Mulberry Component Kuwanon C Exerts Potent Therapeutic Efficacy In Vitro against COVID-19 by Blocking the SARS-CoV-2 Spike S1 RBD:ACE2 Receptor Interaction
Abstract
1. Introduction
2. Results
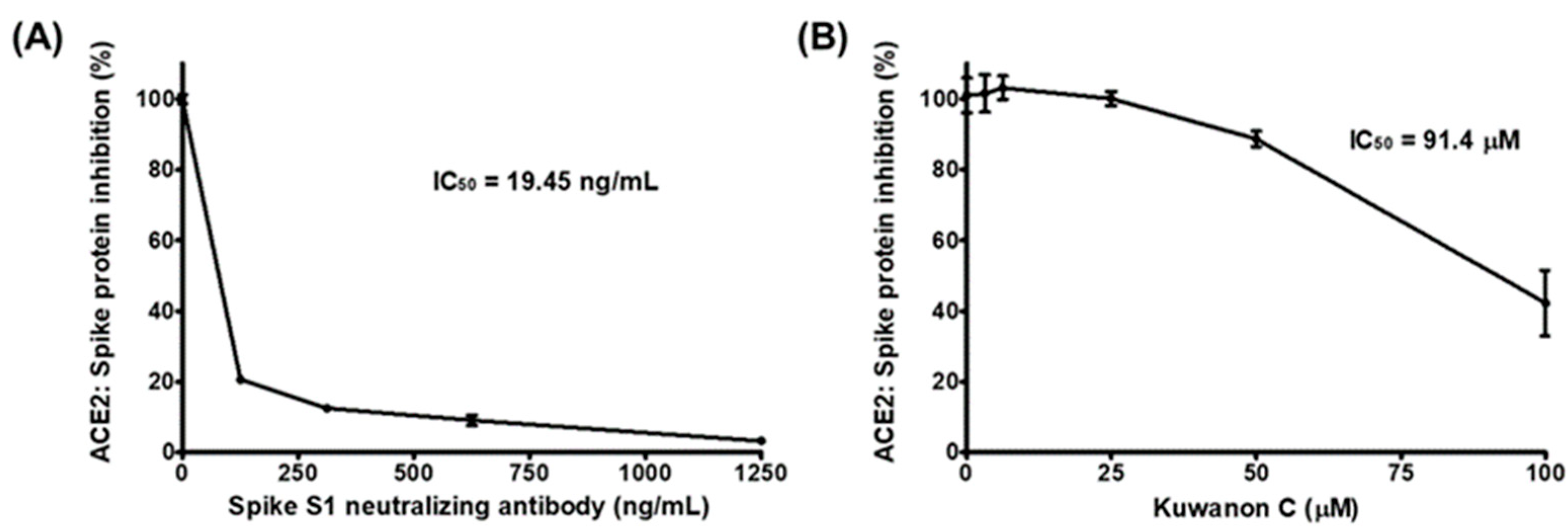
2.1. KC Blocks Binding between Spike S1 RBD and ACE2 Receptor
2.2. Biolayer Interferometry-Based Kinetic Analysis for the Binding of KC to Spike S1 RBD/ACE2 Receptor
2.3. In Silico Docking Simulation and Pharmacophore Analysis of KC with Spike Protein and ACE2 Receptor
2.4. KC Prevents SARS-CoV-2 Lentiviral Pseudovirus from Infecting the ACE2/TMPRSS2-Overexpressing HEK293T Cells
2.5. KC Prevents a Clinical Isolate of SARS-CoV-2 from Infecting Vero Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. SARS-CoV-2 Spike/ACE2 Inhibitor Screening Assay
4.3. Kinetic Analysis of the Binding between KC and Spike Protein/ACE2 Receptor Based on Biolayer Interferometry
4.4. In Silico Docking Simulation and Pharmacophore Analysis
4.5. Cell Viability Assay
4.6. SARS-CoV-2 Lentiviral Pseudovirus Infection Assay
4.7. RNA Isolation and Real-Time PCR
4.8. The Prediction of Physicochemical Properties of KC
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gubareva, L.V.; Kaiser, L.; Hayden, F.G. Influenza virus neuraminidase inhibitors. Lancet 2000, 355, 827–835. [Google Scholar] [CrossRef]
- Kim, C.U.; Lew, W.; Williams, M.A.; Liu, H.; Zhang, L.; Swaminathan, S.; Bischofberger, N.; Chen, M.S.; Mendel, D.B.; Tai, C.Y.; et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: Design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 1997, 119, 681–690. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Gandhi, S.; Klein, J.; Robertson, A.J.; Pena-Hernandez, M.A.; Lin, M.J.; Roychoudhury, P.; Lu, P.; Fournier, J.; Ferguson, D.; Mohamed Bakhash, S.A.K.; et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: A case report. Nat. Commun. 2022, 13, 1547. [Google Scholar] [CrossRef]
- Hu, Y.; Lewandowski, E.M.; Tan, H.; Morgan, R.T.; Zhang, X.; Jacobs, L.M.C.; Butler, S.G.; Mongora, M.V.; Choy, J.; Chen, Y.; et al. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Jochmans, D.; Liu, C.; Donckers, K.; Stoycheva, A.; Boland, S.; Stevens, S.K.; Vita, C.D.; Vanmechelen, B.; Maes, P.; Trüeb, B.; et al. The substitutions L50F, E166A and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Marzolini, C.; Kuritzkes, D.R.; Marra, F.; Boyle, A.; Gibbons, S.; Flexner, C.; Pozniak, A.; Boffito, M.; Waters, L.; Burger, D.; et al. Recommendations for the Management of Drug–Drug Interactions Between the COVID-19 Antiviral Nirmatrelvir/Ritonavir (Paxlovid) and Comedications. Clin. Pharmacol. Ther. 2022. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Singh, A.; Singh, R.; Misra, A. An updated practical guideline on use of molnupiravir and comparison with agents having emergency use authorization for treatment of COVID-19. Diabetes Metab. Syndr. 2022, 16, 102396. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.-T.; Hsu, W.-C.; Lin, C.-C. Antiviral Natural Products and Herbal Medicines. J. Tradit. Complement. Med. 2014, 4, 24–35. [Google Scholar] [CrossRef]
