Mother–Fetus Immune Cross-Talk Coordinates “Extrinsic”/“Intrinsic” Embryo Gene Expression Noise and Growth Stability
Abstract
:1. Introduction
2. Results
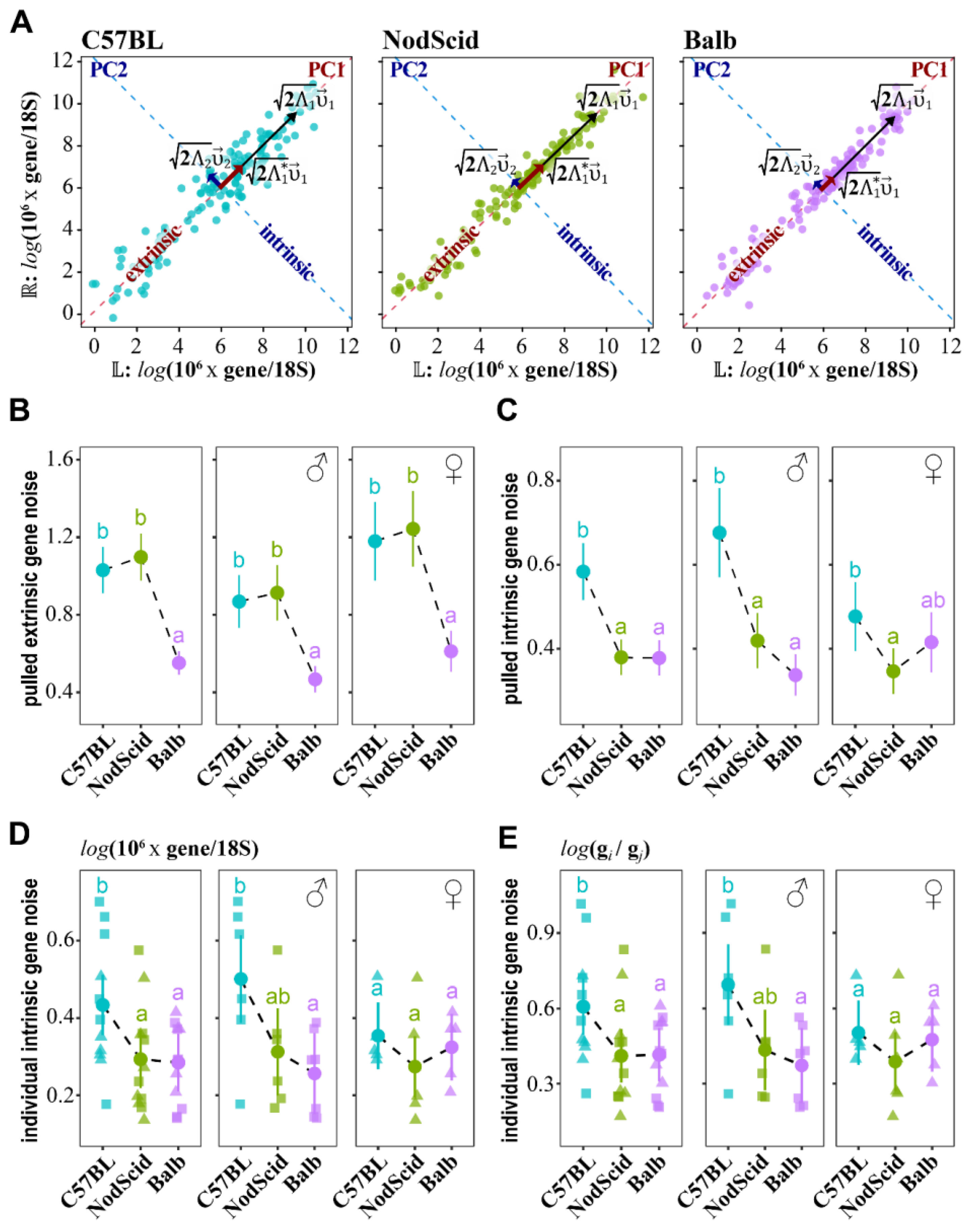
2.1. Estimation of “Extrinsic”/“Intrinsic” Noise from the Log-Transformed Gene/Trait Expressions
2.2. Estimation of Pulled “Extrinsic”/“Intrinsic” Noise
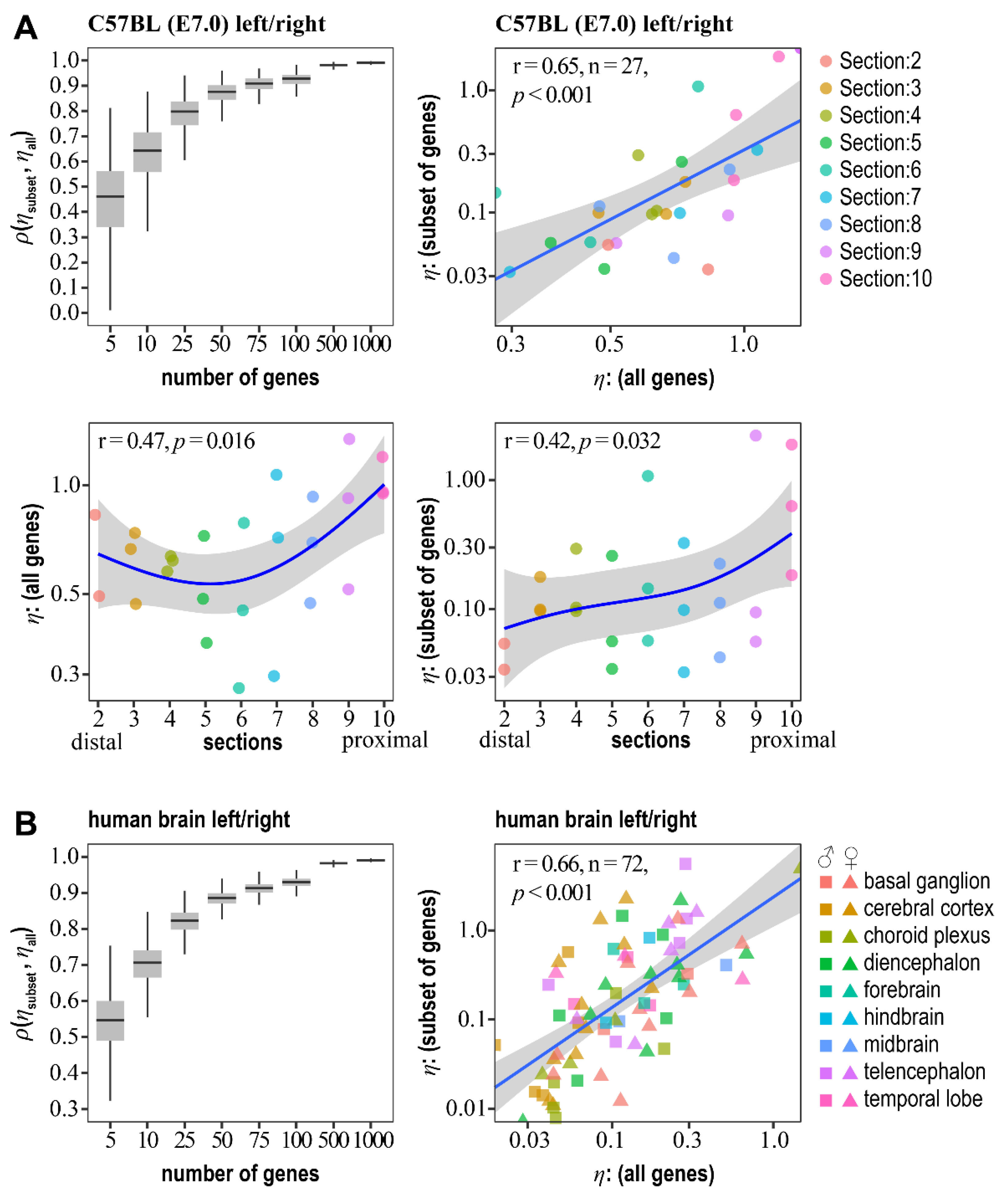
2.3. Estimation of Pulled “Intrinsic” Gene Noise from FA for A Subset of Genes and the Formation of the Distal–Proximal “Intrinsic” Gene Noise Gradient in Mouse Embryo Epiblast
2.4. A Model of Mother–Fetus MHC-Mediated Immune Cross-Talk
2.5. Mother–Fetus Immune Cross-Talk Modulates “Extrinsic”/“Intrinsic” Gene Noise
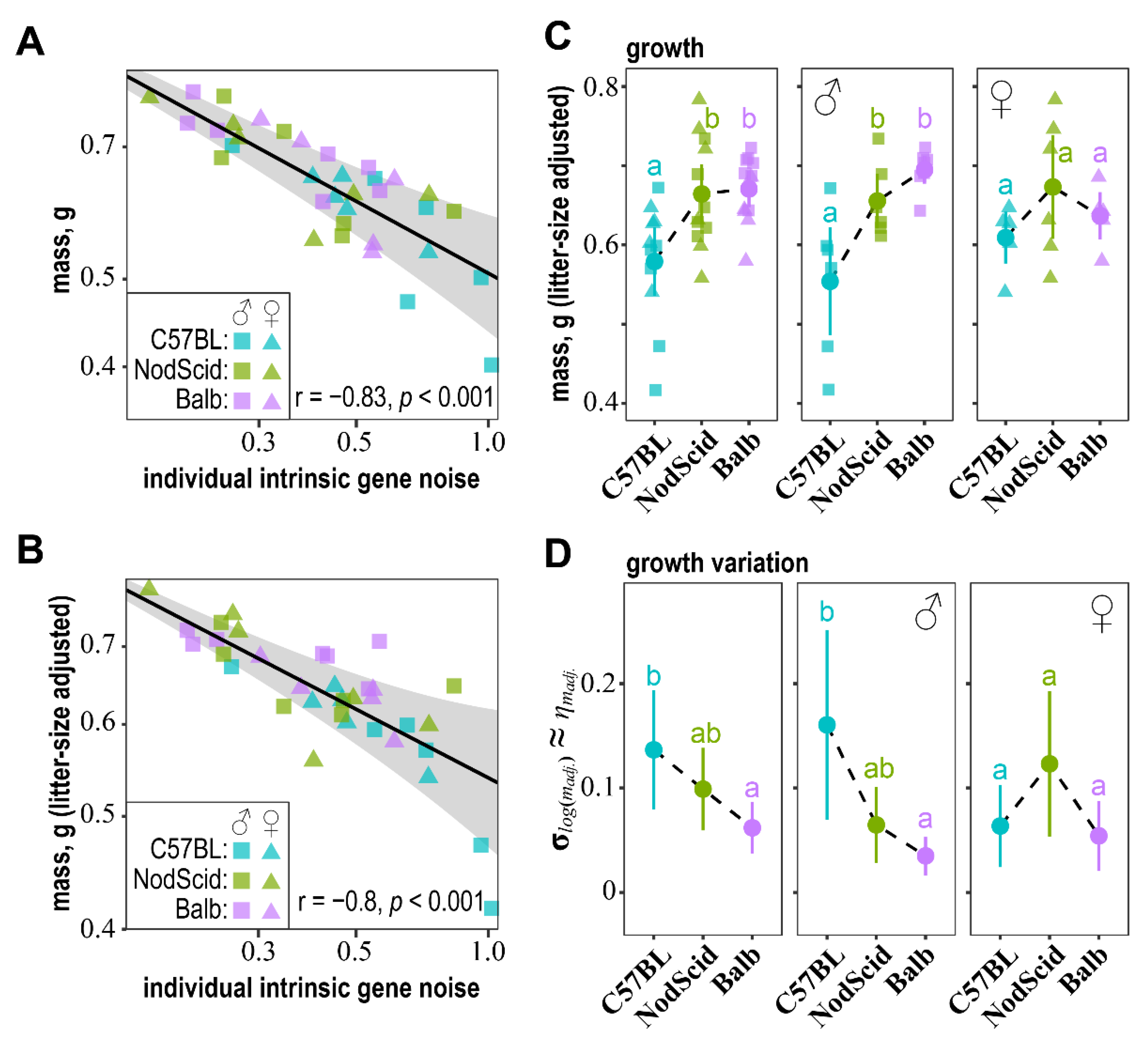
2.6. Association of “Intrinsic” Gene Noise with Embryo Mass and Its Phenotypic Stability
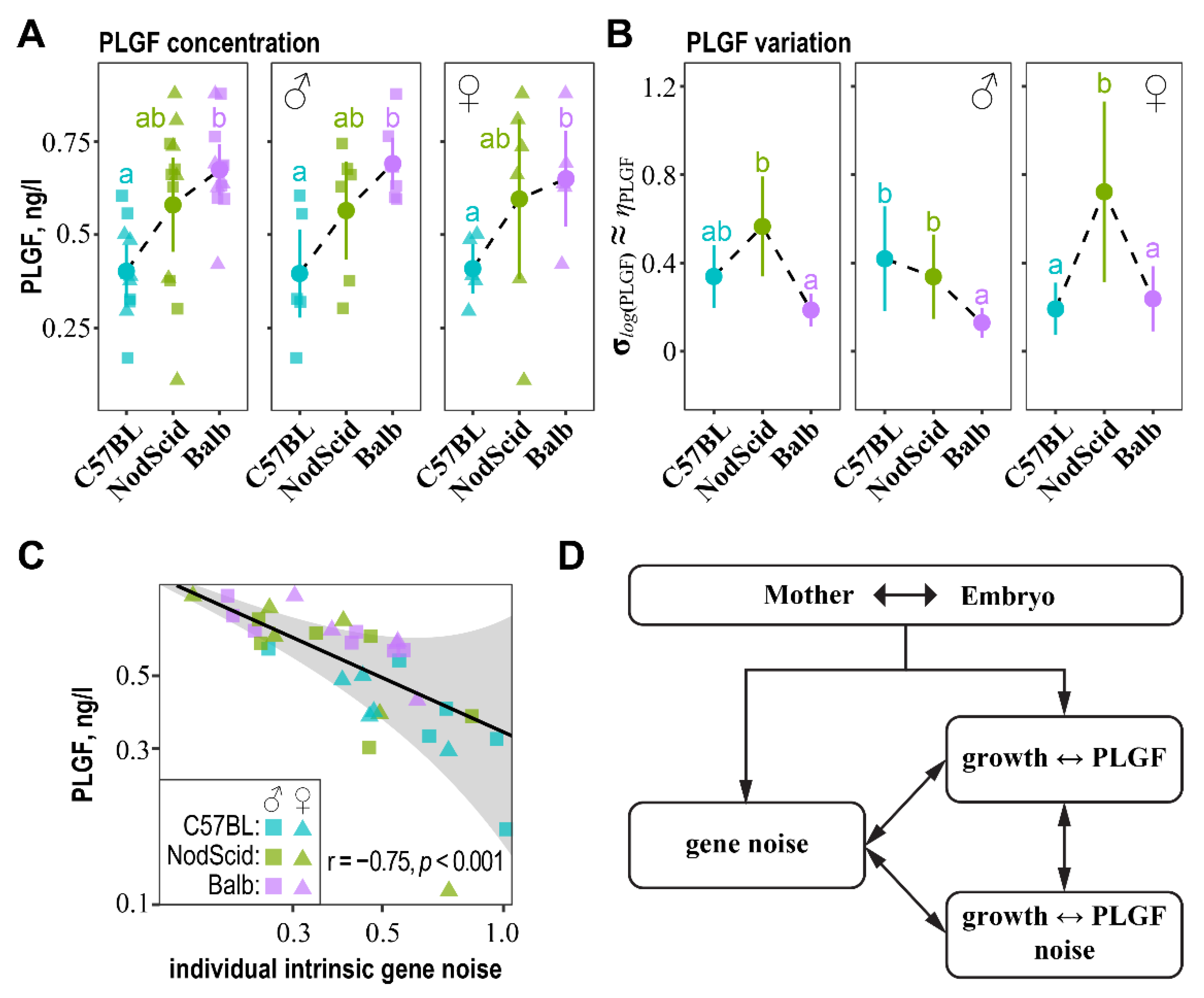
2.7. Association of “Intrinsic” Gene Noise with PLGF Expression and PLGF Noise
3. Discussion
4. Materials and Methods
4.1. Embryo Transfer
4.2. Characteristics of Pregnancy, Embryo Growth, and PLGF and VEGF Immunoassays
4.3. Evaluation of Gene Expression by RT-qPCR
4.4. Statistical Analysis of Noise
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klingenberg, C.P. Phenotypic plasticity, developmental instability, and robustness: The concepts and how they are connected. Front. Ecol. Evol. 2019, 7, 56. [Google Scholar] [CrossRef] [Green Version]
- Dongen, S.V. Fluctuating asymmetry and developmental instability in evolutionary biology: Past, present and future. J. Evol. Biol. 2006, 19, 1727–1743. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.H. Nature, nurture, and noise: Developmental instability, fluctuating asymmetry, and the causes of phenotypic variation. Symmetry 2021, 13, 1204. [Google Scholar] [CrossRef]
