The PSI Domain of the MET Oncogene Encodes a Functional Disulfide Isomerase Essential for the Maturation of the Receptor Precursor
Abstract
:1. Introduction
2. Results
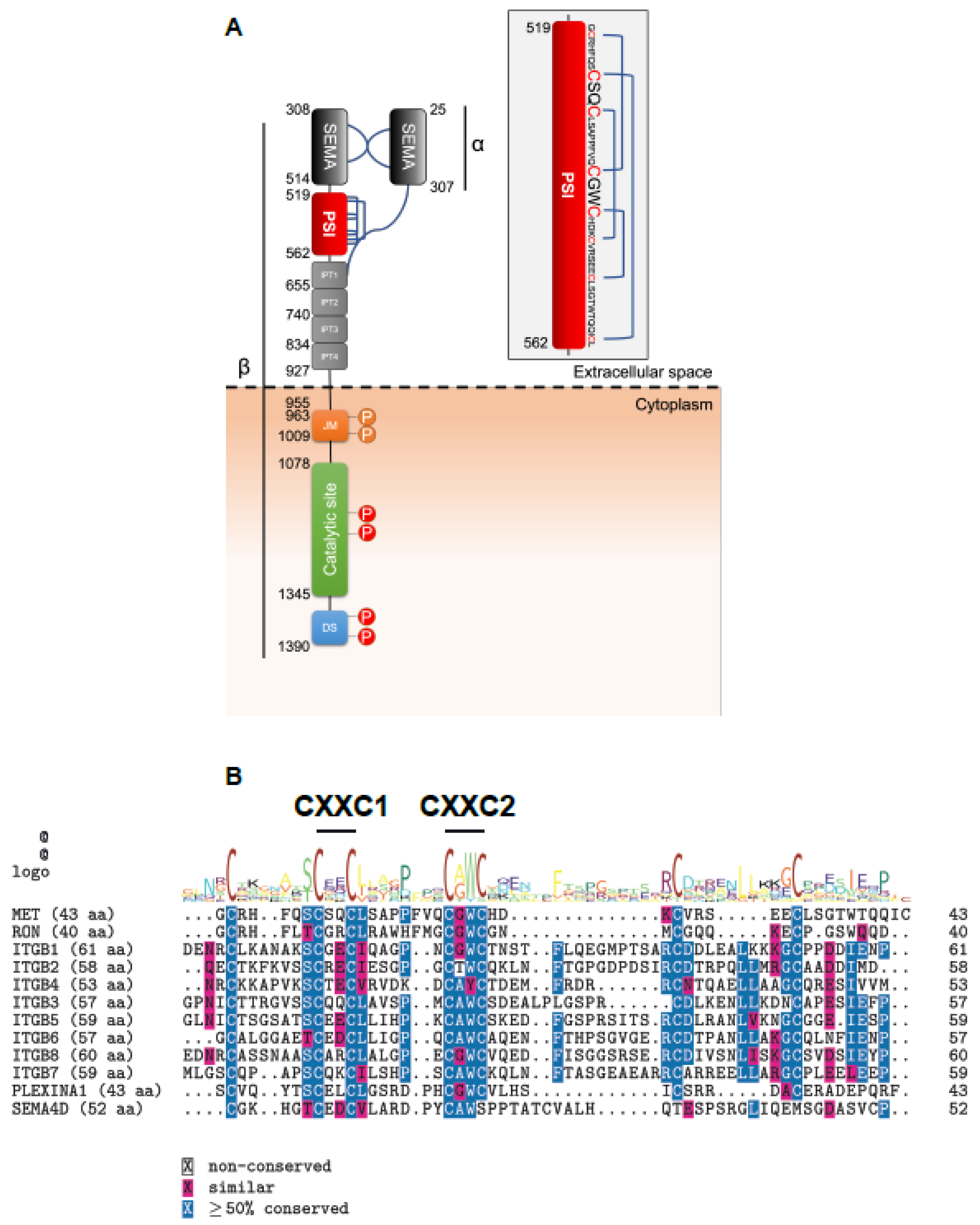
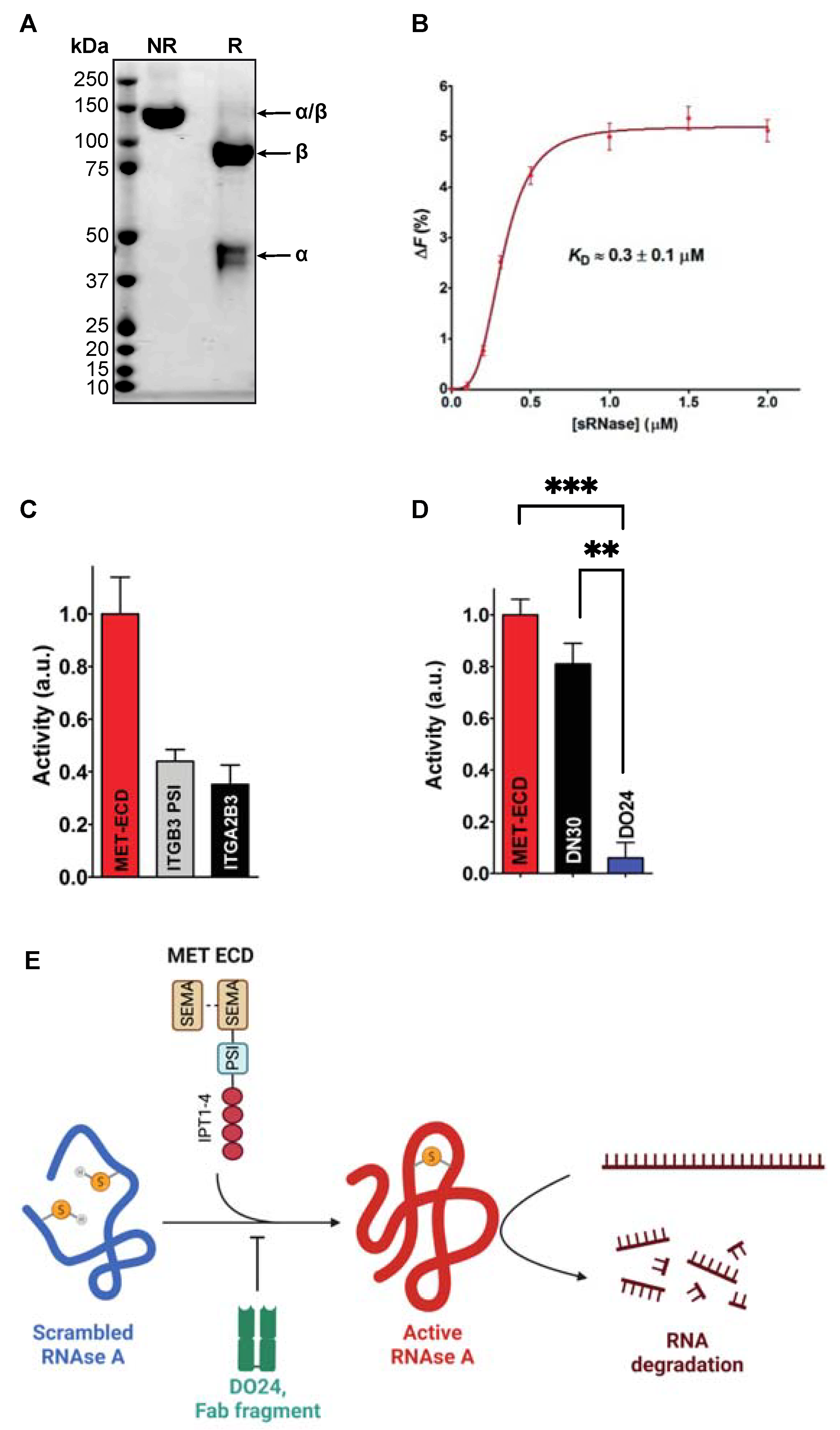
2.1. The MET Extracellular Domain Displays Disulfide Isomerase Activity In Vitro
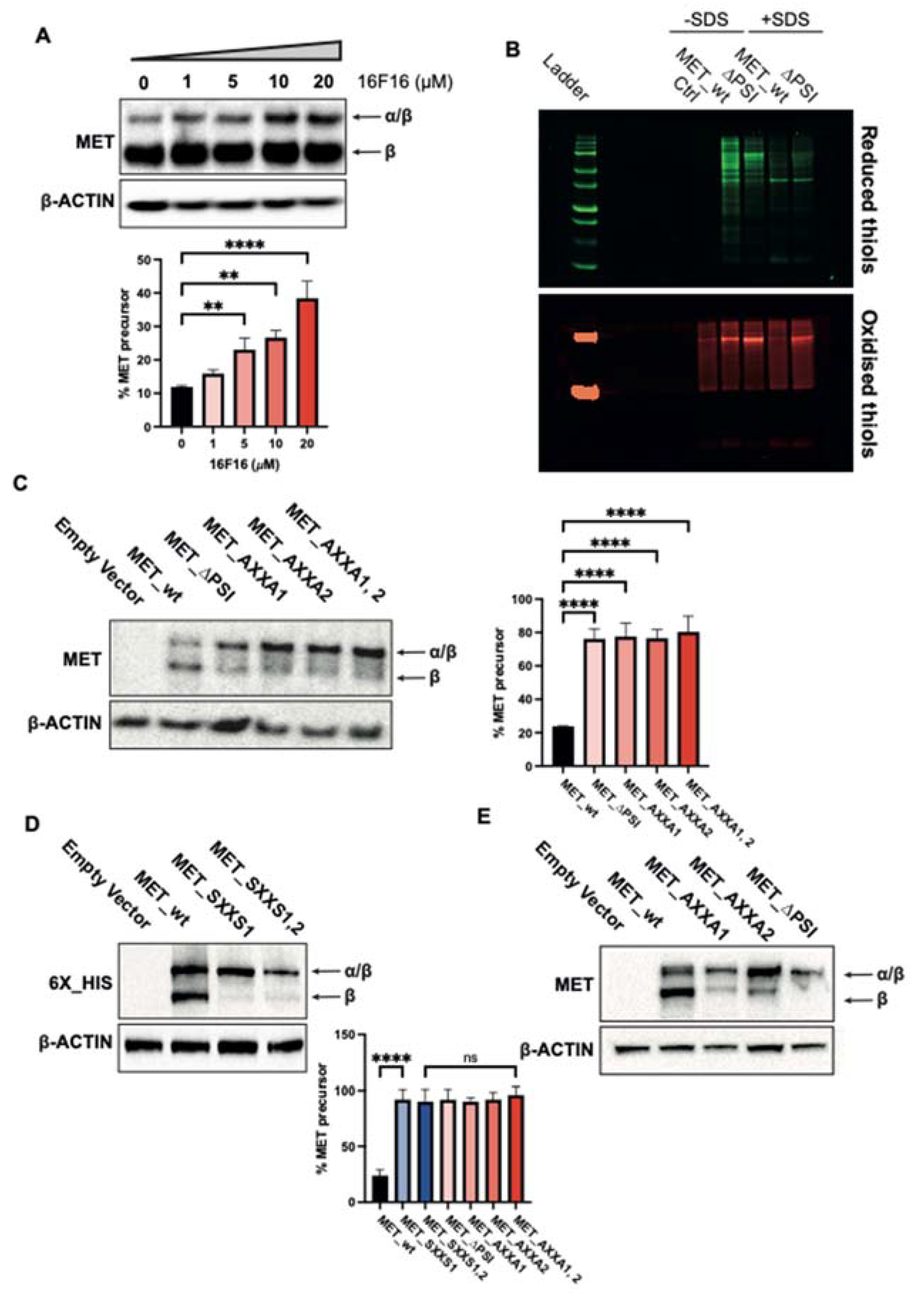
2.2. The PSI Domain Is Crucial for MET Maturation
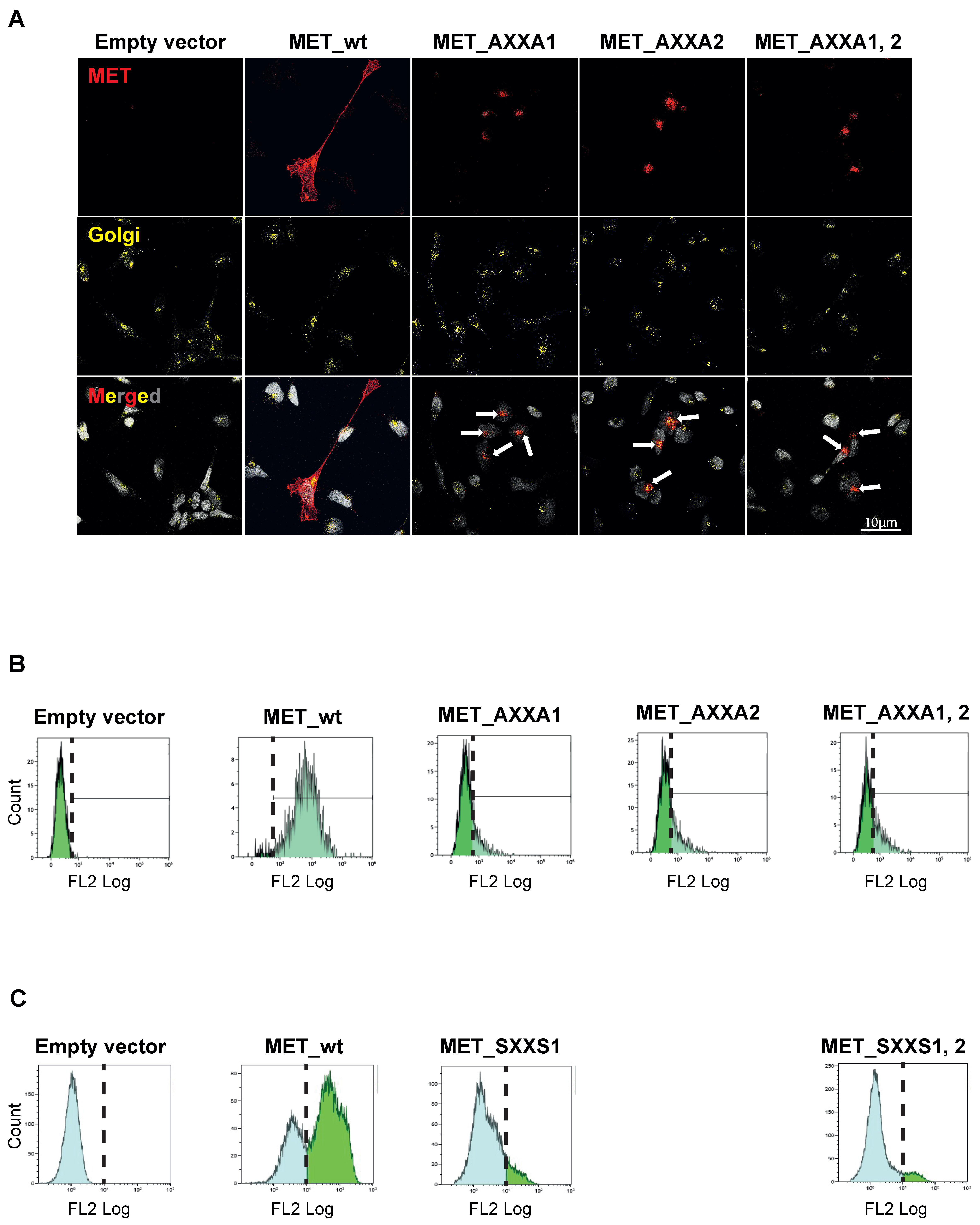
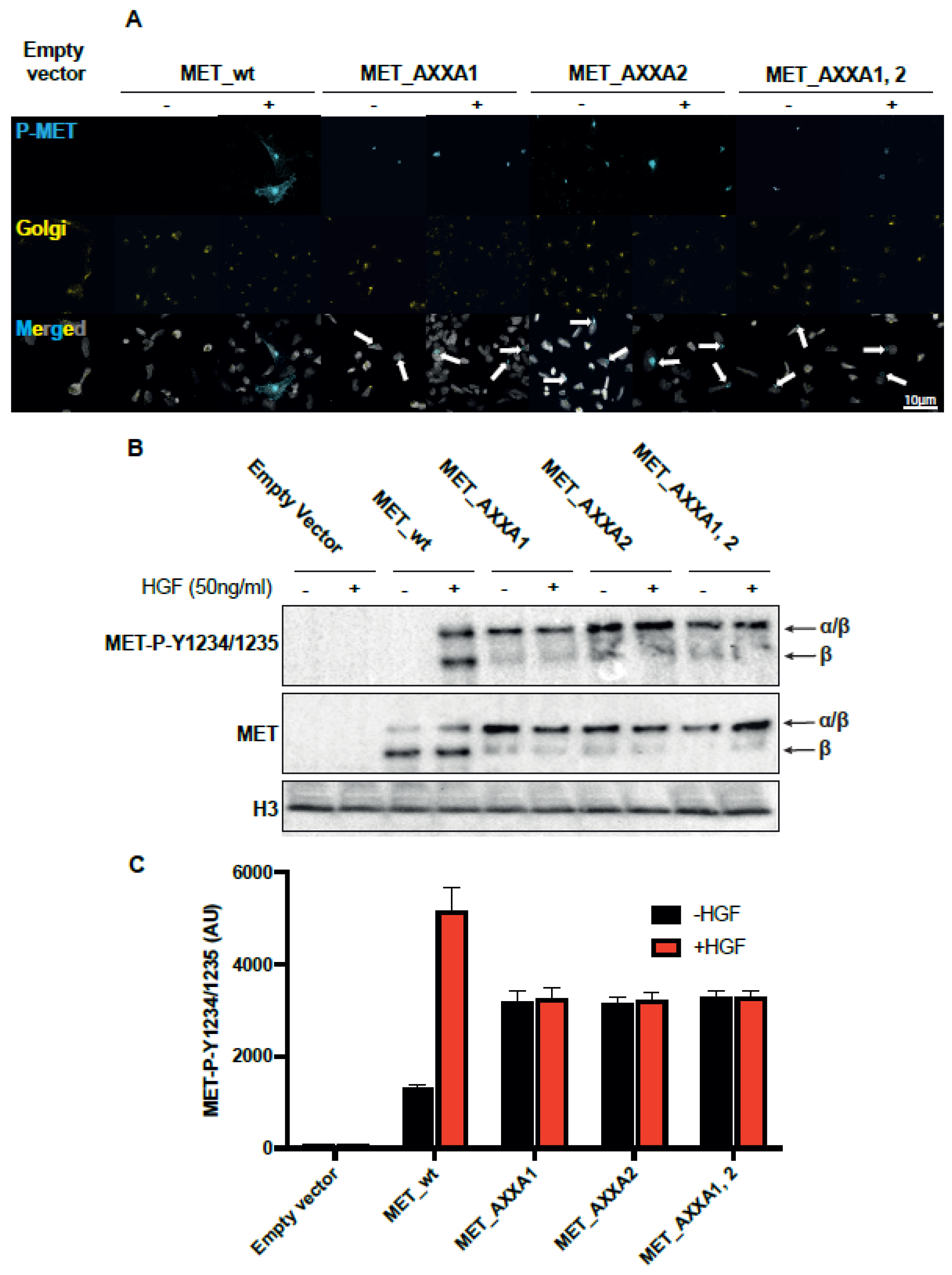
2.3. MET_CXXC Mutants Are Trapped in the Golgi Apparatus and Constitutively Phosphorylated
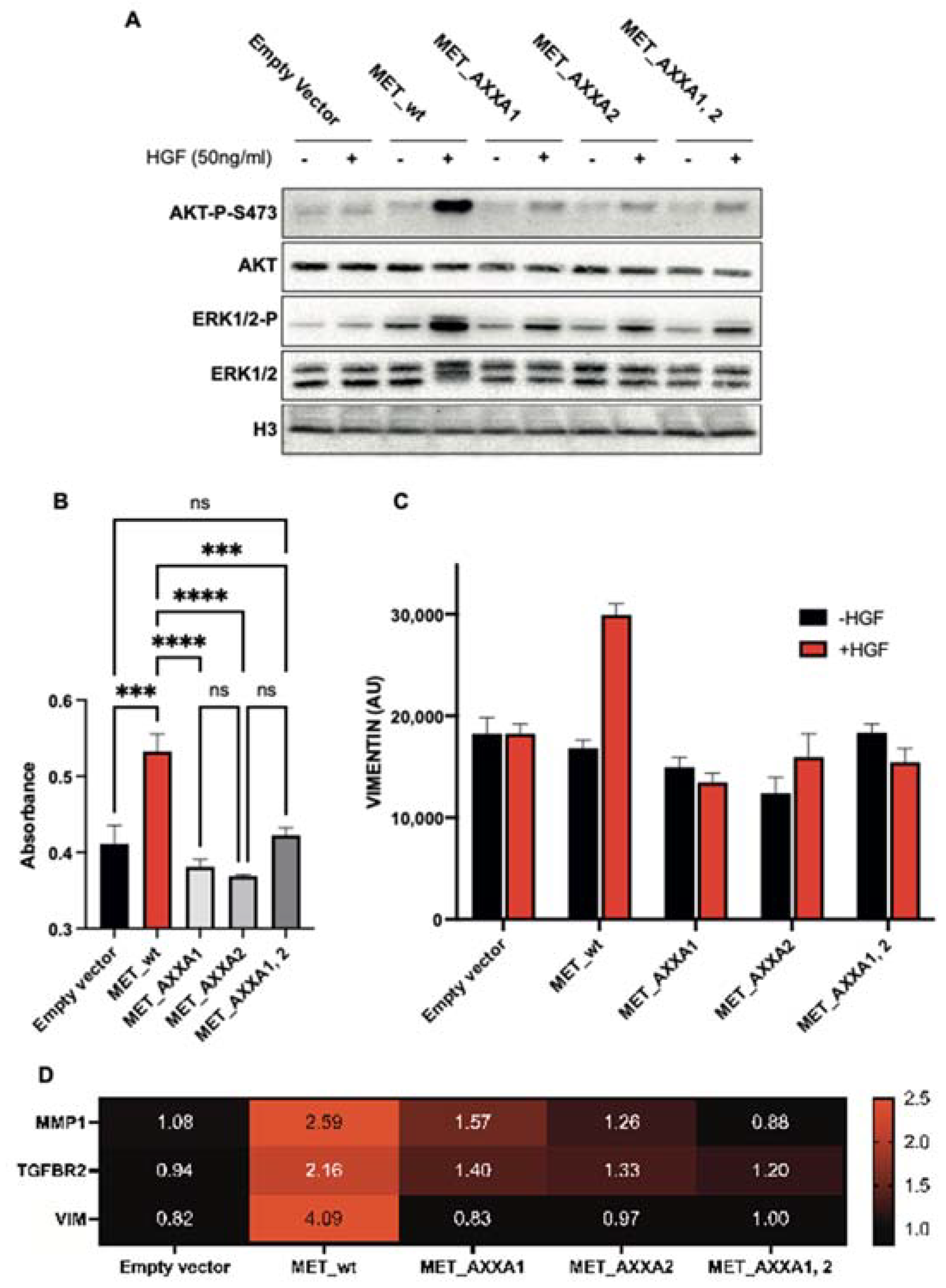
2.4. The Misplaced MET Is Biologically Inactive
3. Materials and Methods
3.1. Chemicals and Antibodies
3.2. Generation, Expression and Purification of the MET Ectodomain
3.3. Preparation of Scrambled RNAse and RNAse Activity Assay
3.4. Fluorescence Measurements
3.5. Cell Culture
3.6. Lentiviral Vectors
3.7. Plasmids
3.8. Western Blot Analyses
3.9. Flow Cytometry
3.10. Immunofluorescence Analysis
3.11. Viability Assay
3.12. Statistical Analyses
3.13. Determination of Cellular Thiol–Disulfide Redox Equilibrium
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uehara, Y.; Minowa, O.; Mori, C.; Shiota, K.; Kuno, J.; Noda, T.; Kitamura, N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 1995, 373, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Bussolino, F.; Di Renzo, M.F.; Ziche, M.; Bocchietto, E.; Olivero, M.; Naldini, L.; Gaudino, G.; Tamagnone, L.; Coffer, A.; Comoglio, P. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth. J. Cell Biol. 1992, 119, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trusolino, L.; Bertotti, A.; Comoglio, P.M. MET signalling: Principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Gherardi, E.; Birchmeier, W.; Birchmeier, C.; Woude, G.V. Targeting MET in cancer: Rationale and progress. Nat. Rev. Cancer 2012, 12, 89–103. [Google Scholar] [CrossRef]
