Complementary Dual Approach for In Silico Target Identification of Potential Pharmaceutical Compounds in Cystic Fibrosis
Abstract
1. Introduction
2. Results
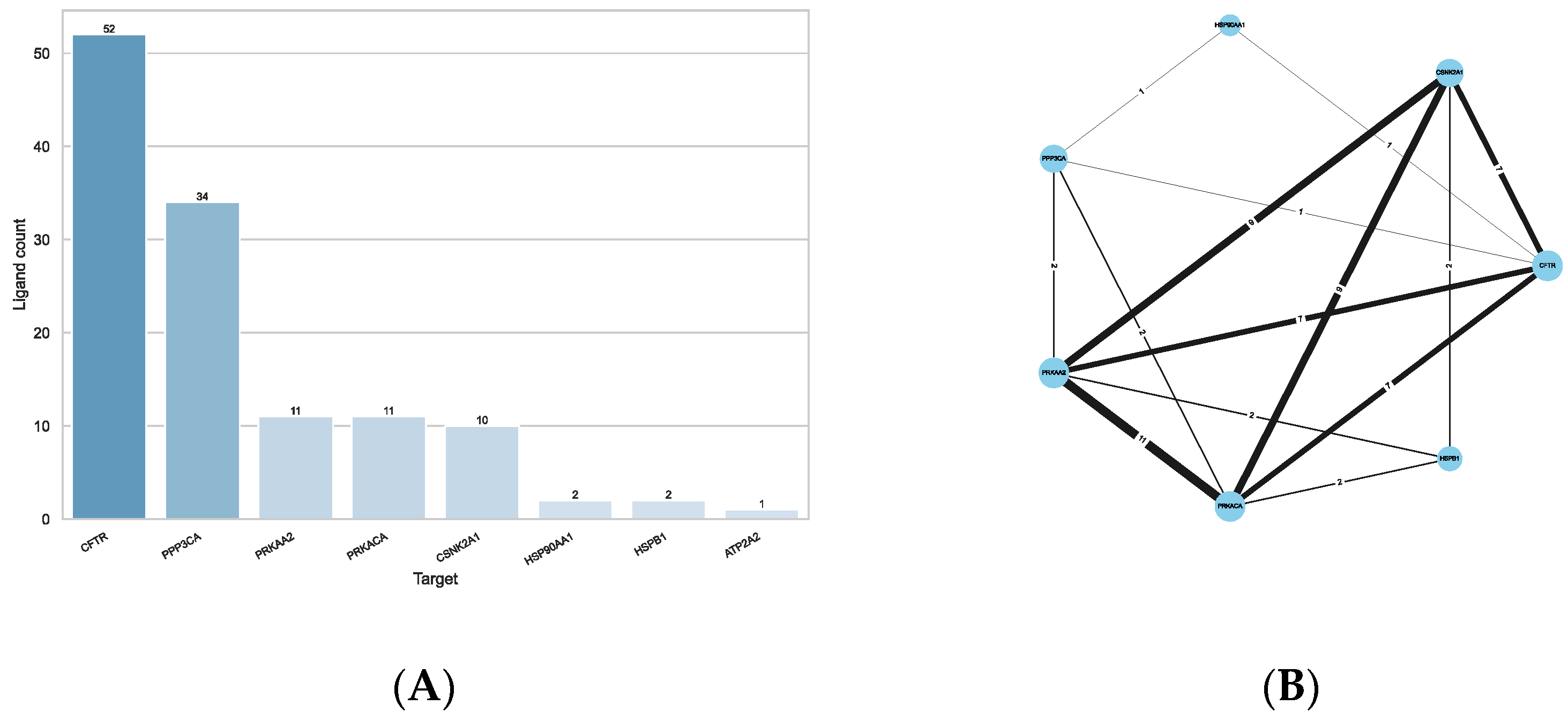
2.1. PDB Targets
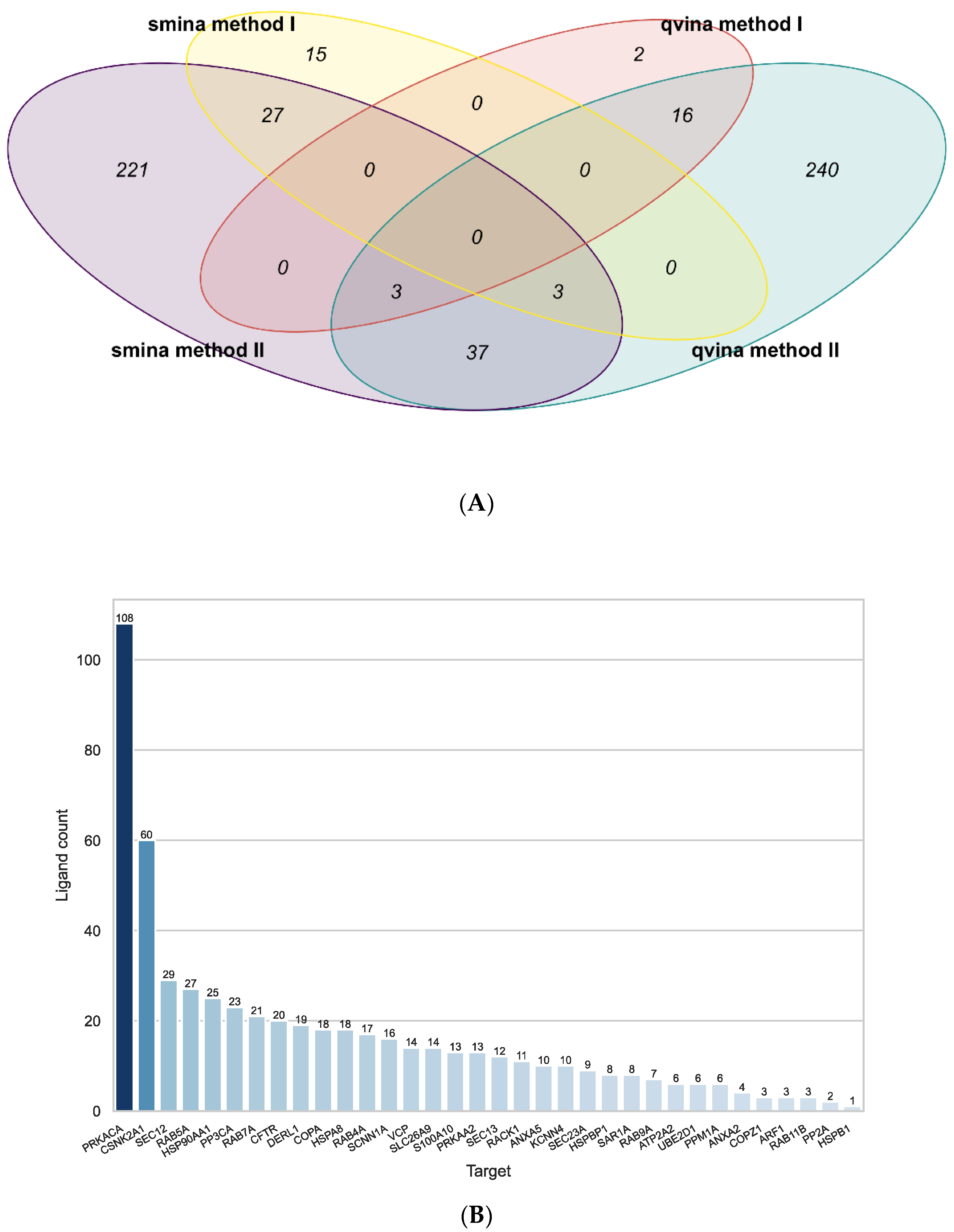
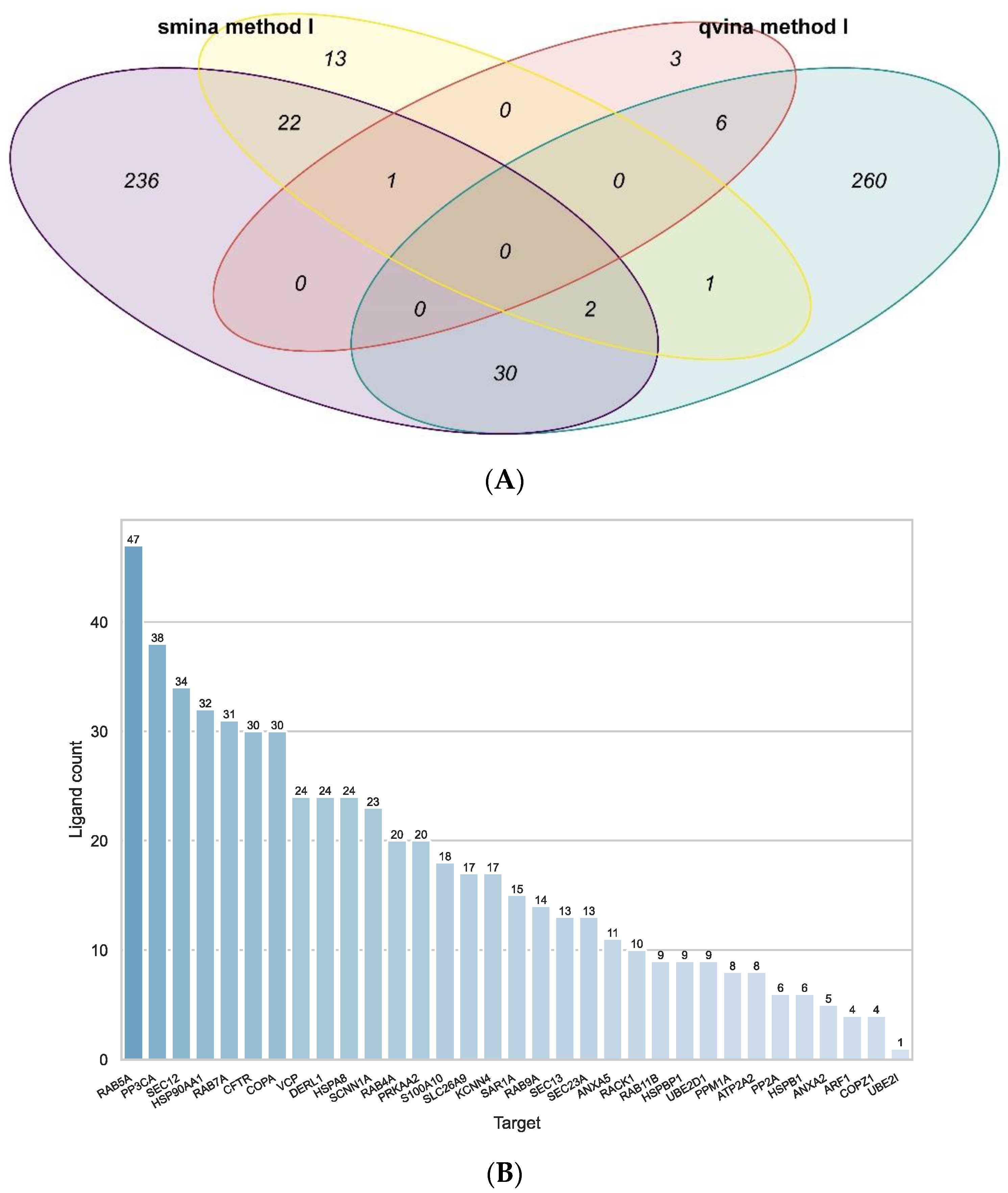
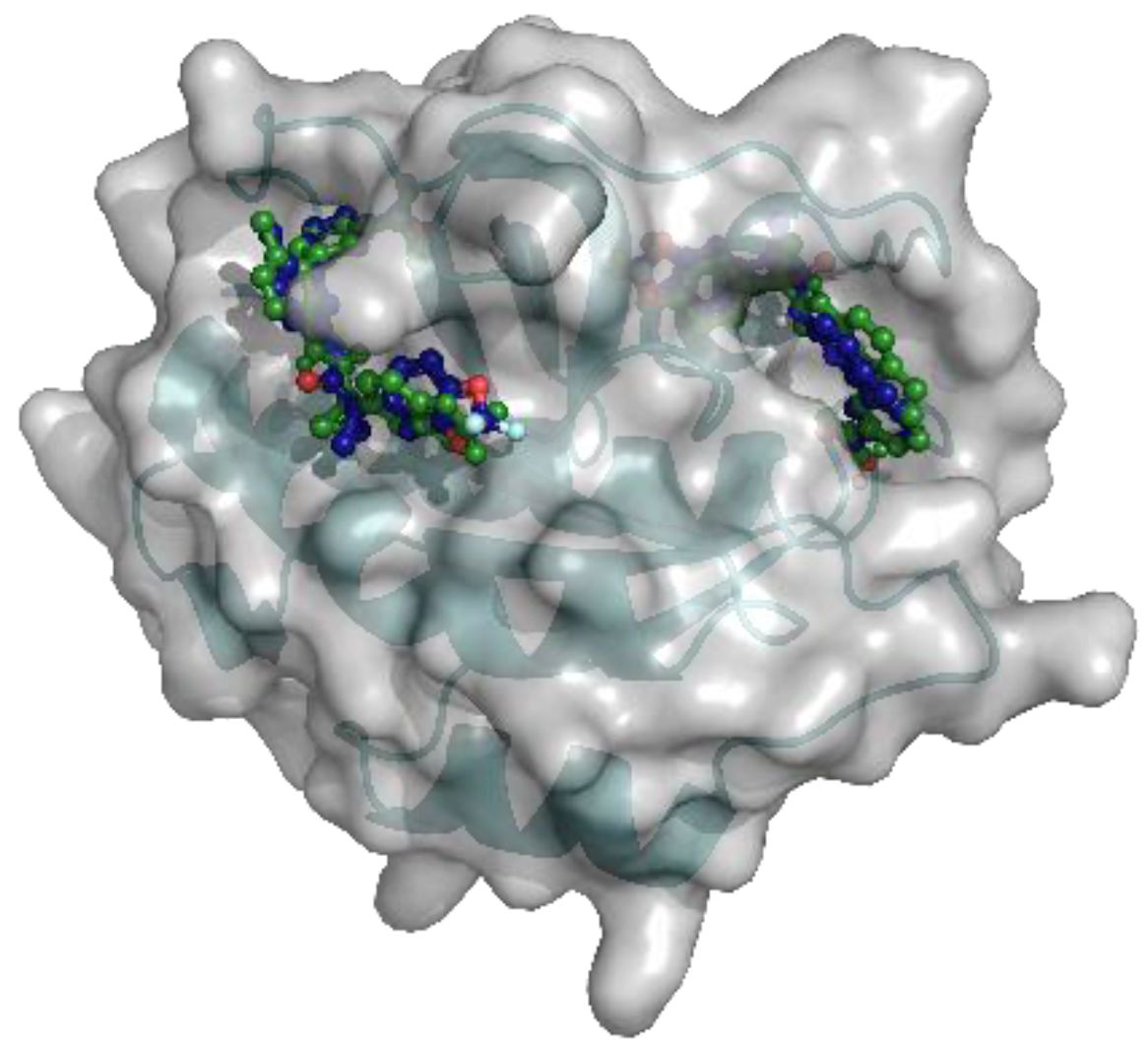
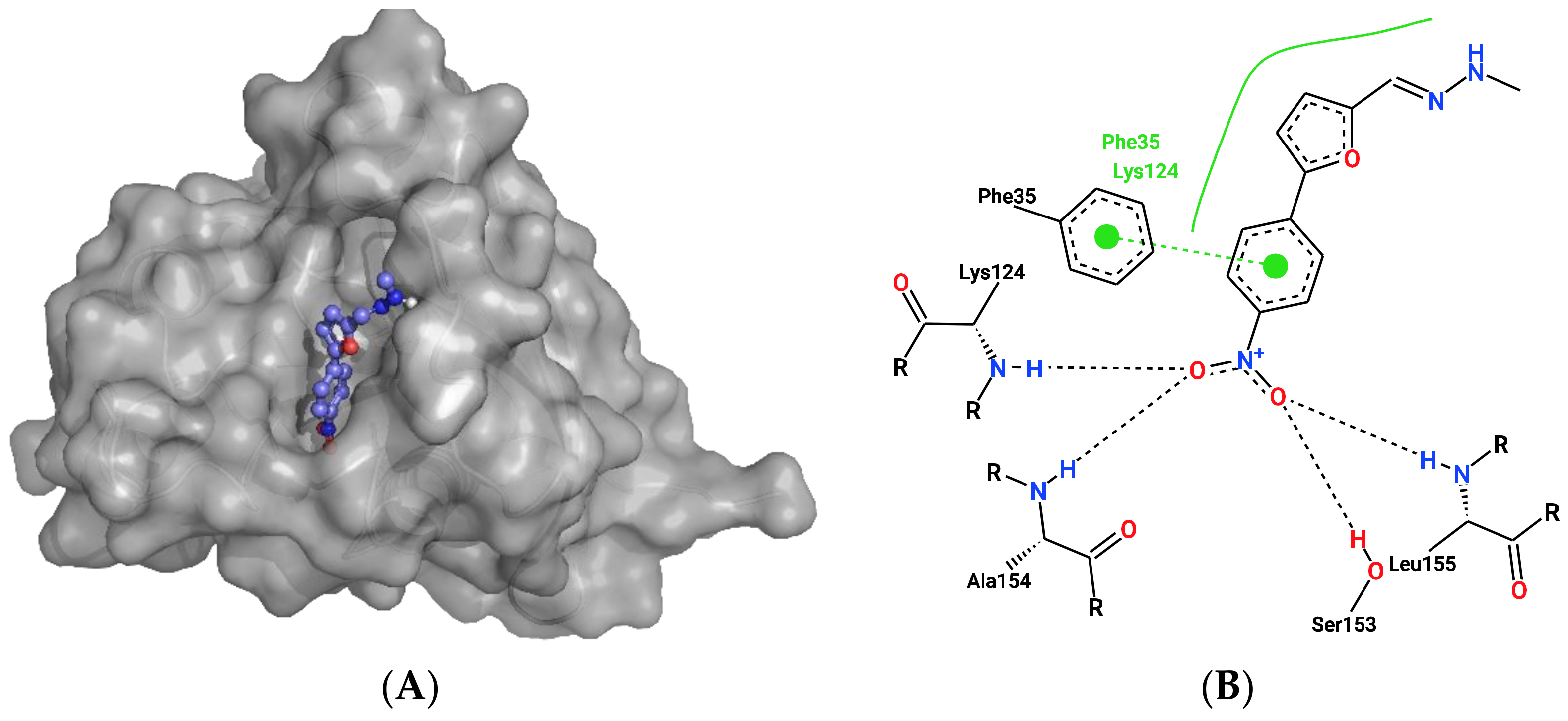
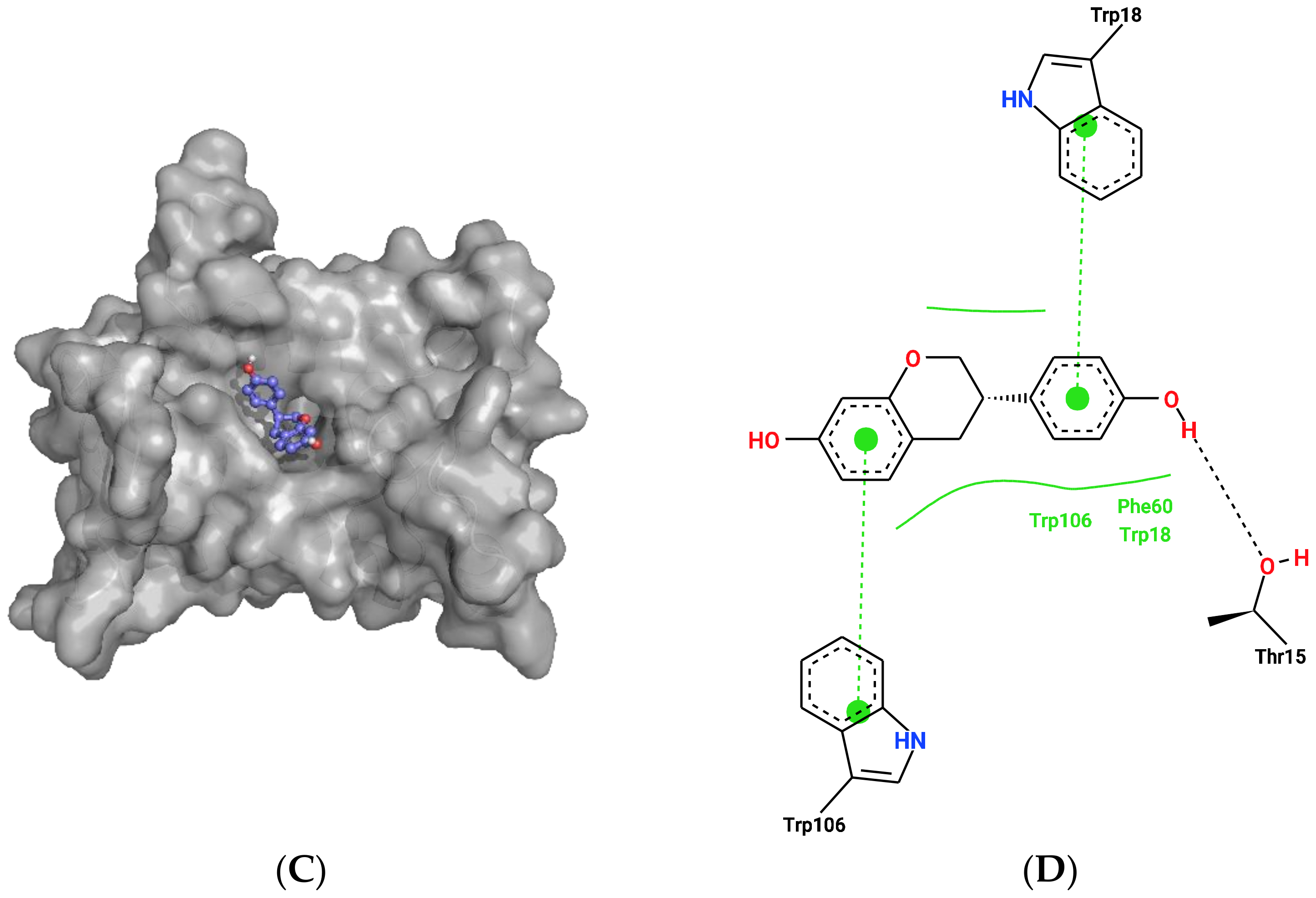
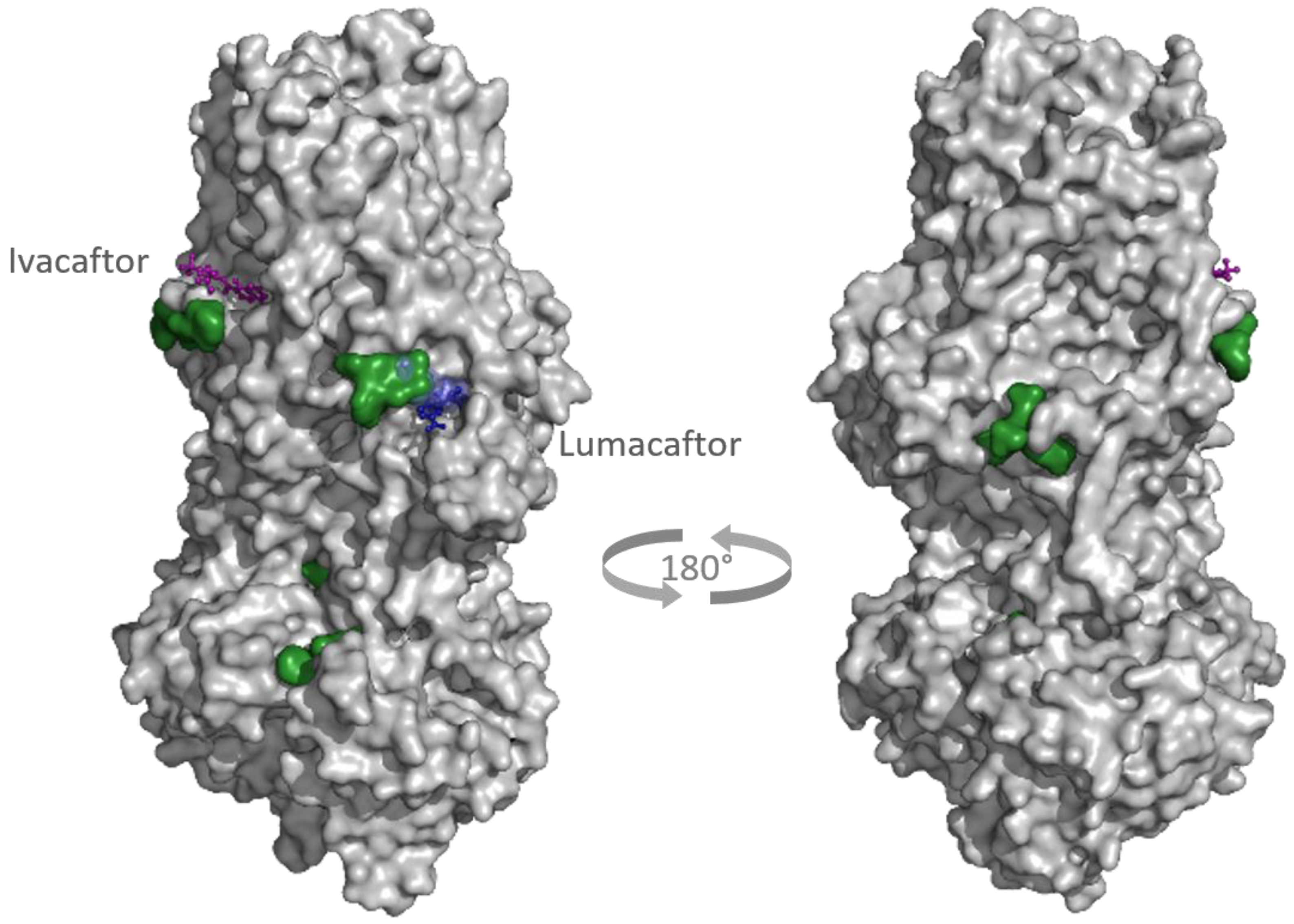
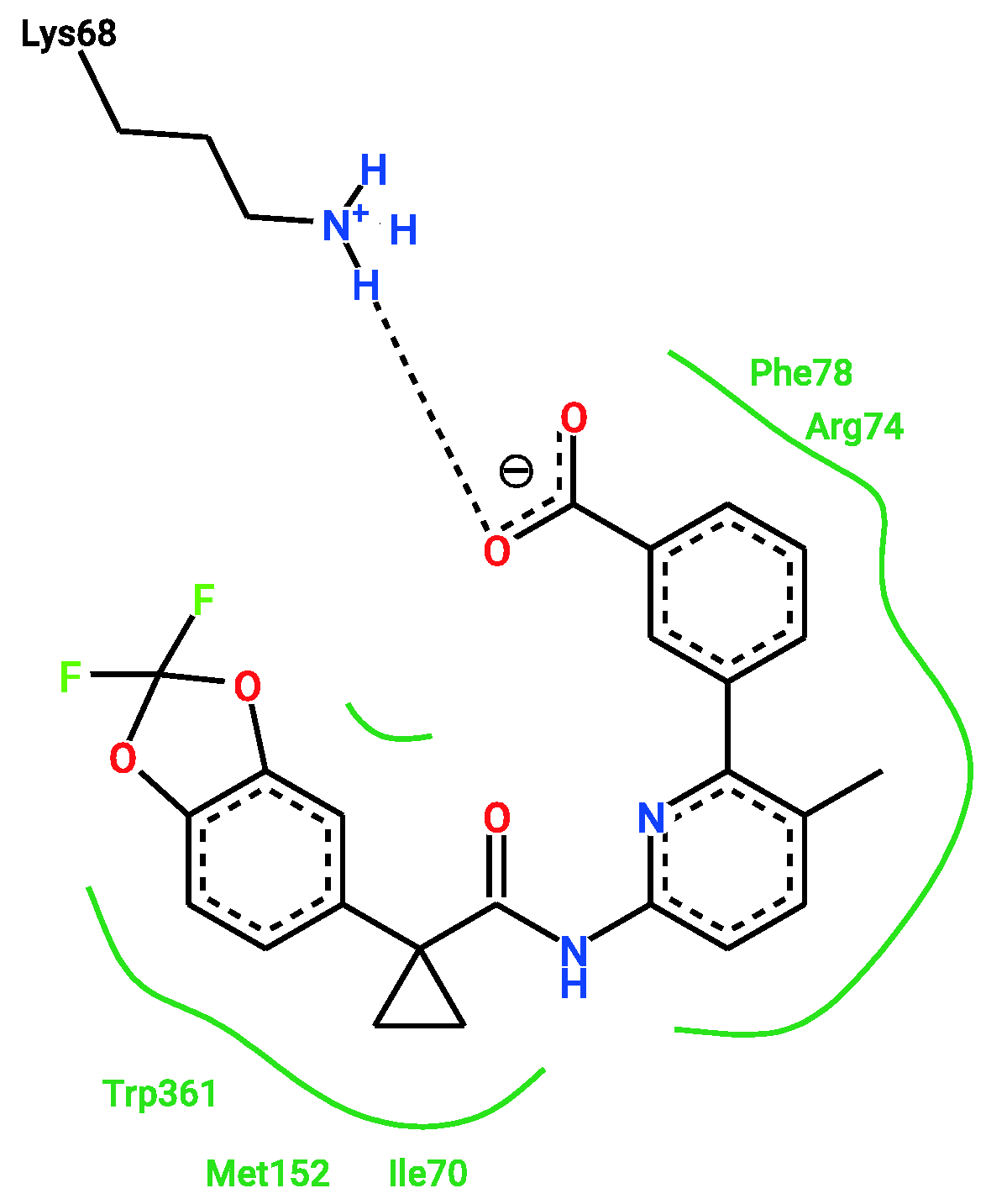
2.2. Docking
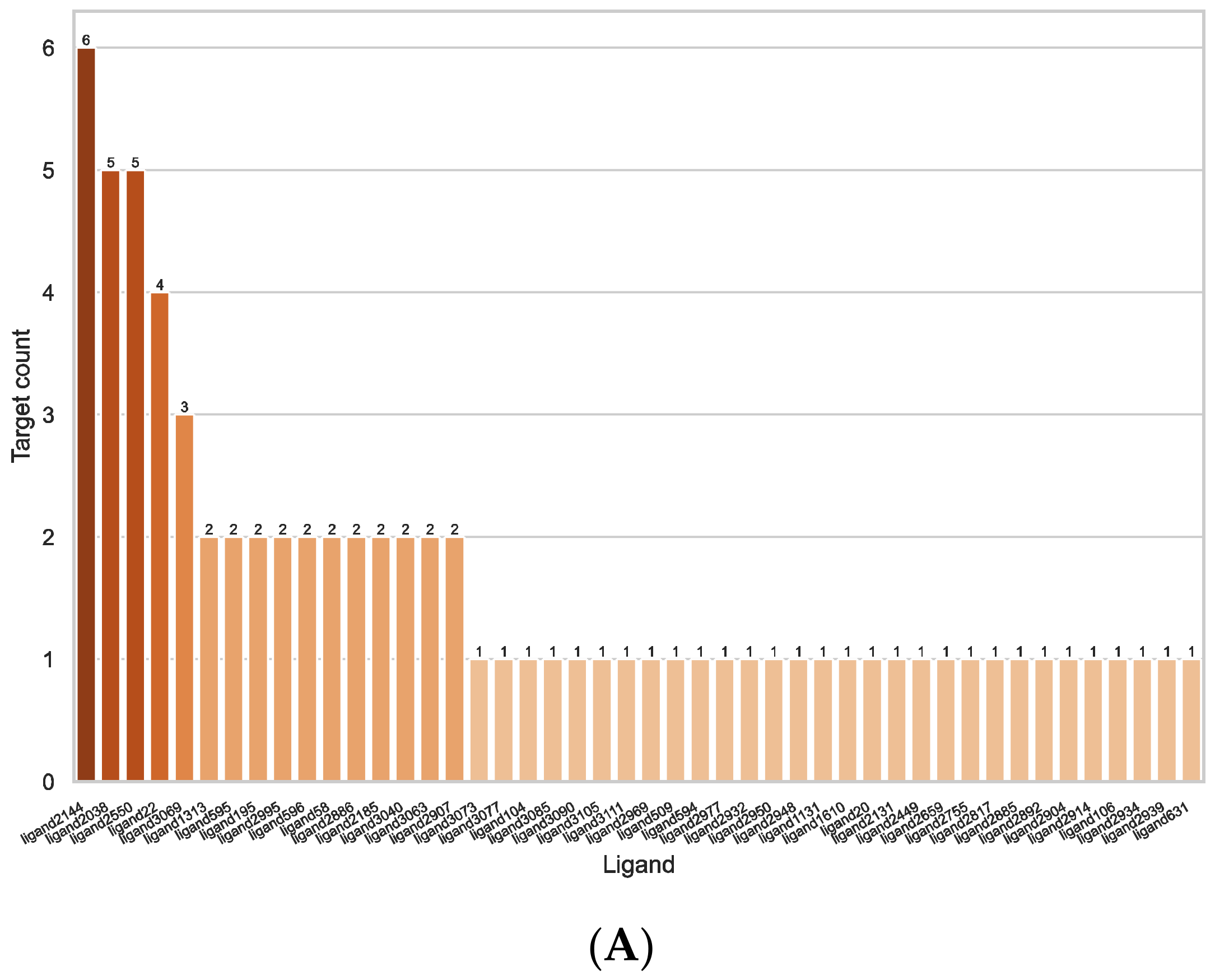
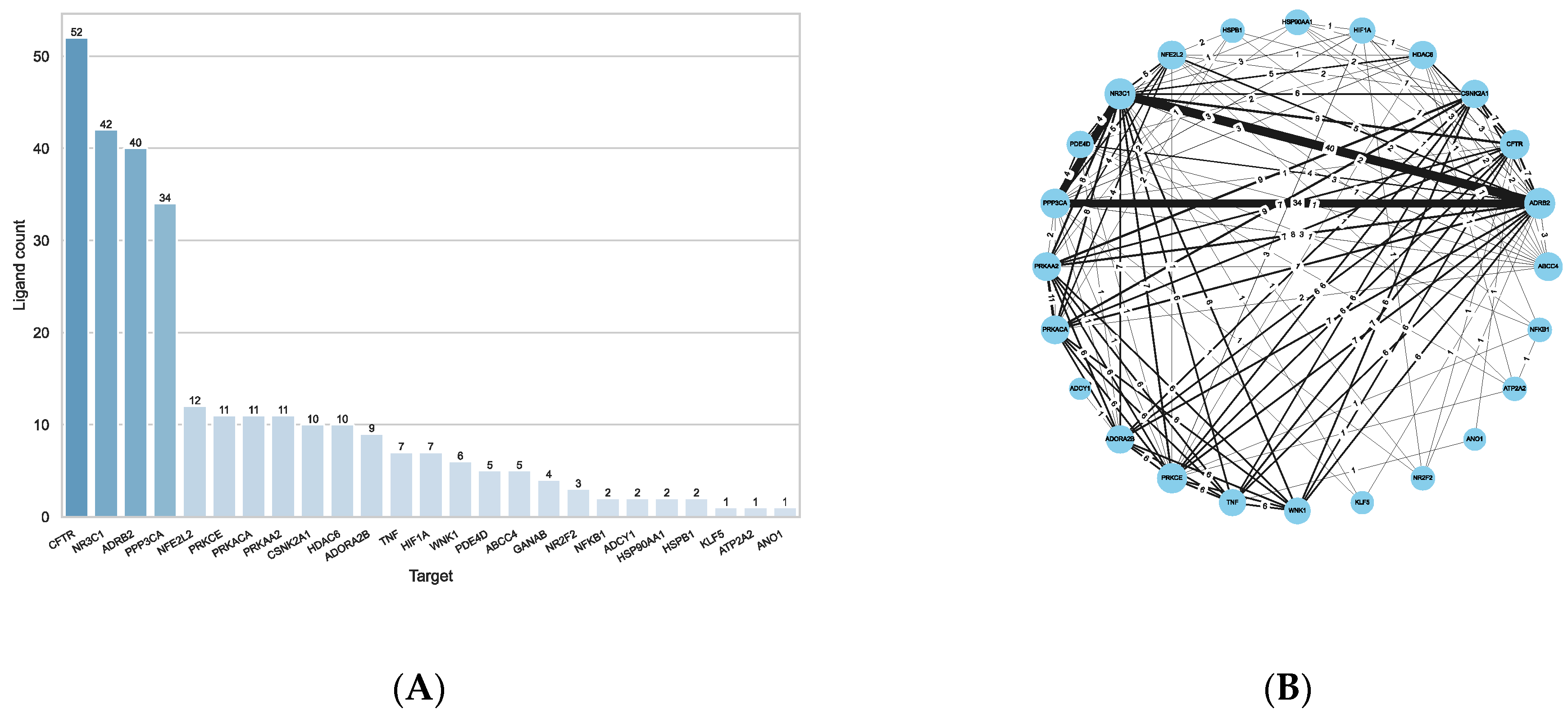
2.3. Ligand Similarity Approach