- Rozhon, E.; Albin, R.; Schwartz, J. Strategies for Discovering Antiviral Agents from Natural Products. Biotechnology 1994, 26, 223–245. [Google Scholar] [CrossRef] [PubMed]
- Alkhatib, A. Antiviral Functional Foods and Exercise Lifestyle Prevention of Coronavirus. Nutrients 2020, 12, 2633. [Google Scholar] [CrossRef]
- Farzana, M.; Shahriar, S.; Jeba, F.R.; Tabassum, T.; Araf, Y.; Ullah, A.; Tasnim, J.; Chakraborty, A.; Naima, T.A.; Marma, K.K.S.; et al. Functional food: Complementary to fight against COVID-19. Benisuef Univ. J. Basic Appl. Sci. 2022, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.I.; Kim, J.; Kim, J.Y.; Kwon, O. Acute intake of mulberry leaf aqueous extract affects postprandial glucose response after maltose loading: Randomized double-blind placebo-controlled pilot study. J. Funct. Foods 2013, 5, 1502–1506. [Google Scholar] [CrossRef]
- Enkhmaa, B.; Shiwaku, K.; Katsube, T.; Kitajima, K.; Anuurad, E.; Yamasaki, M.; Yamane, Y. Mulberry (Morus alba L.) Leaves and Their Major Flavonol Quercetin 3-(6-Malonylglucoside) Attenuate Atherosclerotic Lesion Development in LDL Receptor-Deficient Mice. J. Nutr. 2005, 135, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Jeszka-Skowron, M.; Flaczyk, E.; Jeszka, J.; Krejpcio, Z.; Król, E.; Buchowski, M.S. Mulberry leaf extract intake reduces hyperglycaemia in streptozotocin (STZ)-induced diabetic rats fed high-fat diet. J. Funct. Foods 2014, 8, 9–17. [Google Scholar] [CrossRef]
- Hunyadi, A.; Liktor-Busa, E.; Márki, Á.; Martins, A.; Jedlinszki, N.; Hsieh, T.J.; Báthori, M.; Hohmann, J.; Zupkó, I. Metabolic Effects of Mulberry Leaves: Exploring Potential Benefits in Type 2 Diabetes and Hyperuricemia. Evid. Based Complement. Altern. Med. 2013, 2013, 948627. [Google Scholar] [CrossRef]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Katsube, T.; Tsurunaga, Y.; Sugiyama, M.; Furuno, T.; Yamasaki, Y. Effect of air-drying temperature on antioxidant capacity and stability of polyphenolic compounds in mulberry (Morus alba L.) leaves. Food Chem. 2009, 113, 964–969. [Google Scholar] [CrossRef]
- Thabti, I.; Albert, Q.; Philippot, S.; Dupire, F.; Westerhuis, B.; Fontanay, S.; Risler, A.; Kassab, T.; Elfalleh, W.; Aferchichi, A.; et al. Advances on Antiviral Activity of Morus spp. Plant Extracts: Human Coronavirus and Virus-Related Respiratory Tract Infections in the Spotlight. Molecules 2020, 25, 1876. [Google Scholar] [CrossRef]
- Lee, J.-H.; Bae, S.Y.; Oh, M.; Kim, K.H.; Chung, M.S. Antiviral Effects of Mulberry (Morus alba) Juice and Its Fractions on Foodborne Viral Surrogates. Foodborne Pathog. Dis. 2014, 11, 224–229. [Google Scholar] [CrossRef]
