Inhibition of Viral RNA-Dependent RNA Polymerases by Nucleoside Inhibitors: An Illustration of the Unity and Diversity of Mechanisms
Abstract
1. Introduction
| Virus | Inhibitory Analog (Drug Name) |
|---|---|
| Order Mononegavirales (−sense RNA genome): | |
| Family Paramyxoviridae | |
| Parainfluenza virus (PIV) | Remdesivir [19] |
| Hendra virus (HeV) | Remdesivir [19] |
| Nipah virus (NiV) | Remdesivir [19] |
| Family Filoviridae | |
| Ebola virus (EBOV) | Remdesivir [8,9]; favipiravir [20] |
| Marburg virus (MARV) | Remdesivir [21]; favipiravir [22] |
| Family Pneumoviridae | |
| Respiratory syncytial virus (RSV) | Remdesivir [8,9] |
| Order Amarillovirales (+sense RNA genome): | |
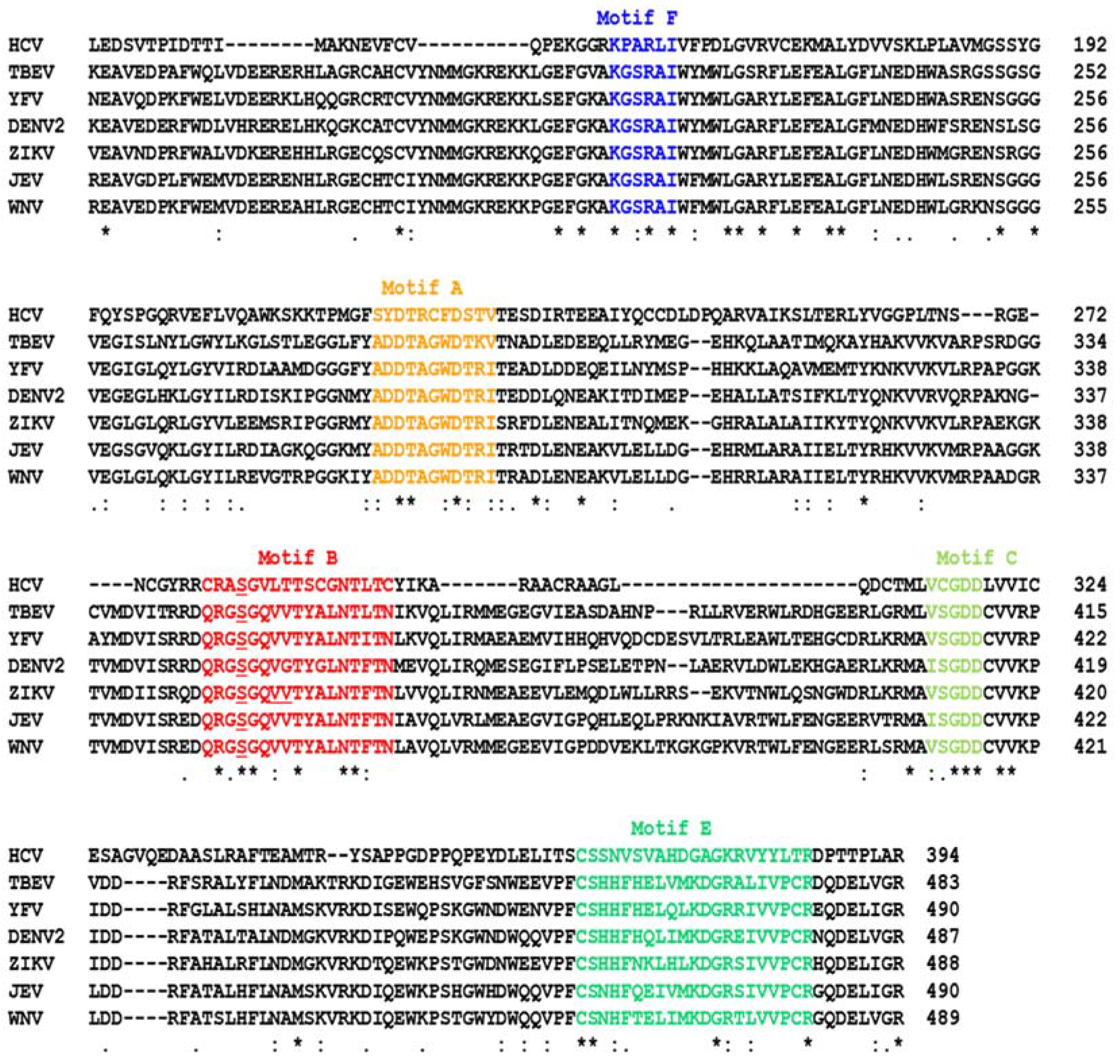
| Family Flaviviridae (NS5; processed from polyprotein) [23] | |
| Hepatitis C virus (HCV) | Remdesivir, sofosbuvir [24] |
| Dengue virus (DENV) | Cordycepin [3] |
| Yellow fever virus (YFV) | Remdesivir [25] |
| Tick-borne encephalitis virus (TBEV) | Remdesivir [23] |
| Zika virus (ZIKV) | Sofosbuvir [24] |
| West Nile virus (WNV) | Sofosbuvir [26] |
| Order Nidovirales (+sense RNA genome): | |
| Family Coronaviridae | |
| SARS-CoV | Remdesivir [12,13]; cordycepin [4]; EIDD-1931 [6] |