- Zakharov, V.M.; Shadrina, E.G.; Trofimov, I.E. Fluctuating asymmetry, developmental noise and developmental stability: Future prospects for the population developmental biology approach. Symmetry 2020, 12, 1376. [Google Scholar] [CrossRef]
- Swain, P.S.; Elowitz, M.B.; Siggia, E.D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. USA 2002, 99, 12795–12800. [Google Scholar] [CrossRef] [Green Version]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef] [Green Version]
- de Jong, T.V.; Moshkin, Y.M.; Guryev, V. Gene expression variability: The other dimension in transcriptome analysis. Physiol. Genom. 2019, 51, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Dattani, J.; Barahona, M. Stochastic models of gene transcription with upstream drives: Exact solution and sample path characterization. J. R. Soc. Interface 2017, 14, 20160833. [Google Scholar] [CrossRef] [Green Version]
- Paulsson, J. Models of stochastic gene expression. Phys. Life Rev. 2005, 2, 157–175. [Google Scholar] [CrossRef]
- Sherman, M.S.; Lorenz, K.; Lanier, M.H.; Cohen, B.A. Cell-to-cell variability in the propensity to transcribe explains correlated fluctuations in gene expression. Cell Syst. 2015, 1, 315–325. [Google Scholar] [CrossRef]
- Graham, J.H. Fluctuating asymmetry and developmental instability, a guide to best practice. Symmetry 2021, 13, 9. [Google Scholar] [CrossRef]
- Palmer, A.R. Fluctuating asymmetry analyses: A primer. In Developmental Instability: Its Origins and Evolutionary Implications: Proceedings of the International Conference on Developmental Instability: Its Origins and Evolutionary Implications, Tempe, Arizona, 14–15 June 1993; Markow, T.A., Ed.; Springer: Dordrecht, The Netherlands, 1994; pp. 335–364. [Google Scholar] [CrossRef]