- Du, J.; He, Z.-D.; Jiang, R.-W.; Ye, W.-C.; Xu, H.-X.; But, P.P.-H. Antiviral flavonoids from the root bark of Morus alba L. Phytochemistry 2003, 62, 1235–1238. [Google Scholar] [CrossRef]
- Chi, Y.S.; Jong, H.G.; Son, K.H.; Chang, H.W.; Kang, S.S.; Kim, H.P. Effects of naturally occurring prenylated flavonoids on enzymes metabolizing arachidonic acid: Cyclooxygenases and lipoxygenases. Biochem. Pharmacol. 2001, 62, 1185–1191. [Google Scholar] [CrossRef]
- Mazimba, O.; Majinda, R.R.T.; Motlhanka, D. Antioxidant and antibacterial constituents from Morus nigra. Afr. J. Pharm. Pharmacol. 2011, 5, 751–754. [Google Scholar] [CrossRef]
- Butt, M.S.; Nazir, A.; Sultan, M.T.; Schroën, K. Morus alba L. nature’s functional tonic. Trends Food Sci. Technol. 2008, 19, 505–512. [Google Scholar] [CrossRef]
- Wei, H.; Zhu, J.-J.; Liu, X.-Q.; Feng, W.-H.; Wang, Z.-M.; Yan, L.-H. Review of bioactive compounds from root barks of Morus plants (Sang-Bai-Pi) and their pharmacological effects. Cogent Chem. 2016, 2, 1212320. [Google Scholar] [CrossRef]
- Chang, L.-W.; Juang, L.-J.; Wang, B.-S.; Wang, M.-Y.; Tai, H.-M.; Hung, W.-J.; Chen, Y.-J.; Huang, M.-H. Antioxidant and antityrosinase activity of mulberry (Morus alba L.) twigs and root bark. Food Chem. Toxicol. 2011, 49, 785–790. [Google Scholar] [CrossRef]
- Ko, W.; Yoon, C.S.; Kim, K.W.; Lee, H.; Kim, N.; Woo, E.R.; Kim, Y.C.; Kang, D.G.; Lee, H.S.; Oh, H.; et al. Neuroprotective and Anti-Inflammatory Effects of Kuwanon C from Cudrania tricuspidata Are Mediated by Heme Oxygenase-1 in HT22 Hippocampal Cells, RAW264.7 Macrophage, and BV2 Microglia. Int. J. Mol. Sci. 2020, 21, 4839. [Google Scholar] [CrossRef]
- Zelová, H.; Hanáková, Z.; Čermáková, Z.; Šmejkal, K.; Acqua, S.D.; Babula, P.; Cvačka, J.; Hošek, J. Evaluation of Anti-Inflammatory Activity of Prenylated Substances Isolated from Morus alba and Morus nigra. J. Nat. Prod. 2014, 77, 1297–1303. [Google Scholar] [CrossRef]
- Kumari, K.; Kumar, D.; Kumar, R.V.; Singh, P. Kuwanons, promising inhibitors against the ACE-2, main protease of SARS-CoV-2 and falcipan-2 using molecular docking. Res. Sq. 2020, 1–15. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Martínez, J.A.; Bolivar, F.; Escalante, A. Shikimic Acid Production in Escherichia coli: From Classical Metabolic Engineering Strategies to Omics Applied to Improve Its Production. Front. Bioeng. Biotechnol. 2015, 3, 145. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Bose, S.; Banerjee, S.; Vishnuprasad, C.N.; Del Pilar Rodriguez-Torres, M.; Shin, H.S. Star anise (Illicium verum): Chemical compounds, antiviral properties, and clinical relevance. Phytother. Res. 2020, 34, 1248–1267. [Google Scholar] [CrossRef]
- Tu, Y. Artemisinin-A Gift from Traditional Chinese Medicine to the World (Nobel Lecture). Angew. Chem. Int. Ed. 2016, 55, 10210–10226. [Google Scholar] [CrossRef]