| MERS-CoV | Remdesivir [27] |
| Order Picornavirales (+sense RNA genome): | |
| Family Picornaviridae | |
| EV71 (+sense RNA genome) | Remdesivir [28] |
2. Major Nucleoside Analogs with an Antiviral Effect
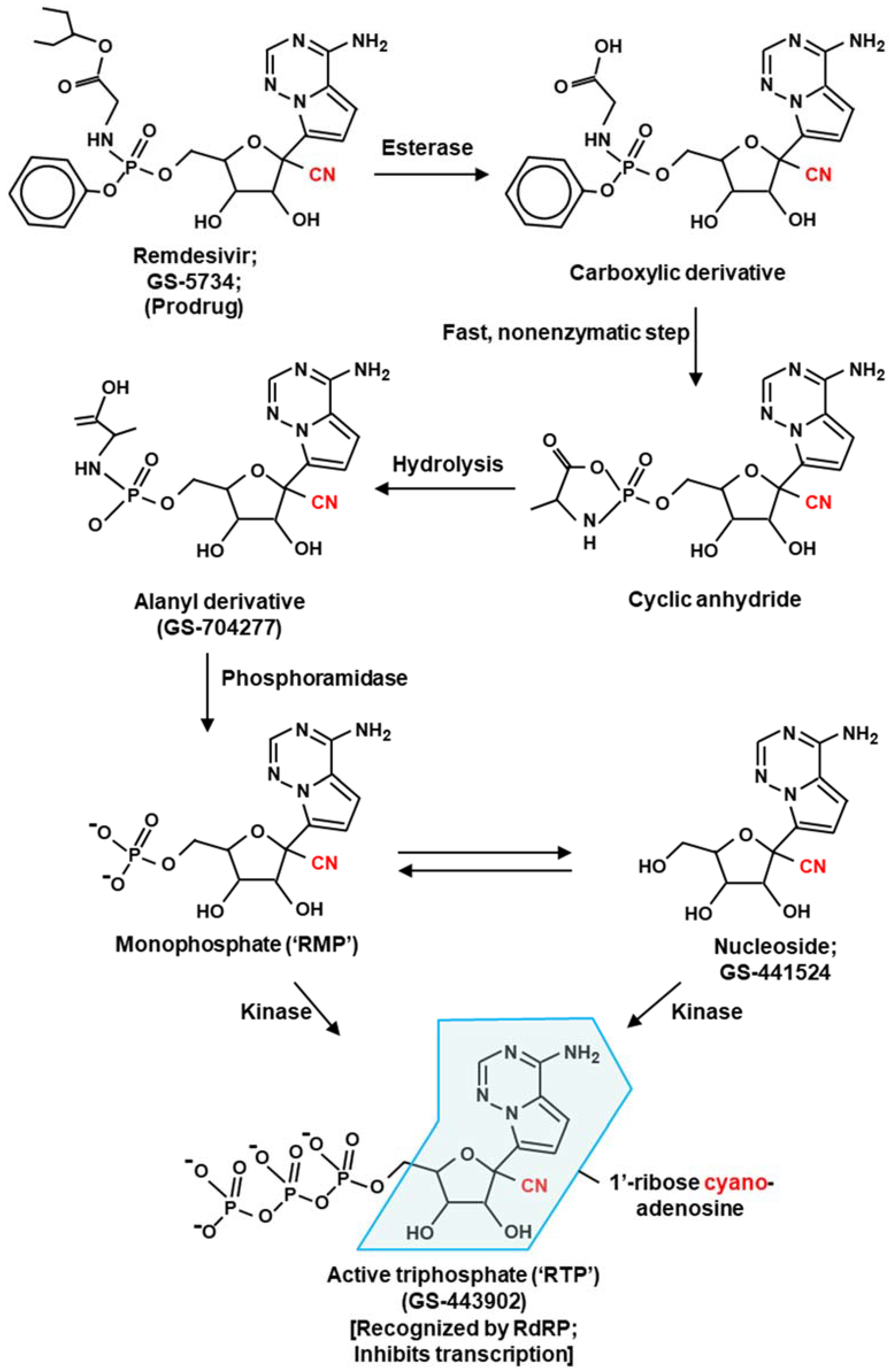
2.1. Remdesivir: Broad-Spectrum Antiviral Role
Remdesivir—Molecular Mechanism of Action
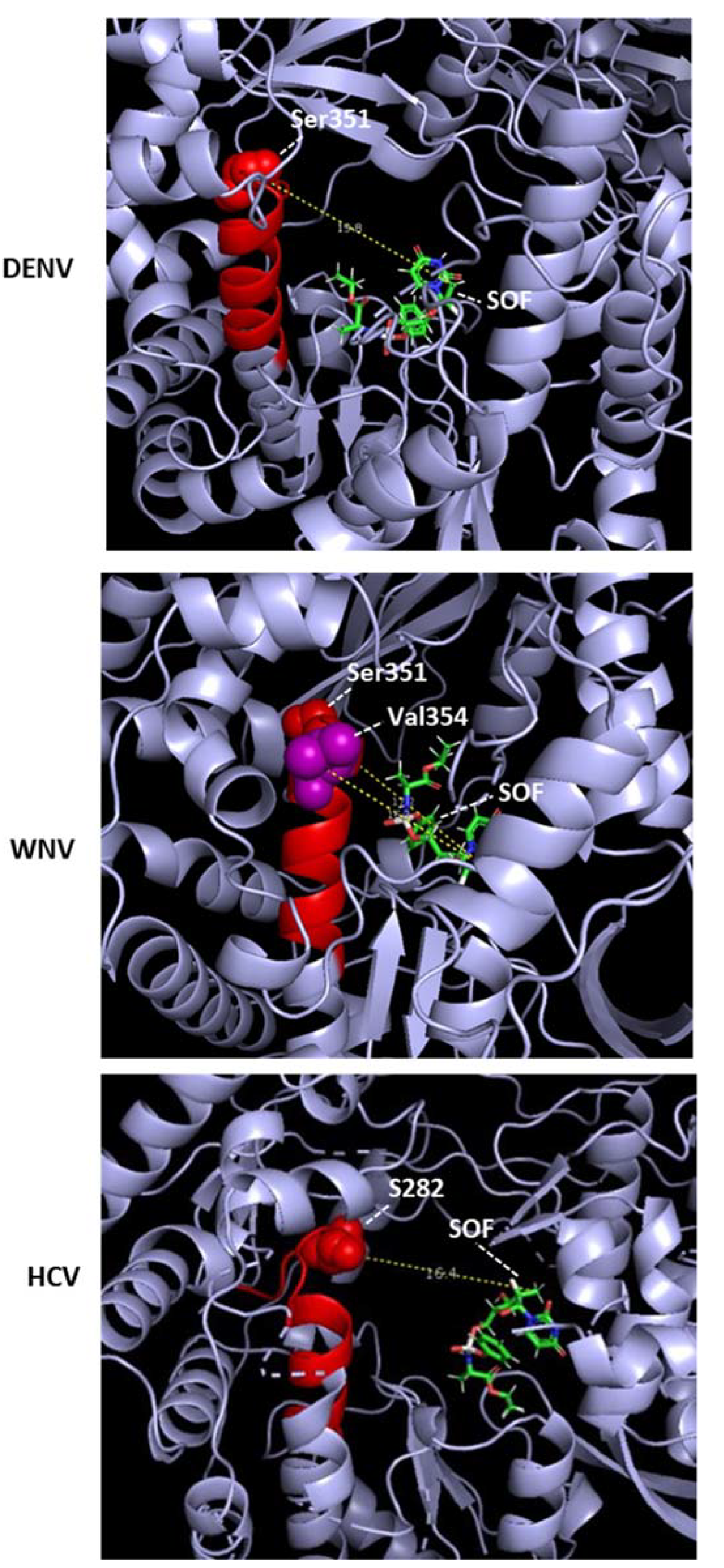
2.2. Sofosbuvir
2.2.1. Sofosbuvir: Molecular Mechanism of Action
2.2.2. Sofosbuvir: Potential Side Effect
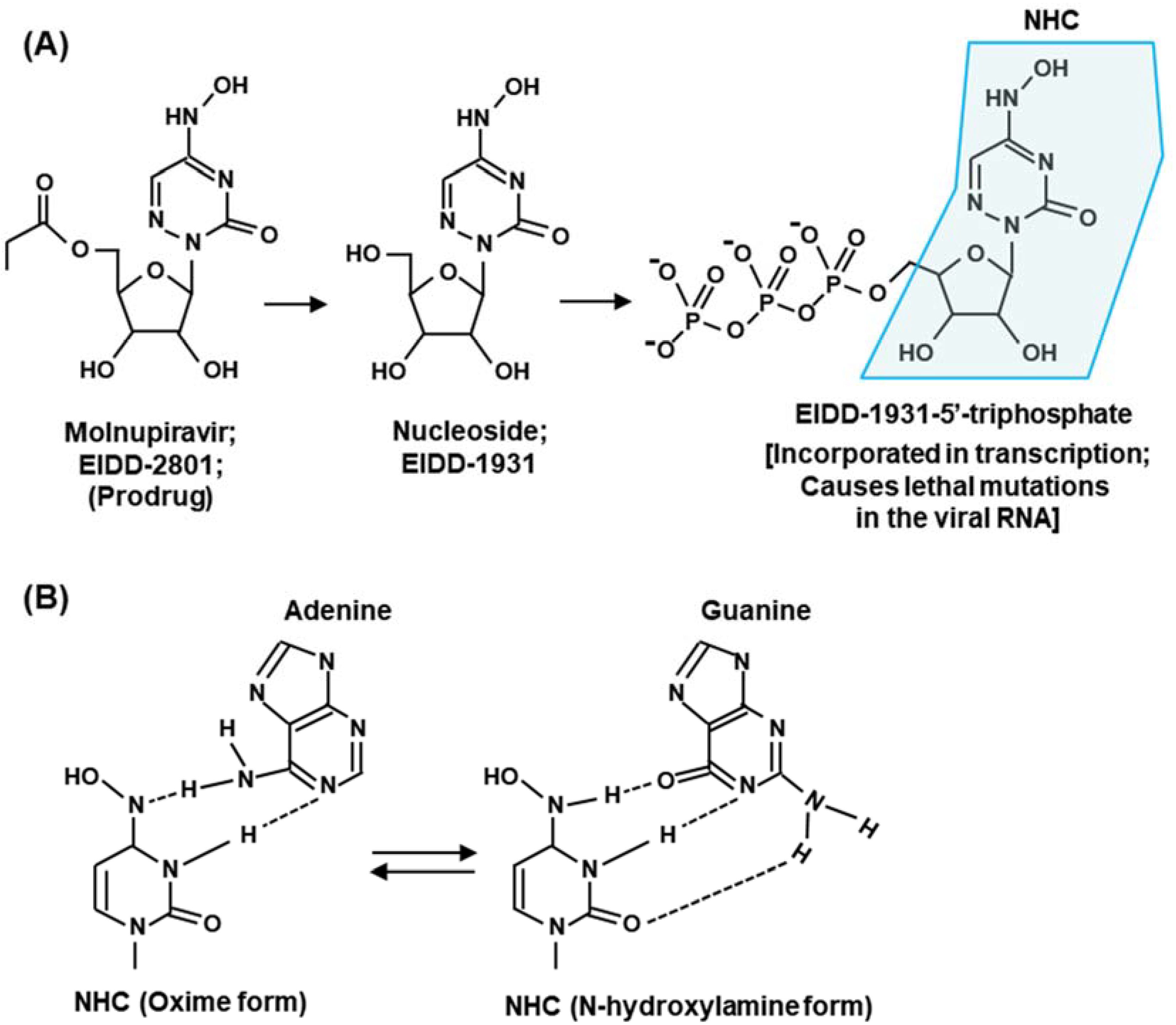
2.3. Molnupiravir
3. Summary Rules and Future Directions
3.1. Summary Rules for Antiviral Nucleoside Design
3.2. Relevant Tissue Culture System: A Suggestion for Future Research
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Afonso, C.L.; Amarasinghe, G.K.; Bányai, K.; Bào, Y.; Basler, C.F.; Bavari, S.; Bejerman, N.; Blasdell, K.R.; Briand, F.-X.; Briese, T.; et al. Taxonomy of the order Mononegavirales: Update 2016. Arch. Virol. 2016, 161, 2351–2360. [Google Scholar] [CrossRef] [PubMed]
- Barik, S.; Ghosh, B.; Whalen, W.; Lazinski, D.; Das, A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell 1987, 50, 885–899. [Google Scholar] [CrossRef]