- Comoglio, M.P.; Trusolino, L.; Boccaccio, C. Known and novel roles of the MET oncogene in cancer: A coherent approach to targeted therapy. Nat. Rev. Cancer 2018, 18, 341–358. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Gonçalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Komada, M.; Hatsuzawa, K.; Shibamoto, S.; Ito, F.; Nakayama, K.; Kitamura, N. Proteolytic processing of the hepatocyte growth factor/scatter factor receptor by furin. FEBS Lett. 1993, 328, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Giordano, S.; Di Renzo, M.F.; Narsimhan, R.P.; Cooper, C.S.; Rosa, C.; Comoglio, P. Biosynthesis of the protein encoded by the c-met proto-oncogene. Oncogene 1989, 4, 1383–1388. [Google Scholar]
- Gravee, C.R.; Tolbert, D.; Woude, G.F.V. MET: A critical player in tumorigenesis and therapeutic target. Cold Spring Harb. Perspect. Biol. 2013, 5, a009209. [Google Scholar]
- Comoglio, M.P.; Giordano, S.; Trusolino, L. Drug development of MET inhibitors: Targeting oncogene addiction and expedience. Nat. Rev. Drug Discov. 2008, 7, 504–516. [Google Scholar] [CrossRef]
- Basilico, C.; Arnesano, A.; Galluzzo, M.; Comoglio, P.; Michieli, P. A High Affinity Hepatocyte Growth Factor-binding Site in the Immunoglobulin-like Region of Met. J. Biol. Chem. 2008, 283, 21267–21277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarnegar, R.; DeFrances, M.C.; Oliver, L.; Michalopoulos, G. Identification and partial characterization of receptor binding sites for HGF on rat hepatocytes. Biochem. Biophys. Res. Commun. 1990, 173, 1179–1185. [Google Scholar] [CrossRef]