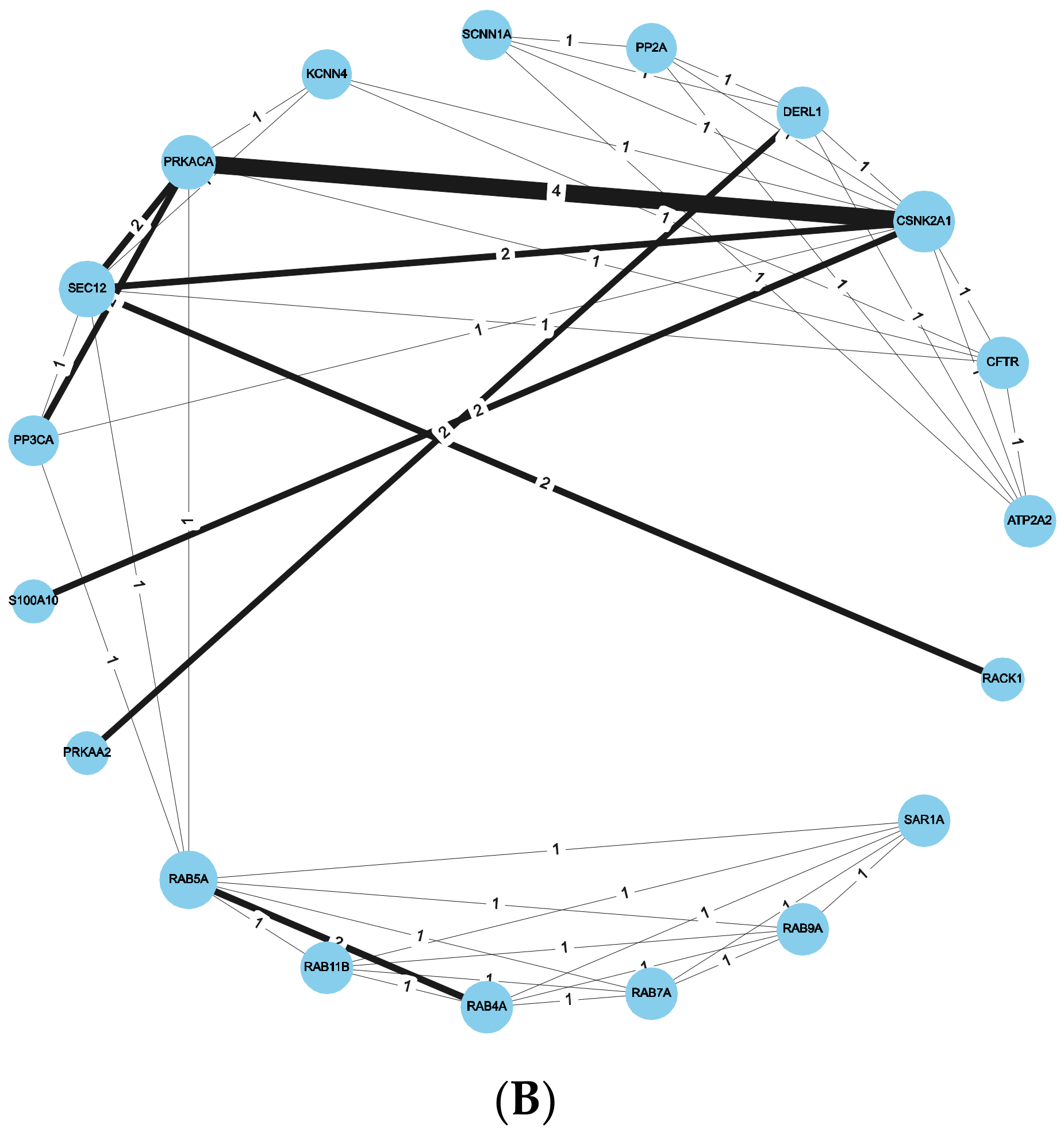
2.4. Combined Approach
3. Discussion
4. Materials and Methods
4.1. Reverse Docking
4.2. Ligand Based Approach
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References and Note
- Bobadilla, J.L.; Macek, M.; Fine, J.P.; Farrell, P.M. Cystic fibrosis: A worldwide analysis of CFTR mutations—Correlation with incidence data and application to screening. Hum. Mutat. 2002, 19, 575–606. [Google Scholar] [CrossRef] [PubMed]
- Farrell, P.M. The prevalence of cystic fibrosis in the European Union. J. Cyst. Fibros. 2008, 7, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.C.; Mall, M.A.; Gutierrez, H.; Macek, M.; Madge, S.; Davies, J.C.; Burgel, P.R.; Tullis, E.; Castaños, C.; Castellani, C.; et