ERAP/HLA-C and KIR Genetic Profile in Couples with Recurrent Implantation Failure
Abstract
:1. Introduction
2. Results
2.1. Frequency of a Female KIR and Male HLA-C Genotype Combinations
2.2. Combinations of a Female ERAP and Male HLA-C Genotypes
2.3. Combinations of Female ERAP/KIR with Male HLA-C Genotypes
2.4. Combinations of Female KIR/HLA-C with Male ERAP Genotypes
2.5. Combinations of Female KIR with Male HLA-C and ERAP Genotypes
2.6. Combinations of Female ERAP/HLA-C with Male HLA-C Genotypes
2.7. Combinations of Male ERAP/HLA-C with Female Partner’s HLA-C Genotypes
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. DNA Preparation and Genotyping
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bashiri, A.; Halper, K.I.; Orvieto, R. Recurrent Implantation Failure-update overview on etiology, diagnosis, treatment and future directions. Reprod. Biol. Endocrinol. 2018, 16, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coughlan, C.; Ledger, W.; Wang, Q.; Liu, F.; Demirol, A.; Gurgan, T.; Cutting, R.; Ong, K.; Sallam, H.; Li, T.C. Recurrent implantation failure: Definition and management. Reprod. Biomed. Online 2014, 28, 14–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busnelli, A.; Reschini, M.; Cardellicchio, L.; Vegetti, W.; Somigliana, E.; Vercellini, P. How common is real repeated implantation failure? An indirect estimate of the prevalence. Reprod. Biomed. Online 2020, 40, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Pollheimer, J.; Vondra, S.; Baltayeva, J.; Beristain, A.G.; Knöfler, M. Regulation of Placental Extravillous Trophoblasts by the Maternal Uterine Environment. Front. Immunol. 2018, 9, 2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manaster, I.; Mandelboim, O. The unique properties of uterine NK cells. Am. J. Reprod. Immunol. 2010, 63, 434–444. [Google Scholar] [CrossRef]
- Faas, M.M.; de Vos, P. Uterine NK cells and macrophages in pregnancy. Placenta 2017, 56, 44–52. [Google Scholar] [CrossRef]