- Kozlov, G.; Perreault, A.; Schrag, J.D.; Park, M.; Cygler, M.; Gehring, K.; Ekiel, I. Insights into function of PSI domains from structure of the Met receptor PSI domain. Biochem. Biophys. Res. Commun. 2004, 321, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Uchikawa, E.; Chen, Z.; Xiao, G.-Y.; Zhang, X.; Bai, X.-C. Structural basis of the activation of c-MET receptor. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ookura, T.; Kainuma, K.; Kim, H.; Otaka, A.; Fujii, N.; Kawamura, Y. Active Site Peptides with CXXC Motif on Map-Resin Can Mimic Protein Disulfide Isomerase Activity. Biochem. Biophys. Res. Commun. 1995, 213, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Hillson, A.D.; Lambert, N.; Freedman, R.B. Formation and isomerization of disulfide bonds in proteins: Protein disulfide-isomerase. Methods Enzym. 1984, 107, 281–294. [Google Scholar]
- Thompson, D.J.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kunitz, M. A Spectrophotometric Method for the Measurement of Ribonuclease Activity. J. Biol. Chem. 1946, 164, 563–568. [Google Scholar] [CrossRef]
- Denisov, A.Y.; Määttänen, P.; Dabrowski, C.; Kozlov, G.; Thomas, D.Y.; Gehring, K. Solution structure of the bb′ domains of human protein disulfide isomerase. FEBS J. 2009, 276, 1440–1449. [Google Scholar] [CrossRef]
- Nyborg, K.J.; Peersen, O.B. That zincing feeling: The effects of EDTA on the behaviour of zinc-binding transcriptional regulators. Biochem J. 2004, 381, e3–e4. [Google Scholar] [CrossRef]
- Desole, C.; Gallo, S.; Vitacolonna, A.; Vigna, E.; Basilico, C.; Montarolo, F.; Zuppini, F.; Casanova, E.; Miggiano, R.; Ferraris, D.M.; et al. Engineering, Characterization, and Biological Evaluation of an Antibody Targeting the HGF Receptor. Front. Immunol. 2021, 12, 775151. [Google Scholar] [CrossRef] [PubMed]
