Veerman, R.E.; Winqvist, O.; Sherif, A.; et al. Proteomic Profiling of Tissue Exosomes Indicates Continuous Release of Malignant Exosomes in Urinary Bladder Cancer Patients, Even with Pathologically Undetectable Tumour. Cancers 2021, 13, 3242. [Google Scholar] [CrossRef]
- Czernek, L.; Düchler, M. Exosomes as Messengers Between Mother and Fetus in Pregnancy. Int. J. Mol. Sci. 2020, 21, 4264. [Google Scholar] [CrossRef]
- Sheller-Miller, S.; Trivedi, J.; Yellon, S.M.; Menon, R. Exosomes Cause Preterm Birth in Mice: Evidence for Paracrine Signaling in Pregnancy. Sci. Rep. 2019, 9, 608. [Google Scholar] [CrossRef]
- Chiarello, D.I.; Salsoso, R.; Toledo, F.; Mate, A.; Vázquez, C.M.; Sobrevia, L. Foetoplacental communication via extracellular vesicles in normal pregnancy and preeclampsia. Mol. Asp. Med. 2018, 60, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Esfandyari, S.; Elkafas, H.; Chugh, R.M.; Park, H.S.; Navarro, A.; Al-Hendy, A. Exosomes as Biomarkers for Female Reproductive Diseases Diagnosis and Therapy. Int. J. Mol. Sci. 2021, 22, 2165. [Google Scholar] [CrossRef] [PubMed]
- Morrell, A.E.; Brown, G.N.; Robinson, S.T.; Sattler, R.L.; Baik, A.D.; Zhen, G.; Cao, X.; Bonewald, L.F.; Jin, W.; Kam, L.C.; et al. Mechanically induced Ca(2+) oscillations in osteocytes release extracellular vesicles and enhance bone formation. Bone Res. 2018, 6, 6. [Google Scholar] [CrossRef]
- Trovato, E.; di Felice, V.; Barone, R. Extracellular Vesicles: Delivery Vehicles of Myokines. Front. Physiol. 2019, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; He, Y.; Hou, X.; Zhang, Z.; Wang, R.; Wu, Q. Endothelial Cells Can Regulate Smooth Muscle Cells in Contractile Phenotype through the miR-206/ARF6&NCX1/Exosome Axis. PLoS ONE 2016, 11, e0152959. [Google Scholar]
- Chen, L.; Wang, L.; Luo, Y.; Huang, Q.; Ji, K.; Bao, J.; Liu, H. Integrated Proteotranscriptomics of Human Myometrium in Labor Landscape Reveals the Increased Molecular Associated With Inflammation Under Hypoxia Stress. Front. Immunol. 2021, 12, 722816. [Google Scholar] [CrossRef]
- Whitham, M.; Parker, B.L.; Friedrichsen, M.; Hingst, J.R.; Hjorth, M.; Hughes, W.E.; Egan, C.L.; Cron, L.; Watt, K.I.; Kuchel, R.P.; et al. Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell Metab. 2018, 27, 237–251.e4. [Google Scholar] [CrossRef] [PubMed]
- Mytidou, C.; Koutsoulidou, A.; Katsioloudi, A.; Prokopi, M.; Kapnisis, K.; Michailidou, K.; Anayiotos, A.; Phylactou, L.A. Muscle-derived exosomes encapsulate myomiRs and are involved in local skeletal muscle tissue communication. FASEB J. 2021, 35, e21279. [Google Scholar] [CrossRef] [PubMed]
- Matejovič, A.; Wakao, S.; Kitada, M.; Kushida, Y.; Dezawa, M. Comparison of separation methods for tissue-derived extracellular vesicles in the liver, heart, and skeletal muscle. FEBS Open Bio 2021, 11, 482–493. [Google Scholar] [CrossRef] [PubMed]
- Hadley, E.E.; Sheller-Miller, S.; Saade, G.; Salomon, C.; Mesiano, S.; Taylor, R.N.; Taylor, B.D.; Menon, R. Amnion epithelial cell-derived exosomes induce inflammatory changes in uterine cells. Am. J. Obstet. Gynecol. 2018, 219, e471–e478. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Dvorakova, L.; Kotlabova, K.; Krofta, L. The Prediction of Gestational Hypertension, Preeclampsia and Fetal Growth Restriction via the First Trimester Screening of Plasma Exosomal C19MC microRNAs. Int. J. Mol. Sci. 2019, 20, 2972. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Jacob, V.; Singal, A.; Lei, S.; Park, M.S.; Lima, M.R.N.; Li, C.; Dhall, S.; Sathyamoorthy, M.; Kohn, J. Exosomes Secreted from Amniotic Membrane Contribute to Its Anti-Fibrotic Activity. Int. J. Mol. Sci. 2021, 22, 2055. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.G.; He, Z.Y.; Liang, S.; Yang, Q.; Cheng, P.; Chen, A.M. Comprehensive proteomic analysis of exosomes derived from human bone marrow, adipose tissue, and umbilical cord mesenchymal stem cells. Stem Cell Res. Ther. 2020, 11, 511. [Google Scholar] [CrossRef]
- Cosme, J.; Guo, H.; Hadipour-Lakmehsari, S.; Emili, A.; Gramolini, A.O. Hypoxia-Induced Changes in the Fibroblast Secretome, Exosome, and Whole-Cell Proteome Using Cultured, Cardiac-Derived Cells Isolated from Neonatal Mice. J. Proteome Res. 2017, 16, 2836–2847. [Google Scholar] [CrossRef] [PubMed]