- Leung, B.; Forbes, M.R.; Houle, D. Fluctuating asymmetry as a bioindicator of stress: Comparing efficacy of analyses involving multiple traits. Am. Nat. 2000, 155, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Leung, B.; Forbes, M.R. Modelling fluctuating asymmetry in relation to stress and fitness. Oikos 1997, 78, 397–405. [Google Scholar] [CrossRef]
- Rantala, M.J.; Ahtiainen, J.J.; Suhonen, J. Fluctuating asymmetry and immune function in a field cricket. Oikos 2004, 107, 479–484. [Google Scholar] [CrossRef]
- Palestis, B.G.; Trivers, R. A longitudinal study of changes in fluctuating asymmetry with age in jamaican youth. Symmetry 2016, 8, 123. [Google Scholar] [CrossRef] [Green Version]
- de Jong, T.V.; Guryev, V.; Moshkin, Y.M. Estimates of gene ensemble noise highlight critical pathways and predict disease severity in H1N1, COVID-19 and mortality in sepsis patients. Sci. Rep. 2021, 11, 10793. [Google Scholar] [CrossRef]
- Juarez-Carreno, S.; Morante, J.; Dominguez, M. Systemic signalling and local effectors in developmental stability, body symmetry, and size. Cell Stress 2018, 2, 340–361. [Google Scholar] [CrossRef]
- Debat, V.; Peronnet, F. Asymmetric flies: The control of developmental noise in Drosophila. Fly 2013, 7, 70–77. [Google Scholar] [CrossRef] [Green Version]
- Dardalhon-Cumenal, D.; Deraze, J.; Dupont, C.A.; Ribeiro, V.; Coleno-Costes, A.; Pouch, J.; Le Crom, S.; Thomassin, H.; Debat, V.; Randsholt, N.B.; et al. Cyclin G and the Polycomb Repressive complexes PRC1 and PR-DUB cooperate for developmental stability. PLoS Genet. 2018, 14, e1007498. [Google Scholar] [CrossRef]
- Vissers, J.H.; van Lohuizen, M.; Citterio, E. The emerging role of Polycomb repressors in the response to DNA damage. J. Cell Sci. 2012, 125, 3939–3948. [Google Scholar] [CrossRef] [Green Version]