- Vigna, E.; Chiriaco, C.; Cignetto, S.; Fontani, L.; Basilico, C.; Petronzelli, F.; Comoglio, P.M. Inhibition of ligand-independent constitutive activation of the Met oncogenic receptor by the engineered chemically-modified antibody DN30. Mol. Oncol. 2015, 9, 1760–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffstrom, B.G.; Kaplan, A.; Letso, R.; Schmid, R.S.; Turmel, G.J.; Lo, D.C.; Stockwell, B.R. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat. Chem. Biol. 2010, 6, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Pacello, F.; D’Orazio, M.; Battistoni, A. An ERp57-mediated disulphide exchange promotes the interaction between Burkholderia cenocepacia and epithelial respiratory cells. Sci. Rep. 2016, 6, 21140. [Google Scholar] [CrossRef] [Green Version]
- Young, H.S.; McGowan, L.M.; Jepson, K.A.; Adams, J.C. Impairment of cell adhesion and migration by inhibition of protein disulphide isomerases in three breast cancer cell lines. Biosci. Rep. 2020, 40, BSR20193271. [Google Scholar] [CrossRef]
- Horowitz, M.P.; Milanese, C.; Di Maio, R.; Hu, X.; Montero, L.M.; Sanders, L.H.; Tapias, V.; Sepe, S.; Van Cappellen, W.A.; Burton, E.; et al. Single-Cell Redox Imaging Demonstrates a Distinctive Response of Dopaminergic Neurons to Oxidative Insults. Antioxidants Redox Signal. 2011, 15, 855–871. [Google Scholar] [CrossRef] [Green Version]
- Milanese, C.; Tapias, V.; Gabriels, S.; Cerri, S.; Levandis, G.; Blandini, F.; Tresini, M.; Shiva, S.; Greenamyre, J.T.; Gladwin, M.T.; et al. Mitochondrial Complex I Reversible S-Nitrosation Improves Bioenergetics and Is Protective in Parkinson’s Disease. Antioxidants Redox Signal. 2018, 28, 44–61. [Google Scholar] [CrossRef]
- Crepaldi, T.; Pollack, A.L.; Prat, M.; Zborek, A.; Mostov, K.; Comoglio, P. Targeting of the SF/HGF receptor to the basolateral domain of polarized epithelial cells. J. Cell Biol. 1994, 125, 313–320. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Matsubara, D.; Goto, A.; Ota, S.; Sachiko, O.; Ishikawa, S.; Aburatani, H.; Miyazawa, K.; Fukayama, M.; Niki, T. Constitutive activation of c-Met is correlated with c-Met overexpression and dependent on cell–matrix adhesion in lung adenocarcinoma cell lines. Cancer Sci. 2007, 99, 14–22. [Google Scholar] [CrossRef]