- Panya, A.; Songprakhon, P.; Panwong, S.; Jantakee, K.; Kaewkod, T.; Tragoolpua, Y.; Sawasdee, N.; Lee, V.S.; Nimmanpipug, P.; Yenchitsomanus, P.T. Cordycepin inhibits virus replication in dengue virus-infected Vero cells. Molecules 2021, 26, 3118. [Google Scholar] [CrossRef]
- Rabie, A.M. Potent inhibitory activities of the adenosine analogue Cordycepin on SARS-CoV-2 replication. ACS Omega 2022, 7, 2960–2969. [Google Scholar] [CrossRef]
- Ju, J.; Li, X.; Kumar, S.; Jockusch, S.; Chien, M.; Tao, C.; Morozova, I.; Kalachikov, S.; Kirchdoerfer, R.N.; Russo, J.J. Nucleotide analogues as inhibitors of SARS-CoV polymerase. Pharmacol. Res. Perspect. 2020, 8, e00674. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Qiang, T.; Liang, C.; Ren, X.; Jia, M.; Zhang, J.; Li, J.; Wan, M.; YuWen, X.; Li, H.; et al. RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021, 213, 113201. [Google Scholar] [CrossRef]
- Zandi, K.; Amblard, F.; Musall, K.; DownsBowen, J.; Keinbard, R.; Oo, A.; Cao, D.; Liang, B.; Russell, O.O.; McBrayer, T.; et al. Repurposing nucleoside analogs for human coronaviruses. Antimicrob. Agents Chemother. 2021, 65, e01652-20. [Google Scholar] [CrossRef] [PubMed]