- Hergenreider, E.; Heydt, S.; Tréguer, K.; Boettger, T.; Horrevoets, A.J.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012, 14, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Boer, K.; den Hollander, I.A.; Meijers, J.C.; Levi, M. Tissue factor-dependent blood coagulation is enhanced following delivery irrespective of the mode of delivery. J. Thromb. Haemost. 2007, 5, 2415–2420. [Google Scholar] [CrossRef] [PubMed]
- Peltier, M.R. Immunology of term and preterm labor. Reprod. Biol. Endocrinol. 2003, 1, 122. [Google Scholar] [CrossRef]
- Malik, M.; Roh, M.; England, S.K. Uterine contractions in rodent models and humans. Acta Physiol. 2021, 231, e13607. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Tong, D.; Lu, X.; Wang, H.X.; Plante, I.; Lui, E.; Laird, D.W.; Bai, D.; Kidder, G.M. A dominant loss-of-function GJA1 (Cx43) mutant impairs parturition in the mouse. Biol. Reprod. 2009, 80, 1099–1106. [Google Scholar] [CrossRef]
- Jenkitkasemwong, S.; Akinyode, A.; Paulus, E.; Weiskirchen, R.; Hojyo, S.; Fukada, T.; Giraldo, G.; Schrier, J.; Garcia, A.; Janus, C.; et al. SLC39A14 deficiency alters manganese homeostasis and excretion resulting in brain manganese accumulation and motor deficits in mice. Proc. Natl. Acad. Sci. USA 2018, 115, e1769–e1778. [Google Scholar] [CrossRef]
- Prajapati, M.; Conboy, H.L.; Hojyo, S.; Fukada, T.; Budnik, B.; Bartnikas, T.B. Biliary excretion of excess iron in mice requires hepatocyte iron import by Slc39a14. J. Biol. Chem. 2021, 297, 100835. [Google Scholar] [CrossRef]
- Mosher, A.A.; Rainey, K.J.; Bolstad, S.S.; Lye, S.J.; Mitchell, B.F.; Olson, D.M.; Wood, S.L.; Slater, D.M. Development and validation of primary human myometrial cell culture models to study pregnancy and labour. BMC Pregnancy Childbirth 2013, 13, S7. [Google Scholar] [CrossRef][Green Version]
- Vella, L.J.; Scicluna, B.J.; Cheng, L.; Bawden, E.G.; Masters, C.L.; Ang, C.S.; Willamson, N.; McLean, C.; Barnham, K.J.; Hill, A.F. A rigorous method to enrich for exosomes from brain tissue. J. Extracell. Vesicles 2017, 6, 1348885. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- van Ooijen, M.P.; Jong, V.L.; Eijkemans, M.J.C.; Heck, A.J.R.; Andeweg, A.C.; Binai, N.A.; van den Ham, H.J. Identification of differentially expressed peptides in high-throughput proteomics data. Brief Bioinform. 2018, 19, 971–981. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]


| Antibody Name | Catalog No. | Company | Concentration |
|---|---|---|---|
| Tsg101 | Ab125011 | Abcam, Cambridge, UK | 1:1000 |
| CD9 | 60232-I-Ig | Proteintech, Rosemont, IL, USA | 1:1000 |
| HSP70 | ab181606 | Abcam, Cambridge, UK | 1:1000 |
| Calnexin | 10427-2-AP | Proteintech, Rosemont, IL, USA | 1:500 |
| CD9 | ab198702 | Abcam, Cambridge, UK | 1:2000 |
| CD63 | ab134045 | Abcam, Cambridge, UK | 1:1000 |
| HSP70 | ab181606 | Abcam, Cambridge, UK | 1:2000 |
| Calnexin | ab133615 | Abcam, Cambridge, UK | 1:1000 |
| GJA1 | ab217676 | Abcam, Cambridge, UK | 1:1000 |
| SLC39A14 | A10413 | Affinity Biosciences, Cincinnati, OH, USA | 1:1000 |
| Transferrin | ab277635 | Abcam, Cambridge, UK | 1:1000 |
| Gene | Sequence (5′-3′) | |
|---|---|---|
| GJA1 | F: GGAGATGAGCAGTCTGCCTTTC | R: TGAGCCAGGTACAAGAGTGTGG |
| SLC39A14 | F: CTGGACCACATGATTCCTCAGC | R: AGAGTAGCGGACACCTTTCAGC |
| ACTB | F: CACCATTGGCAATGAGCGGTTC | R: AGGTCTTTGCGGATGTCCACGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, W.; Wang, X.; Chen, L.; Wen, B.; Chen, Y.; Ji, K.; Liu, H. Proteomic and miRNA Profiles of Exosomes Derived from Myometrial Tissue in Laboring Women. Int. J. Mol. Sci. 2022, 23, 12343. https://doi.org/10.3390/ijms232012343
Deng W, Wang X, Chen L, Wen B, Chen Y, Ji K, Liu H. Proteomic and miRNA Profiles of Exosomes Derived from Myometrial Tissue in Laboring Women. International Journal of Molecular Sciences. 2022; 23(20):12343. https://doi.org/10.3390/ijms232012343
Chicago/Turabian StyleDeng, Wenfeng, Xiaodi Wang, Lina Chen, Bolun Wen, Yunshan Chen, Kaiyuan Ji, and Huishu Liu. 2022. "Proteomic and miRNA Profiles of Exosomes Derived from Myometrial Tissue in Laboring Women" International Journal of Molecular Sciences 23, no. 20: 12343. https://doi.org/10.3390/ijms232012343
APA StyleDeng, W., Wang, X., Chen, L., Wen, B., Chen, Y., Ji, K., & Liu, H. (2022). Proteomic and miRNA Profiles of Exosomes Derived from Myometrial Tissue in Laboring Women. International Journal of Molecular Sciences, 23(20), 12343. https://doi.org/10.3390/ijms232012343