- Gentile, A.; Trusolino, L.; Comoglio, P.M. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008, 27, 85–94. [Google Scholar] [CrossRef]
- Naran, S.; Zhang, X.; Hughes, S.J. Inhibition of HGF/MET as therapy for malignancy. Expert Opin. Ther. Targets 2009, 13, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, C.H.; Blackburn, E.C.; Freedman, R.B. Comparison of the activities of protein disulphide-isomerase and thioredoxin in catalysing disulphide isomerization in a protein substrate. Biochem. J. 1991, 275 Pt 2, 349–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, L.D. Measurement of ligand binding to proteins by fluorescence spectroscopy. Methods Enzymol. 1985, 117, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Cerqua, M.; Botti, O.; Arigoni, M.; Gioelli, N.; Serini, G.; Calogero, R.; Boccaccio, C.; Comoglio, P.M.; Altintas, D.M. 1MET14 promotes a ligand-dependent, AKT-driven invasive growth. Life Sci. Alliance 2022, 5, e202201409. [Google Scholar] [CrossRef] [PubMed]
- Czibik, G.; Mezdari, Z.; Altintas, D.M.; Bréhat, J.; Pini, M.; D’Humières, T.; Delmont, T.; Radu, C.; Breau, M.; Liang, H.; et al. Dysregulated Phenylalanine Catabolism Plays a Key Role in the Trajectory of Cardiac Aging. Circulation 2021, 144, 559–574. [Google Scholar] [CrossRef]
- Cai, H.; Wang, C.C.; Tsou, C.L. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J. Biol. Chem. 1994, 269, 24550–24552. [Google Scholar] [CrossRef]
- Mondino, A.; Giordano, S.; Comoglio, P.M. Defective posttranslational processing activates the tyrosine kinase encoded by the MET proto-oncogene (hepatocyte growth factor receptor). Mol. Cell Biol. 1991, 11, 6084–6092. [Google Scholar]