- Kar, G.; Kim, J.K.; Kolodziejczyk, A.A.; Natarajan, K.N.; Torlai Triglia, E.; Mifsud, B.; Elderkin, S.; Marioni, J.C.; Pombo, A.; Teichmann, S.A. Flipping between Polycomb repressed and active transcriptional states introduces noise in gene expression. Nat. Commun. 2017, 8, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarosz, D.F.; Taipale, M.; Lindquist, S. Protein homeostasis and the phenotypic manifestation of genetic diversity: Principles and mechanisms. Annu. Rev. Genet. 2010, 44, 189–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutherford, S.L.; Lindquist, S. Hsp90 as a capacitor for morphological evolution. Nature 1998, 396, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Debat, V.; Milton, C.C.; Rutherford, S.; Klingenberg, C.P.; Hoffmann, A.A. Hsp90 and the quantitative variation of wing shape in Drosophila melanogaster. Evol. Int. J. Org. Evol. 2006, 60, 2529–2538. [Google Scholar] [CrossRef]
- Garelli, A.; Gontijo, A.M.; Miguela, V.; Caparros, E.; Dominguez, M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 2012, 336, 579–582. [Google Scholar] [CrossRef]
- Indrasamy, H.; Woods, R.E.; McKenzie, J.A.; Batterham, P. Fluctuating asymmetry for specific bristle characters in notch mutants of Drosophila melanogaster. Genetica 2000, 109, 151–159. [Google Scholar] [CrossRef]
- Leamy, L.J.; Klingenberg, C.P. The genetics and evolution of fluctuating asymmetry. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Pertoldi, C.; Kristensen, T.N.; Andersen, D.H.; Loeschcke, V. Developmental instability as an estimator of genetic stress. Heredity 2006, 96, 122–127. [Google Scholar] [CrossRef] [Green Version]