al. The future of cystic fibrosis care: A global perspective. Lancet Respir. Med. 2020, 8, 65–124. [Google Scholar] [CrossRef]
- O’Sullivan, B.P.; Freedman, S.D. Cystic fibrosis. Lancet 2009, 373, 1891–1904. [Google Scholar] [CrossRef]
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- Cystic Fibrosis Mutation Database. Available online: http://www.genet.sickkids.on.ca/ (accessed on 26 January 2021).
- Welcome to CFTR2|CFTR2. Available online: https://www.cftr2.org/ (accessed on 26 January 2021).
- Sosnay, P.R.; Siklosi, K.R.; van Goor, F.; Kaniecki, K.; Yu, H.; Sharma, N.; Ramalho, A.S.; Amaral, M.D.; Dorfman, R.; Zielenski, J.; et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 2013, 45, 1160–1167. [Google Scholar] [CrossRef]
- Pranke, I.M.; Sermet-Gaudelus, I. Biosynthesis of cystic fibrosis transmembrane conductance regulator. Int. J. Biochem. Cell Biol. 2014, 52, 26–38. [Google Scholar] [CrossRef]
- Welsh, M.J.; Smith, A.E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell 1993, 73, 1251–1254. [Google Scholar] [CrossRef]
- Rowe, S.M.; Miller, S.; Sorscher, E.J. Cystic fibrosis. N. Engl. J. Med. 2005, 352, 1992–2001. [Google Scholar] [CrossRef]