- Bian, Q.; Fu, B. Immunological microenvironment at the maternal-fetal interface. J. Reprod. Immunol. 2022, 151, 103632. [Google Scholar] [CrossRef]
- Acar, N.; Ustunel, I.; Demir, R. Uterine natural killer (uNK) cells and their missions during pregnancy: A review. Acta Histochem. 2011, 113, 82–91. [Google Scholar] [CrossRef]
- Roe, D.; Kuang, R. Accurate and Efficient KIR Gene and Haplotype Inference from Genome Sequencing Reads with Novel K-mer Signatures. Front. Immunol. 2020, 11, 583013. [Google Scholar] [CrossRef]
- Djaoud, Z.; Parham, P. HLAs, TCRs, and KIRs, a Triumvirate of Human Cell-Mediated Immunity. Annu. Rev. Biochem. 2020, 89, 717–739. [Google Scholar] [CrossRef]
- Maxwell, L.D.; Wallace, A.; Middleton, D.; Curran, M.D. A common KIR2DS4 deletion variant in the human that predicts a soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens 2002, 60, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Middleton, D.; Gonzelez, F. The extensive polymorphism of KIR genes. Immunology 2010, 129, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; del Zotto, G.; Pietra, G.; et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Peña, R.; de Los Santos, M.J.; Lucia, A.; Castro-Santos, P. Understanding the role of killer cell immunoglobulin-like receptors in pregnancy complications. J. Assist. Reprod. Genet. 2019, 36, 827–835. [Google Scholar] [CrossRef]
- Papúchová, H.; Meissner, T.B.; Li, Q.; Strominger, J.L.; Tilburgs, T. The Dual Role of HLA-C in Tolerance and Immunity at the Maternal-Fetal Interface. Front. Immunol. 2019, 10, 2730. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, S.; Prakash, S. Significance of KIR like natural killer cell receptors in autoimmune disorders. Clin. Immunol. 2020, 216, 108449. [Google Scholar] [CrossRef]
- Admon, A. ERAP1 shapes just part of the immunopeptidome. Hum. Immunol. 2019, 80, 296–301. [Google Scholar] [CrossRef]
- Babaie, F.; Hosseinzadeh, R.; Ebrazeh, M.; Seyfizadeh, N.; Aslani, S.; Salimi, S.; Hemmatzadeh, M.; Azizi, G.; Jadidi-Niaragh, F.; Mohammadi, H. The roles of ERAP1 and ERAP2 in autoimmunity and cancer immunity: New insights and perspective. Mol. Immunol. 2020, 121, 7–19. [Google Scholar] [CrossRef]