- Depierreux, D.M.; Kieckbusch, J.; Shreeve, N.; Hawkes, D.A.; Marsh, B.; Blelloch, R.; Sharkey, A.; Colucci, F. Beyond maternal tolerance: Education of uterine natural killer cells by maternal MHC drives fetal growth. Front. Immunol. 2022, 13, 808227. [Google Scholar] [CrossRef]
- Yang, F.; Zheng, Q.; Jin, L. Dynamic function and composition changes of immune cells during normal and pathological pregnancy at the maternal-fetal interface. Front. Immunol. 2019, 10, 2317. [Google Scholar] [CrossRef] [Green Version]
- Gerlinskaya, L.A.; Litvinova, E.A.; Kontsevaya, G.V.; Feofanova, N.A.; Achasova, K.M.; Anisimova, M.V.; Maslennikova, S.O.; Zolotykh, M.A.; Moshkin, Y.M.; Moshkin, M.P. Phenotypic variations in transferred progeny due to genotype of surrogate mother. Mol. Hum. Reprod. 2019, 25, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Madeja, Z.; Yadi, H.; Apps, R.; Boulenouar, S.; Roper, S.J.; Gardner, L.; Moffett, A.; Colucci, F.; Hemberger, M. Paternal MHC expression on mouse trophoblast affects uterine vascularization and fetal growth. Proc. Natl. Acad. Sci. USA 2011, 108, 4012–4017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, S.A.; Moldenhauer, L.M. Immunological determinants of implantation success. Int. J. Dev. Biol. 2014, 58, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Vrachnis, N.; Kalampokas, E.; Sifakis, S.; Vitoratos, N.; Kalampokas, T.; Botsis, D.; Iliodromiti, Z. Placental growth factor (PlGF): A key to optimizing fetal growth. J. Matern.-Fetal Neonatal Med. 2013, 26, 995–1002. [Google Scholar] [CrossRef]
- Pollheimer, J.; Vondra, S.; Baltayeva, J.; Beristain, A.G.; Knofler, M. Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front. Immunol. 2018, 9, 2597. [Google Scholar] [CrossRef] [Green Version]