- Warren, T.K.; Jordan., R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020, 295, 4773–4779. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.P.K.; Dangerfield, T.L.; Taylor, D.W.; Johnson, K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell 2021, 81, 1548.e4–1552.e4. [Google Scholar] [CrossRef]
- Sourimant, J.; Lieber, C.M.; Aggarwal, M.; Cox, R.M.; Wolf, J.D.; Yoon, J.J.; Toots, M.; Ye, C.; Sticher, Z.; Kolykhalov, A.A.; et al. 4′-Fluorouridine is an oral antiviral that blocks respiratory syncytial virus and SARS-CoV-2 replication. Science 2022, 375, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Pang, Z.; Li, M.; Lou, F.; An, X.; Zhu, S.; Song, L.; Tong, Y.; Fan, H.; Fan, J. Molnupiravir and its antiviral activity against COVID-19. Front. Immunol. 2022, 13, 855496. [Google Scholar] [CrossRef]
- Shannon, A.; Selisko, B.; Le, N.-T.-T.; Huchting, J.; Touret, F.; Piorkowski, G.; Fattorini, V.; Ferron, F.; Decroly, E.; Meier, C.; et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020, 11, 4682. [Google Scholar] [CrossRef]
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L. Decoding molnupiravir-induced mutagenesis in SARS-CoV-2. J. Biol. Chem. 2021, 297, 100867. [Google Scholar] [CrossRef]