- Zielenski, J.; Tsui, L.C. Cystic fibrosis: Genotypic and phenotypic variations. Annu. Rev. Genet. 1995, 29, 777–807. [Google Scholar] [CrossRef]
- Zielenski, J. Genotype and Phenotype in Cystic Fibrosis. Respiration 2000, 67, 117–133. [Google Scholar] [CrossRef] [PubMed]
- De Boeck, K. Cystic fibrosis in the year 2020: A disease with a new face. Acta Paediatr. 2020, 109, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Veit, G.; Avramescu, R.G.; Chiang, A.N.; Houck, S.A.; Cai, Z.; Peters, K.W.; Hong, J.S.; Pollard, H.B.; Guggino, W.B.; Balch, W.E.; et al. From CFTR biology toward combinatorial pharmacotherapy: Expanded classification of cystic fibrosis mutations. Mol. Biol. Cell 2016, 27, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Gentzsch, M.; Mall, M.A. Ion Channel Modulators in Cystic Fibrosis. Chest 2018, 154, 383–393. [Google Scholar] [CrossRef]
- Zaher, A.; ElSaygh, J.; Elsori, D.; ElSaygh, H.; Sanni, A. A Review of Trikafta: Triple Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Modulator Therapy. Cureus 2021, 13, e16144. [Google Scholar] [CrossRef]
- Drug Development Pipeline: CFF Clinical Trials Tool. Available online: https://www.cff.org/Trials/Pipeline (accessed on 26 January 2021).
- Clinical Pipeline. Available online: https://www.glpg.com/clinical-pipelines (accessed on 28 June 2022).