- Beal, J. Biochemical complexity drives log-normal variation in genetic expression. Eng. Biol. 2017, 1, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Moshkin, Y.M. Chromatin—A global buffer for eukaryotic gene control. AIMS Biophys. 2015, 2, 531–554. [Google Scholar] [CrossRef]
- Chereji, R.V.; Kan, T.W.; Grudniewska, M.K.; Romashchenko, A.V.; Berezikov, E.; Zhimulev, I.F.; Guryev, V.; Morozov, A.V.; Moshkin, Y.M. Genome-wide profiling of nucleosome sensitivity and chromatin accessibility in Drosophila melanogaster. Nucleic Acids Res. 2016, 44, 1036–1051. [Google Scholar] [CrossRef] [Green Version]
- McBrian, M.A.; Behbahan, I.S.; Ferrari, R.; Su, T.; Huang, T.W.; Li, K.; Hong, C.S.; Christofk, H.R.; Vogelauer, M.; Seligson, D.B.; et al. Histone acetylation regulates intracellular pH. Mol. Cell 2013, 49, 310–321. [Google Scholar] [CrossRef]
- Peng, G.; Suo, S.; Chen, J.; Chen, W.; Liu, C.; Yu, F.; Wang, R.; Chen, S.; Sun, N.; Cui, G.; et al. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell 2016, 36, 681–697. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, S.J.; Xu, Y.; Lisgo, S.N.; Harkin, L.F.; Copp, A.J.; Gerrelli, D.; Clowry, G.J.; Talbot, A.; Keogh, M.J.; Coxhead, J.; et al. HDBR expression: A unique resource for global and individual gene expression studies during early human brain development. Front. Neuroanat. 2016, 10, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shultz, L.D.; Schweitzer, P.A.; Christianson, S.W.; Gott, B.; Schweitzer, I.B.; Tennent, B.; McKenna, S.; Mobraaten, L.; Rajan, T.V.; Greiner, D.L.; et al. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J. Immunol. 1995, 154, 180–191. [Google Scholar] [PubMed]
- Stasinopoulos, M.D.; Rigby, R.A.; Heller, G.Z.; Voudouris, V.; De Bastiani, F. Flexible Regression and Smoothing: Using GAMLSS in R; Chapman and Hall: London, UK; CRC Press: Boca Raton, FL, USA, 2017; p. 549. [Google Scholar] [CrossRef]