- López de Castro, J.A.; Alvarez-Navarro, C.; Brito, A.; Guasp, P.; Martín-Esteban, A.; Sanz-Bravo, A. Molecular and pathogenic effects of endoplasmic reticulum aminopeptidases ERAP1 and ERAP2 in MHC-I-associated inflammatory disorders: Towards a unifying view. Mol. Immunol. 2016, 77, 193–204. [Google Scholar] [CrossRef]
- Stratikos, E.; Stamogiannos, A.; Zervoudi, E.; Fruci, D. A role for naturally occurring alleles of endoplasmic reticulum aminopeptidases in tumor immunity and cancer pre-disposition. Front. Oncol. 2014, 4, 363. [Google Scholar] [CrossRef]
- Saulle, I.; Vicentini, C.; Clerici, M.; Biasin, M. An Overview on ERAP Roles in Infectious Diseases. Cells 2020, 9, 720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierabracci, A.; Milillo, A.; Locatelli, F.; Fruci, D. The putative role of endoplasmic reticulum aminopeptidases in autoimmunity: Insights from genomic-wide association studies. Autoimmun. Rev. 2012, 12, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Niepiekło-Miniewska, W.; Mpakali, A.; Stratikos, E.; Matusiak, Ł.; Narbutt, J.; Lesiak, A.; Kuna, P.; Wilczyńska, K.; Nowak, I.; Wiśniewski, A.; et al. Endoplasmic reticulum aminopeptidase 1 polymorphism Ile276Met is associated with atopic dermatitis and affects the generation of an HLA-C associated antigenic epitope in vitro. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Paladini, F.; Fiorillo, M.T.; Vitulano, C.; Tedeschi, V.; Piga, M.; Cauli, A.; Mathieu, A.; Sorrentino, R. An allelic variant in the intergenic region between ERAP1 and ERAP2 correlates with an inverse expression of the two genes. Sci. Rep. 2018, 8, 10398. [Google Scholar] [CrossRef] [Green Version]
- Piekarska, K.; Radwan, P.; Tarnowska, A.; Wiśniewski, A.; Radwan, M.; Wilczyński, J.R.; Malinowski, A.; Nowak, I. ERAP, KIR, and HLA-C profile in recurrent implantation failure. Front. Immunol. 2021, 12, 755624. [Google Scholar] [CrossRef]