- Chakraborty, S.; Li, L.; Puliyappadamba, V.T.; Guo, G.; Hatanpaa, K.J.; Mickey, B.; Souza, R.F.; Vo, P.; Herz, J.; Chen, M.-R.; et al. Constitutive and ligand-induced EGFR signalling triggers distinct and mutually exclusive downstream signalling networks. Nat. Commun. 2014, 5, 5811. [Google Scholar] [CrossRef] [Green Version]
- Chung, I.; Akita, R.W.; Vandlen, R.; Toomre, D.; Schlessinger, J.; Mellman, I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 2010, 464, 783–787. [Google Scholar] [CrossRef]
- Endres, N.F.; Das, R.; Smith, A.W.; Arkhipov, A.; Kovacs, E.; Huang, Y.; Pelton, J.G.; Shan, Y.; Shaw, D.E.; Wemmer, D.E.; et al. Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor. Cell 2013, 152, 543–556. [Google Scholar] [CrossRef] [Green Version]
- Gomes, D.A.; Rodrigues, M.A.; Leite, M.F.; Gomez, M.V.; Varnai, P.; Balla, T.; Bennett, A.M.; Nathanson, M.H. c-Met Must Translocate to the Nucleus to Initiate Calcium Signals. J. Biol. Chem. 2008, 283, 4344–4351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomas, A.; Futter, C.E.; Eden, E.R. EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014, 24, 26–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrow-McGee, R.; Kermorgant, S. Met endosomal signalling: In the right place, at the right time. Int. J. Biochem. Cell Biol. 2014, 49, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Kermorgant, S.; Parker, P.J. c-Met signalling: Spatio-temporal decisions. Cell Cycle 2005, 4, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Frazier, N.M.; Brand, T.; Gordan, J.D.; Grandis, J.; Jura, N. Overexpression-mediated activation of MET in the Golgi promotes HER3/ERBB3 phosphorylation. Oncogene 2019, 38, 1936–1950. [Google Scholar] [CrossRef] [PubMed]