- McFarlane, L.; Truong, V.; Palmer, J.S.; Wilhelm, D. Novel PCR assay for determining the genetic sex of mice. Sex. Dev. 2013, 7, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.S.; Foley, J.E.; Limsirichai, P.; Schaffer, D.V.; Arkin, A.P. Orthogonal control of expression mean and variance by epigenetic features at different genomic loci. Mol. Syst. Biol. 2015, 11, 806. [Google Scholar] [CrossRef]
- Chen, J.; Li, E.; Lai, J. The coupled effect of nucleosome organization on gene transcription level and transcriptional plasticity. Nucleus 2017, 8, 605–612. [Google Scholar] [CrossRef] [Green Version]
- Weinberger, L.; Voichek, Y.; Tirosh, I.; Hornung, G.; Amit, I.; Barkai, N. Expression noise and acetylation profiles distinguish HDAC functions. Mol. Cell 2012, 47, 193–202. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, D.; Zoller, B.; Suter, D.M.; Naef, F. Modulation of transcriptional burst frequency by histone acetylation. Proc. Natl. Acad. Sci. USA 2018, 115, 7153–7158. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Zhou, M.; Chen, S.; Li, D.; Cao, X.; Liu, B. Effects of pH alterations on stress- and aging-induced protein phase separation. Cell. Mol. Life Sci. 2022, 79, 380. [Google Scholar] [CrossRef]
- Araos, J.; Silva, L.; Salsoso, R.; Saez, T.; Barros, E.; Toledo, F.; Gutierrez, J.; Pardo, F.; Leiva, A.; Sanhueza, C.; et al. Intracellular and extracellular pH dynamics in the human placenta from diabetes mellitus. Placenta 2016, 43, 47–53. [Google Scholar] [CrossRef]
- Asakura, H. Fetal and neonatal thermoregulation. J. Nippon Med. Sch. 2004, 71, 360–370. [Google Scholar] [CrossRef] [Green Version]