- Hiby, S.E.; Walker, J.J.; O’shaughnessy, K.M.; Redman, C.W.; Carrington, M.; Trowsdale, J. Moffett, A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J. Exp. Med. 2004, 200, 957–965. [Google Scholar] [CrossRef] [Green Version]
- Moffett, A.; Chazara, O.; Colucci, F.; Johnson, M.H. Variation of maternal KIR and fetal HLA-C genes in reproductive failure: Too early for clinical intervention. Reprod. Biomed. Online 2016, 33, 763–769. [Google Scholar] [CrossRef] [Green Version]
- Łuszczek, W.; Majorczyk, E.; Nowak, I.; Pawlik, A.; Jasek, M.; Wiśniewski, A.; Kuśnierczyk, P. Inhibitory and activatory KIR gene frequencies in the Polish population. Int. J. Immunogenet. 2006, 33, 167–170. [Google Scholar] [CrossRef]
- Blunt, M.D.; Khakoo, S.I. Activating killer cell immunoglobulin-like receptors: Detection, function and therapeutic use. Int. J. Immunogenet. 2020, 47, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.Y.; Ren, C.E.; Qiao, P.Y.; Meng, Y.H. Uterine natural killer cells and recurrent spontaneous abortion. Am. J. Reprod. Immunol. 2021, 86, e13433. [Google Scholar] [CrossRef]
- Xiong, S.; Sharkey, A.M.; Kennedy, P.R.; Gardner, L.; Farrell, L.E.; Chazara, O.; Bauer, J.; Hiby, S.E.; Colucci, F.; Moffett, A. Maternal uterine NK cell-activating receptor KIR2DS1 enhances placentation. J. Clin. Invest. 2013, 123, 4264–4272. [Google Scholar] [CrossRef]
- Hiby, S.E.; Apps, R.; Sharkey, A.M.; Farrell, L.E.; Gardner, L.; Mulder, A.; Claas, F.H.; Walker, J.J.; Redman, C.C.; Morgan, L.; et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J. Clin. Invest. 2010, 120, 4102–4110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamogiannos, A.; Koumantou, D.; Papakyriakou, A.; Stratikos, E. Effects of polymorphic variation on the mechanism of Endoplasmic Reticulum Aminopeptidase 1. Mol. Immunol. 2015, 67, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Wilczyńska, K.; Wiśniewski, A.; Malinowski, A.; Barcz, E.; Wilczyński, J.R.; Kuśnierczyk, P.; Nowak, I. ERAP, KIR and HLA-C gene interaction in susceptibility to recurrent spontaneous abortion in the Polish population. Hum. Immunol. 