- Furie, B.; Flaumenhaft, R. Thiol isomerases in thrombus formation. Circ. Res. 2014, 114, 1162–1173. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.; Furie, B.C.; Coughlin, S.R.; Furie, B. A critical role for extracellular protein disulfide isomerase during thrombus formation in mice. J. Clin. Investig. 2008, 118, 1123–1131. [Google Scholar] [CrossRef]
- Reinhardt, C.; Von Brühl, M.-L.; Manukyan, D.; Grahl, L.; Lorenz, M.; Altmann, B.; Dlugai, S.; Hess, S.; Konrad, I.; Orschiedt, L.; et al. Protein disulfide isomerase acts as an injury response signal that enhances fibrin generation via tissue factor activation. J. Clin. Investig. 2008, 118, 1110–1122. [Google Scholar] [CrossRef]
- Jasuja, R.; Passam, F.H.; Kennedy, D.R.; Kim, S.H.; Van Hessem, L.; Lin, L.; Bowley, S.R.; Joshi, S.S.; Dilks, J.R.; Furie, B.; et al. Protein disulfide isomerase inhibitors constitute a new class of antithrombotic agents. J. Clin. Investig. 2012, 122, 2104–2113. [Google Scholar] [CrossRef]
- Karala, R.A.; Ruddock, L.W. Bacitracin is not a specific inhibitor of protein disulfide isomerase. FEBS J. 2010, 277, 2454–2462. [Google Scholar] [CrossRef]
- Lovat, P.E.; Corazzari, M.; Armstrong, J.L.; Martin, S.; Pagliarini, V.; Hill, D.; Brown, A.M.; Piacentini, M.; Birch-Machin, M.A.; Redfern, C.P. Increasing Melanoma Cell Death Using Inhibitors of Protein Disulfide Isomerases to Abrogate Survival Responses to Endoplasmic Reticulum Stress. Cancer Res. 2008, 68, 5363–5369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.M.G.; Simizu, S.; Lai, N.S.; Kawatani, M.; Shimizu, T.; Osada, H. Discovery of a Small Molecule PDI Inhibitor That Inhibits Reduction of HIV-1 Envelope Glycoprotein gp120. ACS Chem. Biol. 2010, 6, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Kim, D.; Kim, E.; Sa, J.K.; Lee, H.W.; Yu, S.; Oh, J.; Kim, S.