- Ma, N.; Zhang, Z.; Liao, F.; Jiang, T.; Tu, Y. The birth of artemisinin. Pharmacol. Ther. 2020, 216, 107658. [Google Scholar] [CrossRef]
- Gotaskie, G.E.; Andreassi, B.F. Paclitaxel: A new antimitotic chemotherapeutic agent. Cancer Pract. 1994, 2, 27–33. [Google Scholar]
- Foa, R.; Norton, L.; Seidman, A.D. Taxol (paclitaxel): A novel anti-microtubule agent with remarkable anti-neoplastic activity. Int. J. Clin. Lab Res. 1994, 24, 6–14. [Google Scholar] [CrossRef]
- Shakya, A.; Chikhale, R.V.; Bhat, H.R.; Alasmary, F.A.; Almutairi, T.M.; Ghosh, S.K.; Alhajri, H.M.; Alissa, S.A.; Nagar, S.; Islam, M.A. Pharmacoinformatics-based identification of transmembrane protease serine-2 inhibitors from Morus alba as SARS-CoV-2 cell entry inhibitors. Mol. Divers. 2022, 26, 265–278. [Google Scholar] [CrossRef]
- Shah, A.; Patel, V.; Parmar, B. Discovery of some antiviral natural products to fight against novel coronavirus (SARS-CoV-2) using an in silico approach. Comb. Chem. High Throughput Screen 2021, 24, 1271–1280. [Google Scholar] [CrossRef]
- Eskandari, V. Repurposing the natural compounds as potential therapeutic agents for COVID-19 based on the molecular docking study of the main protease and the receptor-binding domain of spike protein. J. Mol. Model. 2022, 28, 1–11. [Google Scholar] [CrossRef]
- Chen, G.Y.; Pan, Y.C.; Wu, T.Y.; Yao, T.Y.; Wang, W.J.; Shen, W.J.; Ahmed, A.; Chan, S.T.; Tang, C.H.; Huang, W.C.; et al. Potential natural products that target the SARS-CoV-2 spike protein identified by structure-based virtual screening, isothermal titration calorimetry and lentivirus particles pseudotyped (Vpp) infection assay. J. Tradit. Complement. Med. 2022, 12, 73–89. [Google Scholar] [CrossRef]
- Basu, A.; Sarkar, A.; Maulik, U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020, 10, 17699. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Stock, P.; Liefeldt, L.; Paul, M.; Ganten, D. Local reninangiotensin systems in cardiovascular tissues: Localization and functional role. Cardiology 1995, 86 (Suppl. S1), 2–8. [Google Scholar] [CrossRef]
- Donoghue, M.; Hsieh, F.; Baronas, E.; Godbout, K.; Gosselin, M.; Stagliano, N.; Donovan, M.; Woolf, B.; Robison, K.; Jeyaseelan, R.; et al. A Novel Angiotensin-Converting Enzyme–Related Carboxypeptidase (ACE2) Converts Angiotensin I to Angiotensin 1-9. Circ. Res. 2000, 87, e1–e9. [Google Scholar] [CrossRef]
- Keidar, S.; Kaplan, M.; Gamliel-Lazarovich, A. ACE2 of the heart: From angiotensin I to angiotensin (1–7). Cardiovasc. Res. 2007, 73, 463–469. [Google Scholar] [CrossRef]
- Cheng, Q.; Zhou, L.; Zhou, J.; Wan, H.; Li, Q.; Feng, Y. ACE2 overexpression inhibits acquired platinum resistance-induced tumor angiogenesis in NSCLC. Oncol. Rep. 2016, 36, 1403–1410. [Google Scholar] [CrossRef]