2019, 80, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Cifaldi, L.; Romania, P.; Lorenzi, S.; Locatelli, F.; Fruci, D. Role of endoplasmic reticulum aminopeptidases in health and disease: From infection to cancer. Int. J. Mol. Sci. 2012, 13, 8338–8352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, N.; Nakayama, J.; Yamakawa-Kobayashi, K.; Hamaguchi, H.; Miyazaki, R.; Arinami, T. Identification of 33 polymorphisms in the adipocyte-derived leucine aminopeptidase (ALAP) gene and possible association with hypertension. Hum. Mutat. 2002, 19, 251–257. [Google Scholar] [CrossRef]
- Goto, Y.; Hattori, A.; Ishii, Y.; Mizutani, S.; Tsujimoto, M. Enzymatic properties of human aminopeptidase A. Regulation of its enzymatic activity by calcium and angiotensin IV. J. Biol. Chem. 2006, 281, 23503–23513. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, L.C.; Gomes, C.E.M.; Duggal, P.; Holanda, I.D.P.; de Lima, A.S.; do Nascimento, P.R.P.; Bezerra Jeronimo, S.M. Genetic association of ERAP1 and ERAP2 with eclampsia and preeclampsia in northeastern Brazilian women. Sci. Rep. 2021, 11, 6764. [Google Scholar] [CrossRef]
- Wiśniewski, A.; Kasprzyk, S.; Majorczyk, E.; Nowak, I.; Wilczyńska, K.; Chlebick, A.; Zoń-Giebel, A.; Kuśnierczyk, P. ERAP1-ERAP2 haplotypes are associated with ankylosing spondylitis in Polish patients. Hum. Immunol. 2019, 80, 339–343. [Google Scholar] [CrossRef]
- Masi, S.; Uliana, M.; Virdis, A. Angiotensin II and vascular damage in hypertension: Role of oxidative stress and sympathetic activation. Vascul. Pharmacol. 2019, 115, 13–17. [Google Scholar] [CrossRef]
- Bui, A.D.; Sharma, R.; Henkel, R.; Agarwal, A. Reactive oxygen species impact on sperm DNA and its role in male infertility. Andrologia 2018, 50, e13012. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Chandra, A.; Chakraborty, J.; Chattopadhyay, A.; Senapati, S.; Chatterjee, G.; Chatterjee, R. Associations of ERAP1 coding variants and domain specific interaction with HLA-C∗06 in the early onset psoriasis patients of India. Hum. Immunol. 