- Swanstrom, R.; Schinazi, R.F. Lethal mutagenesis as an antiviral strategy. Science 2022, 375, 497–498. [Google Scholar] [CrossRef]
- Lo, M.K.; Jordan, R.; Arvey, A.; Sudhamsu, J.; Shrivastava-Ranjan, P.; Hotard, A.L.; Flint, A.; McMullan, L.K.; Siegel, D.; Clarke, M.O.; et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017, 7, 43395. [Google Scholar] [CrossRef]
- Guedj, J.; Piorkowski, G.; Jacquot, F.; Madelain, V.; Nguyen, T.H.T.; Rodallec, A.; Gunther, S.; Carbonnelle, C.; Mentré, F.; Raoul, H.; et al. Antiviral efficacy of favipiravir against Ebola virus: A translational study in cynomolgus macaques. PLoS Med. 2018, 15, e1002535. [Google Scholar] [CrossRef]
- Porter, D.P.; Weidner, J.M.; Gomba, L.; Bannister, R.; Blair, C.; Jordan, R.; Wells, J.; Wetzel, K.; Garza, N.; Van Tongeren, S.; et al. Remdesivir (GS-5734) Is Efficacious in Cynomolgus macaques infected with Marburg virus. J. Infect. Dis. 2020, 222, 1894–1901. [Google Scholar] [CrossRef]
- Bixler, S.L.; Bocan, T.M.; Wells, J.; Wetzel, K.S.; Van Tongeren, S.A.; Dong, L.; Garza, N.L.; Donnelly, G.; Cazares, L.H.; Nuss, J.; et al. Efficacy of favipiravir (T-705) in nonhuman primates infected with Ebola virus or Marburg virus. Antivir. Res. 