- Reinhold, K.; Engqvist, L. The variability is in the sex chromosomes. Evol. Int. J. Org. Evol. 2013, 67, 3662–3668. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, J.J.; Møller, A.P. Phenotypic variation and fluctuating asymmetry in sexually dimorphic feather ornaments in relation to sex and mating system. Biol. J. Linn. Soc. 1999, 68, 505–529. [Google Scholar] [CrossRef]
- Zajitschek, S.R.; Zajitschek, F.; Bonduriansky, R.; Brooks, R.C.; Cornwell, W.; Falster, D.S.; Lagisz, M.; Mason, J.; Senior, A.M.; Noble, D.W.; et al. Sexual dimorphism in trait variability and its eco-evolutionary and statistical implications. eLife 2020, 9, e63170. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babochkina, T.I.; Gerlinskaya, L.A.; Anisimova, M.V.; Kontsevaya, G.V.; Feofanova, N.A.; Stanova, A.K.; Moshkin, M.P.; Moshkin, Y.M. Mother–Fetus Immune Cross-Talk Coordinates “Extrinsic”/“Intrinsic” Embryo Gene Expression Noise and Growth Stability. Int. J. Mol. Sci. 2022, 23, 12467. https://doi.org/10.3390/ijms232012467
Babochkina TI, Gerlinskaya LA, Anisimova MV, Kontsevaya GV, Feofanova NA, Stanova AK, Moshkin MP, Moshkin YM. Mother–Fetus Immune Cross-Talk Coordinates “Extrinsic”/“Intrinsic” Embryo Gene Expression Noise and Growth Stability. International Journal of Molecular Sciences. 2022; 23(20):12467. https://doi.org/10.3390/ijms232012467
Chicago/Turabian StyleBabochkina, Tatyana Ivanovna, Ludmila Alekseevna Gerlinskaya, Margarita Vladimirovna Anisimova, Galina Vladimirovna Kontsevaya, Natalia Aleksandrovna Feofanova, Aliya Konstantinovna Stanova, Mikhail Pavlovich Moshkin, and Yuri Mikhailovich Moshkin. 2022. "Mother–Fetus Immune Cross-Talk Coordinates “Extrinsic”/“Intrinsic” Embryo Gene Expression Noise and Growth Stability" International Journal of Molecular Sciences 23, no. 20: 12467. https://doi.org/10.3390/ijms232012467