2017, 78, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Kuśnierczyk, P. To Be or Not to Be: The Case of Endoplasmic Reticulum Aminopeptidase 2. Front. Immunol. 2022, 13, 902567. [Google Scholar] [CrossRef] [PubMed]
- Saulle, I.; Vanetti, C.; Goglia, S.; Vicentini, C.; Tombetti, E.; Garziano, M.; Clerici, M.; Biasin, M. A New ERAP2/Iso3 Isoform Expression Is Triggered by Different Microbial Stimuli in Human Cells. Could It Play a Role in the Modulation of SARS-CoV-2 Infection? Cells 2020, 9, 1951. [Google Scholar] [CrossRef]
- Nowak, I.; Malinowski, A.; Tchorzewski, H.; Barcz, E.; Wilczynski, J.R.; Grybos, M.; Kurpisz, M.; Luszczek, W.; Banasik, M.; Reszczynska-Slezak, D.; et al. Frequencies of killer immunoglobulin-like receptor genotypes influence susceptibility to spontaneous abortion. J. Appl. Genet. 2009, 50, 391–398. [Google Scholar] [CrossRef]
- Nowak, I.; Malinowski, A.; Tchórzewski, H.; Barcz, E.; Wilczyński, J.R.; Banasik, M.; Gryboś, M.; Kurpisz, M.M.; Luszczek, W.; Majorczyk, E.; et al. HLA-C C1C2 heterozygosity may protect women bearing the killer immunoglobulin-like receptor AA genotype from spontaneous abortion. J. Reprod. Immunol. 2011, 88, 32–37. [Google Scholar] [CrossRef]
- Frohn, C.; Schlenke, P.; Ebel, B.; Dannenberg, C.; Bein, G.; Kirchner, H. DNA typing for natural killer cell inhibiting HLA-Cw groups NK1 and NK2 by PCR-SSP. J. Immunol. Methods 1998, 218, 155–160. [Google Scholar] [CrossRef]

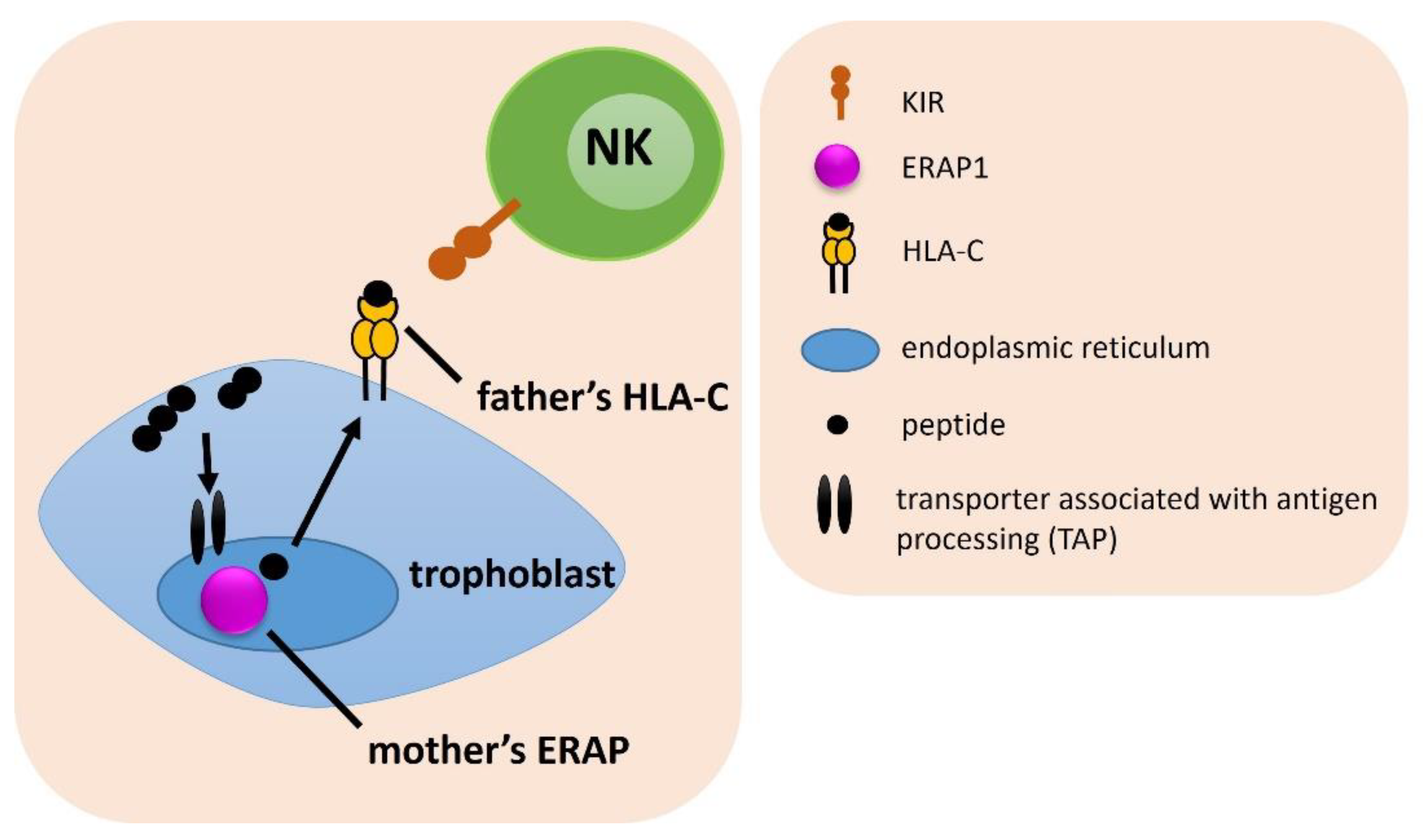



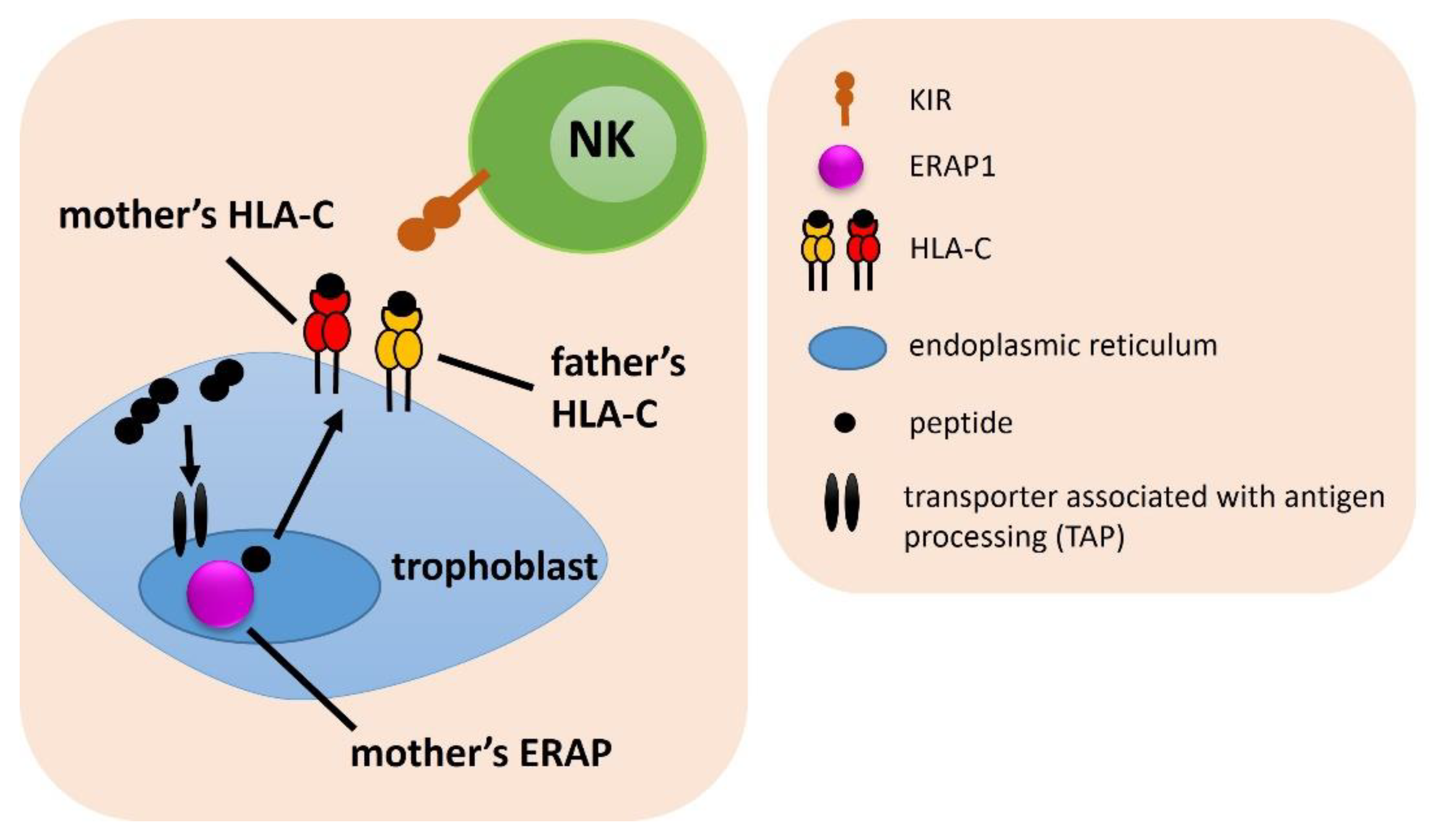
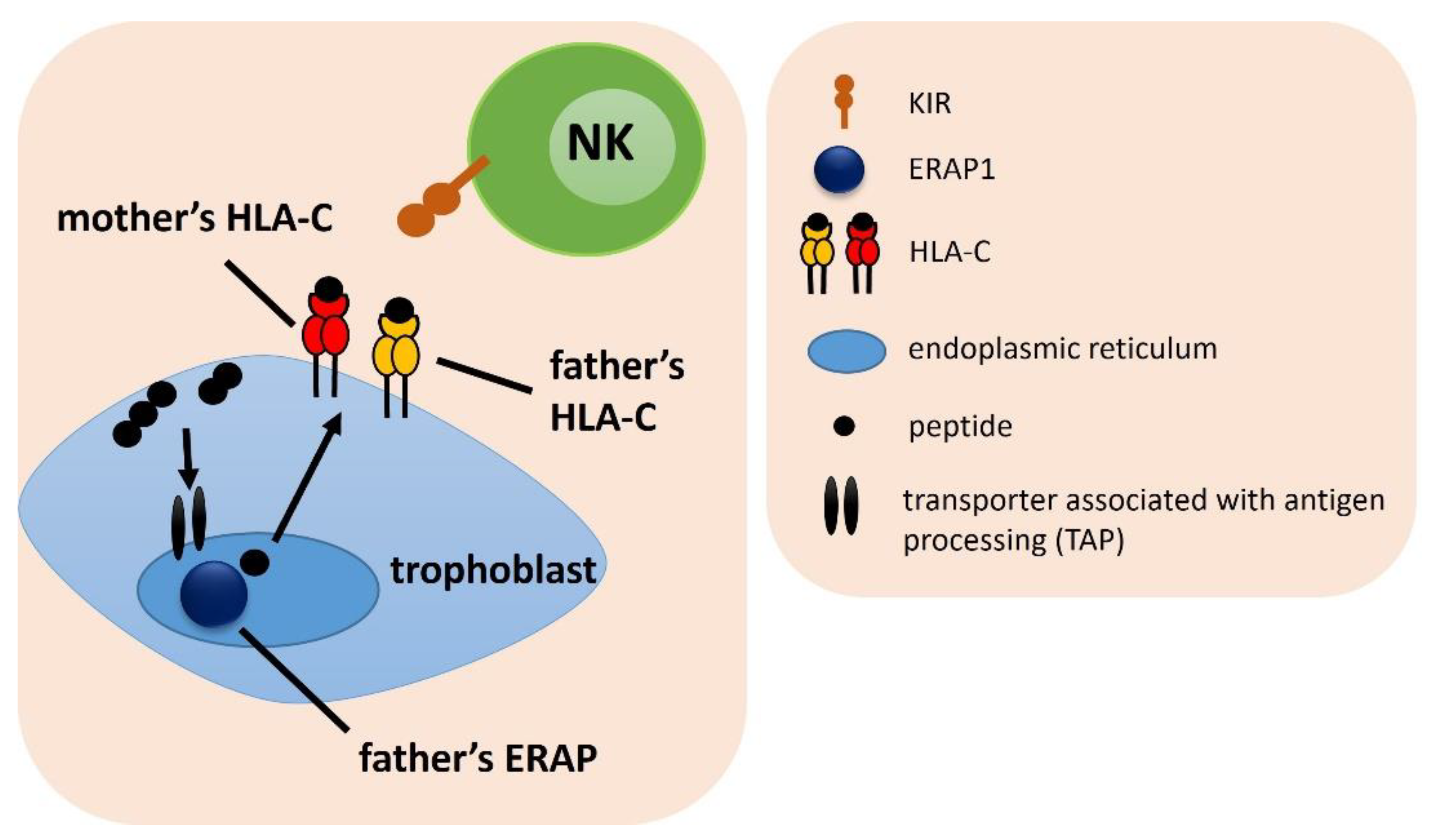

| ERAP, HLA-C, KIR Combination | Associated Combination | Compared Groups | p | pcorr. | OR | 95% CI | Effect | Table |
|---|---|---|---|---|---|---|---|---|
| Female ERAP/male HLA-C | ||||||||
| ERAP1 rs30187 C>T/HLA-C | TT/C1C2 | IVF vs. Fertile | 0.006 | 0.050 | 2.843 | 1.30–6.92 | ↑ | Supplementary Table S2 |