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.J.; Burton, B.; Cao, D.; Neuberger, T.; Turnbull, A.; Singh, A.; Joubran, J.; Hazlewood, A.; et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. USA 2009, 106, 18825–18830. [Google Scholar] [CrossRef]
- Ramsey, B.W.; Davies, J.; McElvaney, N.G.; Tullis, E.; Bell, S.C.; Dřevínek, P.; Griese, M.; McKone, E.F.; Wainwright, C.E.; Konstan, M.W.; et al. A CFTR Potentiator in Patients with Cystic Fibrosis and the G551D Mutation. N. Engl. J. Med. 2011, 365, 1663–1672. [Google Scholar] [CrossRef]
- Clancy, J.P.; Rowe, S.M.; Accurso, F.J.; Aitken, M.L.; Amin, R.S.; Ashlock, M.A.; Ballmann, M.; Boyle, M.P.; Bronsveld, I.; Campbell, P.W.; et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax 2012, 67, 12–18. [Google Scholar] [CrossRef]
- Wainwright, C.E.; Elborn, J.S.; Ramsey, B.W.; Marigowda, G.; Huang, X.; Cipolli, M.; Colombo, C.; Davies, J.C.; de Boeck, K.; Flume, P.A.; et al. Lumacaftor–Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N. Engl. J. Med. 2015, 373, 220–231. [Google Scholar] [CrossRef]
- Taylor-Cousar, J.L.; Munck, A.; McKone, E.F.; van der Ent, C.K.; Moeller, A.; Simard, C.; Wang, L.T.; Ingenito, E.P.; McKee, C.; Lu, Y.; et al. Tezacaftor–Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del. N. Engl. J. Med. 2017, 377, 2013–2023. [Google Scholar] [CrossRef]
- Voelker, R. Patients with Cystic Fibrosis Have New Triple-Drug Combination. JAMA 2019, 322, 2068. [Google Scholar] [CrossRef] [PubMed]
- Ridley, K.; Condren, M. Elexacaftor-tezacaftor-ivacaftor: The first triple-combination cystic fibrosis transmembrane conductance regulator modulating therapy. J. Pediatr. Pharmacol. Ther. 2020, 25, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Goetz, D.M.; Savant, A.P. Review of CFTR modulators 2020. Pediatr. Pulmonol. 2021, 56, 3595–3606. [Google Scholar] [CrossRef] [PubMed]
- Martiniano, S.L.; Sagel, S.D.; Zemanick, E.T. Cystic fibrosis: A model system for precision medicine. Curr. Opin. Pediatr. 2016, 28, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Southern, K.W.; Patel, S.; Sinha, I.P.; Nevitt, S.J. Correctors (specific therapies for class II CFTR mutations) for cystic fibrosis. Cochrane Database Syst. Rev. 2018, 2018, CD010966. [Google Scholar] [CrossRef]
- Pedemonte, N.; Lukacs, G.L.; Du, K.; Caci, E.; Zegarra-Moran, O.; Galietta, L.J.V.; Verkman, A.S. Small-molecule correctors of defective ΔF508-CFTR cellular processing identified by high-throughput screening. J. Clin. Investig. 2005, 115, 2564–2571. [Google Scholar] [CrossRef]
- Berg, A.; Hallowell, S.; Tibbetts, M.; Beasley, C.; Brown-Phillips, T.; Healy, A.; Pustilnik, L.; Doyonnas, R.; Pregel, M. High-Throughput Surface Liquid Absorption and Secretion Assays to Identify F508del CFTR Correctors Using Patient Primary Airway Epithelial Cultures. SLAS Discov. 2019, 24, 724–737. [Google Scholar] [CrossRef]
- De Wilde, G.; Gees, M.; Musch, S.; Verdonck, K.; Jans, M.; Wesse, A.S.; Singh, A.K.; Hwang, T.C.; Christophe, T.; Pizzonero, M.; et al. Identification of GLPG/ABBV-2737, a novel class of corrector, which exerts functional synergy with other CFTR modulators. Front. Pharmacol. 2019, 10, 514. [Google Scholar] [CrossRef]
- Merkert, S.; Schubert, M.; Olmer, R.; Engels, L.; Radetzki, S.; Veltman, M.; Scholte, B.J.; Zöllner, J.; Pedemonte, N.; Galietta, L.J.V.; et al. High-Throughput Screening for Modulators of CFTR Activity Based on Genetically Engineered Cystic Fibrosis Disease-Specific iPSCs. Stem Cell Rep. 2019, 12, 1389–1403. [Google Scholar] [CrossRef]
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.J.; Burton, B.; Stack, J.H.; Straley, K.S.; Decker, C.J.; Miller, M.; McCartney, J.; Olson, E.R.; et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA 2011, 108, 18843–18848. [Google Scholar] [CrossRef]
- Phuan, P.W.; Veit, G.; Tan, J.A.; Finkbeiner, W.E.; Lukacs, G.L.; Verkman, A.S. Potentiators of defective DF508-CFTR gating that do not interfere with corrector action. Mol. Pharmacol. 2015, 88, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Carlile, G.W.; Robert, R.; Goepp, J.; Matthes, E.; Liao, J.; Kus, B.; Macknight, S.D.; Rotin, D.; Hanrahan, J.W.; Thomas, D.Y. Ibuprofen rescues mutant cystic fibrosis transmembrane conductance regulator trafficking. J. Cyst. Fibros. 2015, 14, 16–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liang, F.; Shang, H.; Jordan, N.J.; Wong, E.; Mercadante, D.; Saltz, J.; Mahiou, J.; Bihler, H.J.; Mense, M. High-Throughput Screening for Readthrough Modulators of CFTR PTC Mutations. SLAS Technol. 2017, 22, 315–324. [Google Scholar] [CrossRef]
- Giuliano, K.A.; Wachi, S.; Drew, L.; Dukovski, D.; Green, O.; Bastos, C.; Cullen, M.D.; Hauck, S.; Tait, B.D.; Munoz, B.; et al. Use of a High-Throughput Phenotypic Screening Strategy to Identify Amplifiers, a Novel Pharmacological Class of Small Molecules That Exhibit Functional Synergy with Potentiators and Correctors. SLAS Discov. 2018, 23, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Van der Plas, S.E.; Kelgtermans, H.; de Munck, T.; Martina, S.L.X.; Dropsit, S.; Quinton, E.; de Blieck, A.; Joannesse, C.; Tomaskovic, L.; Jans, M.; et al. Discovery of N-(3-Carbamoyl-5,5,7,7-tetramethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-2-yl)-lH-pyrazole-5-carboxamide (GLPG1837), a Novel Potentiator Which Can Open Class III Mutant Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Channels to a High Extent. J. Med. Chem. 2018, 61, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Veit, G.; Xu, H.; Dreano, E.; Avramescu, R.G.; Bagdany, M.; Beitel, L.K.; Roldan, A.; Hancock, M.A.; Lay, C.; Li, W.; et al. Structure-guided combination therapy to potently improve the function of mutant CFTRs. Nat. Med. 2018, 24, 1732–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, B.; Searle, X.; Yeung, C.; Bogdan, A.; Greszler, S.; Singh, A.; Fan, Y.; Swensen, A.M.; Vortherms, T.; et al. Discovery of 4-[(2R,4R)-4-({[1-(2,2-Difluoro-1,3-benzodioxol-5-yl)cyclopropyl]carbonyl}amino)-7-(difluoromethoxy)-3,4-dihydro-2H-chromen-2-yl]benzoic Acid (ABBV/GLPG-2222), a Potent Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Corrector for the Treatment of Cystic Fibrosis. J. Med. Chem. 2018, 61, 1436–1449. [Google Scholar] [CrossRef]
- Welcome to CandActCFTR. Available online: https://candactcftr.ams.med.uni-goettingen.de/ (accessed on 26 January 2021).
- Nietert, M.M.; Vinhoven, L.; Auer, F.; Hafkemeyer, S.; Stanke, F. Comprehensive Analysis of Chemical Structures That Have Been Tested as CFTR Activating Substances in a Publicly Available Database CandActCFTR. Front. Pharmacol. 2021, 12, 689205. [Google Scholar] [CrossRef]
- CF-Map. Available online: https://cf-map.uni-goettingen.de/ (accessed on 29 June 2022).
- Vinhoven, L.; Stanke, F.; Hafkemeyer, S.; Nietert, M.M. CFTR Lifecycle Map—A Systems Medicine Model of CFTR Maturation to Predict Possible Active Compound Combinations. Int. J. Mol. Sci. 2021, 22, 7590. [Google Scholar] [CrossRef]
- Xu, X.; Huang, M.; Zou, X. Docking-based inverse virtual screening: Methods, applications, and challenges. Biophys. Reports 2018, 4, 1–16. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, G.; Zhou, Y.; Lin, C.; Chen, S.; Lin, Y.; Mai, S.; Huang, Z. Reverse screening methods to search for the protein targets of chemopreventive compounds. Front. Chem. 2018, 6, 138. [Google Scholar] [CrossRef] [PubMed]
- Lim, T.G.; Lee, S.Y.; Huang, Z.; Lim, D.Y.; Chen, H.; Jung, S.K.; Bode, A.M.; Lee, K.W.; Dong, Z. Curcumin suppresses proliferation of colon cancer cells by targeting CDK2. Cancer Prev. Res. 2014, 7, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Buendia-Atencio, C.; Pieffet, G.P.; Montoya-Vargas, S.; Martínez Bernal, J.A.; Rangel, H.R.; Muñoz, A.L.; Losada-Barragán, M.; Segura, N.A.; Torres, O.A.; Bello, F.; et al. Inverse Molecular Docking Study of NS3-Helicase and NS5-RNA Polymerase of Zika Virus as Possible Therapeutic Targets of Ligands Derived from Marcetia taxifolia and Its Implications to Dengue Virus. ACS Omega 2021, 6, 6134–6143. [Google Scholar] [CrossRef] [PubMed]
- Ban, F.; Hu, L.; Zhou, X.H.; Zhao, Y.; Mo, H.; Li, H.; Zhou, W. Inverse molecular docking reveals a novel function of thymol: Inhibition of fat deposition induced by high-dose glucose in Caenorhabditis elegans. Food Sci. Nutr. 2021, 9, 4243–4253. [Google Scholar] [CrossRef] [PubMed]
- Lauro, G.; Romano, A.; Riccio, R.; Bifulco, G. Inverse virtual screening of antitumor targets: Pilot study on a small database of natural bioactive compounds. J. Nat. Prod. 2011, 74, 1401–1407. [Google Scholar] [CrossRef]
- RCSB Research Collaboratory for Structural Bioinformatics (RCSB).