- Yu, C.; Tang, W.; Wang, Y.; Shen, Q.; Wang, B.; Cai, C.; Meng, X.; Zou, F. Downregulation of ACE2/Ang-(1-7)/Mas axis promotes breast cancer metastasis by enhancing store-operated calcium entry. Cancer Lett. 2016, 376, 268–277. [Google Scholar] [CrossRef]
- Narayan, S.S.; Lorenz, K.; Ukkat, J.; Hoang-Vu, C.; Trojanowicz, B. Angiotensin converting enzymes ACE and ACE2 in thyroid cancer progression. Neoplasma 2020, 67, 402–409. [Google Scholar] [CrossRef]
- Niu, X.; Zhu, Z.; Shao, E.; Bao, J. ACE2 Is a Prognostic Biomarker and Associated with Immune Infiltration in Kidney Renal Clear Cell Carcinoma: Implication for COVID-19. J. Oncol. 2021, 2021, 8847307. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, S.; Li, T.; Yu, L.; Zhang, Y.; Zeng, H.; Qian, X.; Bi, J.; Lin, Y. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J. Exp. Clin. Cancer Res. 2019, 38, 173. [Google Scholar] [CrossRef]
- Vittinghoff, E.; Scheer, S.; O’Malley, P.; Colfax, G.; Holmberg, S.D.; Buchbinder, S.P. Combination Antiretroviral Therapy and Recent Declines in AIDS Incidence and Mortality. J. Infect. Dis. 1999, 179, 717–720. [Google Scholar] [CrossRef]
- Kontoghiorghes, G.J.; Fetta, S.; Kontoghiorghe, C.N. The need for a multi-level drug targeting strategy to curb the COVID-19 pandemic. Front. Biosci. 2021, 26, 1723–1736. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]





| KD (M) | ka (M−1 s−1) | kd (s−1) | R2 | |
|---|---|---|---|---|
| Spike S1 RBD | 5.03 × 10−4 | 4.24 × 102 | 2.13 × 10−1 | 0.9924 |
| ACE2 receptor | 8.11 × 10−4 | 3.15 × 102 | 2.56 × 10−1 | 0.9922 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.S.; Kwon, E.-B.; Kim, B.; Chung, H.-S.; Choi, G.; Kim, Y.-H.; Choi, J.-G. Mulberry Component Kuwanon C Exerts Potent Therapeutic Efficacy In Vitro against COVID-19 by Blocking the SARS-CoV-2 Spike S1 RBD:ACE2 Receptor Interaction. Int. J. Mol. Sci. 2022, 23, 12516. https://doi.org/10.3390/ijms232012516
Kim YS, Kwon E-B, Kim B, Chung H-S, Choi G, Kim Y-H, Choi J-G. Mulberry Component Kuwanon C Exerts Potent Therapeutic Efficacy In Vitro against COVID-19 by Blocking the SARS-CoV-2 Spike S1 RBD:ACE2 Receptor Interaction. International Journal of Molecular Sciences. 2022; 23(20):12516. https://doi.org/10.3390/ijms232012516
Chicago/Turabian StyleKim, Young Soo, Eun-Bin Kwon, Buyun Kim, Hwan-Suck Chung, Garam Choi, Yeoun-Hee Kim, and Jang-Gi Choi. 2022. "Mulberry Component Kuwanon C Exerts Potent Therapeutic Efficacy In Vitro against COVID-19 by Blocking the SARS-CoV-2 Spike S1 RBD:ACE2 Receptor Interaction" International Journal of Molecular Sciences 23, no. 20: 12516. https://doi.org/10.3390/ijms232012516
APA StyleKim, Y. S., Kwon, E.-B., Kim, B., Chung, H.-S., Choi, G., Kim, Y.-H., & Choi, J.-G. (2022). Mulberry Component Kuwanon C Exerts Potent Therapeutic Efficacy In Vitro against COVID-19 by Blocking the SARS-CoV-2 Spike S1 RBD:ACE2 Receptor Interaction. International Journal of Molecular Sciences, 23(20), 12516. https://doi.org/10.3390/ijms232012516