| ERAP1 rs27044 C>G/HLA-C | CC/C1C1 | RIF vs. SIVF | 0.005 | 0.044 | 0.343 | 0.15–0.75 | ↓ | Supplementary Table S2 |
| ERAP1 rs27044 C>G/HLA-C | CC/C1C2 | SIVF vs. Fertile | 0.003 | 0.027 | 0.442 | 0.25–0.78 | ↓ | Supplementary Table S2 |
| Female ERAP/male HLA-C/female KIR | ||||||||
| ERAP1 rs27044 C>G/HLA-C/KIR | CC/C1C2/Bx | RIF vs. Fertile | 0.004 | 0.037 | 0.428 | 0.23–0.78 | ↓ | Supplementary Table S3 |
| ERAP1 rs27044 C>G/HLA-C/KIR | CG/C1C2/Bx | RIF vs. Fertile | 0.006 | 0.050 | 2.313 | 1.25–4.33 | ↑ | Supplementary Table S3 |
| Male ERAP/female HLA-C/female KIR | ||||||||
| ERAP1 rs30187 C>T/HLA-C/KIR | CT/C2C2/AA | RIF vs. SIVF | 0.005 | 0.049 | 24.000 | 1.76–1549.16 | ↑ | Supplementary Table S4 |
| Male ERAP/male HLA-C/female KIR | ||||||||
| ERAP1 rs27044 C>G/HLA-C/KIR | GG/C1+/AA | RIF vs. SIVF | 0.005 | 0.031 | 0.114 | 0.01–0.66 | ↓ | Supplementary Table S5 |
| ERAP1 rs27044 C>G/HLA-C/KIR | CG/C2C2/Bx | RIF vs. SIVF | 0.004 | 0.033 | 0.096 | 0.01–0.56 | ↓ | Supplementary Table S5 |
| Parameter | IVF | RIF | SIVF | Unclassified | Fertile Control | |
|---|---|---|---|---|---|---|
| Number of women | N = 491 | N = 278 | N = 161 | N = 52 | N = 322 | |
| Age of women | Mean ± SD | 33.65 ± 4.13 | 34.50 ± 4.10 | 32.11 ± 3.82 | 34.09 ± 3.91 | 32.52 ± 5.99 |
| Range | 22–46 | 23–46 | 22–41 | 25–45 | 22–68 | |
| Number of men | N = 491 | N = 278 | N = 161 | N = 52 | N = 322 | |
| Age of men | Mean ± SD | 35.51 ± 4.89 | 36.27 ± 4.85 | 34.32 ± 4.72 | 36.89 ± 4.89 | 34.15 ± 6.3 |
| Range | 24–53 | 25–53 | 24–53 | 28–51 | 25–70 | |
| Indications for IVF-ET (%) | Only male factor | 145 (29.53) | 78 (28.06) | 58 (36.02) | 9 (17.31) | - |
| Only female factor | 129 (26.27) | 68 (24.46) | 45 (27.95) | 16 (30.77) | - | |
| Both factors | 72 (14.67) | 46 (16.55) | 19 (11.80) | 7 (13.46) | - | |
| Unknown factor | 145 (29.53) | 86 (30.93) | 39 (24.23) | 20 (38.46) | - | |
| Number of IVF-ET | Mean ± SD | 3.37 ± 2.06 | 4.65 ± 1.77 | 1.63 ± 0.71 | 1.62 ± 0.68 | - |
| Range | 1–15 | 3–15 | 1–3 | 1–3 | - | |