- AlphaFold Protein Structure Database. Available online: https://alphafold.ebi.ac.uk/ (accessed on 3 June 2022).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef]
- Hassan, N.M.; Alhossary, A.A.; Mu, Y.; Kwoh, C.K. Protein-Ligand Blind Docking Using QuickVina-W with Inter-Process Spatio-Temporal Integration. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Kim, S.S.; Aprahamian, M.L.; Lindert, S. Improving inverse docking target identification with Z-score selection. Chem. Biol. Drug Des. 2019, 93, 1105–1116. [Google Scholar] [CrossRef]
- Gilson, M.K.; Liu, T.; Baitaluk, M.; Nicola, G.; Hwang, L.; Chong, J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016, 44, D1045–D1053. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.; Nowotka, M.; Papadatos, G.; Dedman, N.; Gaulton, A.; Atkinson, F.; Bellis, L.; Overington, J.P. ChEMBL web services: Streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015, 43, W612–W620. [Google Scholar] [CrossRef] [PubMed]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; de Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- CHEMBL Database Release 30. 2022. Available online: http://chembl.blogspot.com/2022/03/chembl-30-released.html (accessed on 9 September 2022).
- Fiedorczuk, K.; Chen, J. Mechanism of CFTR correction by type I folding correctors. Cell 2022, 185, 158.e11–168.e11. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, R.; Villarreal, M.A. Vinardo: A scoring function based on autodock vina improves scoring, docking, and virtual screening. PLoS ONE 2016, 11, e0155183. [Google Scholar] [CrossRef] [PubMed]
- Hanson, S.M.; Georghiou, G.; Thakur, M.K.; Miller, W.T.; Rest, J.S.; Chodera, J.D.; Seeliger, M.A. What Makes a Kinase Promiscuous for Inhibitors? Cell Chem. Biol. 2019, 26, 390.e5–399.e5. [Google Scholar] [CrossRef]
- Cerisier, N.; Petitjean, M.; Regad, L.; Bayard, Q.; Réau, M.; Badel, A.; Camproux, A.C. High impact: The role of promiscuous binding sites in polypharmacology. Molecules 2019, 24, 2529. [Google Scholar] [CrossRef]
- Ameen, N.; Silvis, M.; Bradbury, N.A. Endocytic trafficking of CFTR in health and disease. J. Cyst. Fibros. 2007, 6, 1–14. [Google Scholar] [CrossRef]
- Farinha, C.M.; Matos, P. Rab GTPases regulate the trafficking of channels and transporters—A focus on cystic fibrosis. Small GTPases 2018, 9, 136–144. [Google Scholar] [CrossRef]
- Hou, X.; Wu, Q.; Rajagopalan, C.; Zhang, C.; Bouhamdan, M.; Wei, H.; Chen, X.; Zaman, K.; Li, C.; Sun, X.; et al. CK19 stabilizes CFTR at the cell surface by limiting its endocytic pathway degradation. FASEB J. 2019, 33, 12602–12615. [Google Scholar] [CrossRef]
- Muthyala, R.S.; Ju, Y.H.; Sheng, S.; Williams, L.D.; Doerge, D.R.; Katzenellenbogen, B.S.; Helferich, W.G.; Katzenellenbogen, J.A. Equol, a natural estrogenic metabolite from soy isoflavones: Convenient preparation and resolution of R- and S-equols and their differing binding and biological activity through estrogen receptors alpha and beta. Bioorganic Med. Chem. 2004, 12, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Pyle, L.C.; Fulton, J.C.; Sloane, P.A.; Backer, K.; Mazur, M.; Prasain, J.; Barnes, S.; Clancy, J.P.; Rowe, S.M. Activation of the cystic fibrosis transmembrane conductance regulator by the flavonoid quercetin: Potential use as a biomarker of ΔF508 cystic fibrosis transmembrane conductance regulator rescue. Am. J. Respir. Cell Mol. Biol. 2010, 43, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Younger, J.M.; Chen, L.; Ren, H.-Y.; Rosser, M.F.N.; Turnbull, E.L.; Fan, C.-Y.; Patterson, C.; Cyr, D.M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 2006, 126, 571–582. [Google Scholar] [CrossRef]
- Grove, D.E.; Fan, C.-Y.; Ren, H.Y.; Cyr, D.M. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTRDeltaF508. Mol. Biol. Cell 2011, 22, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Thiel, K.; Wiswedel, B. KNIME—The Konstanz information miner. ACM SIGKDD Explor. Newsl. 2009, 11, 26–31. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Open Babel. Available online: http://openbabel.org/wiki/Main_Page (accessed on 20 May 2022).
- Gorgulla, C.; Boeszoermenyi, A.; Wang, Z.F.; Fischer, P.D.; Coote, P.W.; Padmanabha Das, K.M.; Malets, Y.S.; Radchenko, D.S.; Moroz, Y.S.; Scott, D.A.; et al. An open-source drug discovery platform enables ultra-large virtual screens. Nature 2020, 580, 663–668. [Google Scholar] [CrossRef]
- SLURM. Available online: https://slurm.schedmd.com (accessed on 9 September 2022).
- Data Analytics Platform: Open Source Software Tools|KNIME. Available online: https://www.knime.com/knime-analytics-platform (accessed on 31 May 2022).
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Southan, C. InChI in the wild: An assessment of InChIKey searching in Google. J. Cheminform. 2013, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.L. The Generation of a Unique Machine Description for Chemical Structures—A Technique Developed at Chemical Abstracts Service. J. Chem. Doc. 1965, 5, 107–113. [Google Scholar] [CrossRef]
- RDKit. Available online: https://www.rdkit.org/ (accessed on 2 June 2022).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vinhoven, L.; Stanke, F.; Hafkemeyer, S.; Nietert, M.M. Complementary Dual Approach for In Silico Target Identification of Potential Pharmaceutical Compounds in Cystic Fibrosis. Int. J. Mol. Sci. 2022, 23, 12351. https://doi.org/10.3390/ijms232012351
Vinhoven L, Stanke F, Hafkemeyer S, Nietert MM. Complementary Dual Approach for In Silico Target Identification of Potential Pharmaceutical Compounds in Cystic Fibrosis. International Journal of Molecular Sciences. 2022; 23(20):12351. https://doi.org/10.3390/ijms232012351
Chicago/Turabian StyleVinhoven, Liza, Frauke Stanke, Sylvia Hafkemeyer, and Manuel Manfred Nietert. 2022. "Complementary Dual Approach for In Silico Target Identification of Potential Pharmaceutical Compounds in Cystic Fibrosis" International Journal of Molecular Sciences 23, no. 20: 12351. https://doi.org/10.3390/ijms232012351
APA StyleVinhoven, L., Stanke, F., Hafkemeyer, S., & Nietert, M. M. (2022). Complementary Dual Approach for In Silico Target Identification of Potential Pharmaceutical Compounds in Cystic Fibrosis. International Journal of Molecular Sciences, 23(20), 12351. https://doi.org/10.3390/ijms232012351