-H.; Yoon, Y.; Nam, D.-H. Tumor Inhibitory Effect of IRCR201, a Novel Cross-Reactive c-Met Antibody Targeting the PSI Domain. Int. J. Mol. Sci. 2017, 18, 1968. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altintas, D.M.; Gallo, S.; Basilico, C.; Cerqua, M.; Bocedi, A.; Vitacolonna, A.; Botti, O.; Casanova, E.; Rancati, I.; Milanese, C.; et al. The PSI Domain of the MET Oncogene Encodes a Functional Disulfide Isomerase Essential for the Maturation of the Receptor Precursor. Int. J. Mol. Sci. 2022, 23, 12427. https://doi.org/10.3390/ijms232012427
Altintas DM, Gallo S, Basilico C, Cerqua M, Bocedi A, Vitacolonna A, Botti O, Casanova E, Rancati I, Milanese C, et al. The PSI Domain of the MET Oncogene Encodes a Functional Disulfide Isomerase Essential for the Maturation of the Receptor Precursor. International Journal of Molecular Sciences. 2022; 23(20):12427. https://doi.org/10.3390/ijms232012427
Chicago/Turabian StyleAltintas, Dogus Murat, Simona Gallo, Cristina Basilico, Marina Cerqua, Alessio Bocedi, Annapia Vitacolonna, Orsola Botti, Elena Casanova, Ilaria Rancati, Chiara Milanese, and et al. 2022. "The PSI Domain of the MET Oncogene Encodes a Functional Disulfide Isomerase Essential for the Maturation of the Receptor Precursor" International Journal of Molecular Sciences 23, no. 20: 12427. https://doi.org/10.3390/ijms232012427
APA StyleAltintas, D. M., Gallo, S., Basilico, C., Cerqua, M., Bocedi, A., Vitacolonna, A., Botti, O., Casanova, E., Rancati, I., Milanese, C., Notari, S., Gambardella, G., Ricci, G., Mastroberardino, P. G., Boccaccio, C., Crepaldi, T., & Comoglio, P. M. (2022). The PSI Domain of the MET Oncogene Encodes a Functional Disulfide Isomerase Essential for the Maturation of the Receptor Precursor. International Journal of Molecular Sciences, 23(20), 12427. https://doi.org/10.3390/ijms232012427