| Number of embryos | Mean ± SD | 3.81 ± 2.56 | 5.35 ± 2.31 | 1.69 ± 0.79 | 1.76 ± 0.65 | - |
| Range | 1–19 | 3–19 | 1–5 | 1–3 | - | |
| SNP | SNP Variation | Amino Acid Change | Gene | Locus | Potential Effect | Assay ID |
|---|---|---|---|---|---|---|
| rs26653 | G > C | Pro127Arg | ERAP1 | 5q15 | Presumably indirectly affects specificity and enzymatic activity [22] | C__794818_30 |
| rs2287987 | T > C | Met349Val | ERAP1 | 5q15 | Interactions with the substrate [22] | C__3056893_20 |
| rs30187 | C > T | Arg528Lys | ERAP1 | 5q15 | Enzymatic activity, expression level [20] | C__3056885_10 |
| rs27044 | C > G | Gln730Glu | ERAP1 | 5q15 | Enzymatic activity, substrate length preference [22] | C__3056870_10 |
| rs26618 | T > C | Ile276Met | ERAP1 | 5q15 | Affects efficiency of a precursor peptide trimming for the HLA-C*05-bound epitope [23] | C__3056894_10 |
| rs6861666 | A > G | - | ERAP1/ ERAP2 | 5q15 | rs6861666 in 100% LD with rs75862629 which influences the expression level of ERAP2 and ERAP1 [24] | C__29091789_20 |
| rs2248374 | A > G | - | ERAP2 | 5q15 | Lack of expression of functional forms of the enzyme [20] | C__25649529_10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piekarska, K.; Radwan, P.; Tarnowska, A.; Radwan, M.; Wilczyński, J.R.; Malinowski, A.; Nowak, I. ERAP/HLA-C and KIR Genetic Profile in Couples with Recurrent Implantation Failure. Int. J. Mol. Sci. 2022, 23, 12518. https://doi.org/10.3390/ijms232012518
Piekarska K, Radwan P, Tarnowska A, Radwan M, Wilczyński JR, Malinowski A, Nowak I. ERAP/HLA-C and KIR Genetic Profile in Couples with Recurrent Implantation Failure. International Journal of Molecular Sciences. 2022; 23(20):12518. https://doi.org/10.3390/ijms232012518
Chicago/Turabian StylePiekarska, Karolina, Paweł Radwan, Agnieszka Tarnowska, Michał Radwan, Jacek R. Wilczyński, Andrzej Malinowski, and Izabela Nowak. 2022. "ERAP/HLA-C and KIR Genetic Profile in Couples with Recurrent Implantation Failure" International Journal of Molecular Sciences 23, no. 20: 12518. https://doi.org/10.3390/ijms232012518