2018, 151, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Konkolova, E.; Dejmek, M.; Hřebabecký, H.; Šála, M.; Böserle, J.; Nencka, R.; Boura, E. Remdesivir triphosphate can efficiently inhibit the RNA-dependent RNA polymerase from various flaviviruses. Antivir. Res. 2020, 182, 104899. [Google Scholar] [CrossRef]
- Boccuto, A.; Dragoni, F.; Picarazzi, F.; Lai, A.; Ventura, C.D.; Veo, C.; Giammarino, F.; Saladini, F.; Zehender, G.; Zazzi, M.; et al. Sofosbuvir selects for drug-resistant amino acid variants in the Zika virus RNA-dependent RNA-polymerase complex in vitro. Int. J. Mol. Sci. 2021, 22, 2670. [Google Scholar] [CrossRef]
- Julander, J.G.; Bunyan, E.; Jordan, R.; Porter, D.P. Remdesivir efficacy against yellow fever in a hamster model. Antivir. Res. 2022, 203, 105331. [Google Scholar] [CrossRef]
- Dragoni, F.; Boccuto, A.; Picarazzi, F.; Giannini, A.; Giammarino, F.; Saladini, F.; Mori, M.; Mastrangelo, E.; Zazzi, M.; Vicenti, I. Evaluation of sofosbuvir activity and resistance profile against West Nile virus in vitro. Antivir. Res. 2020, 175, 104708. [Google Scholar] [CrossRef] [PubMed]
- de Wit, E.; Feldmann, F.; Cronin, J.; Jordan, R.; Okumura, A.; Thomas, T.; Scott, D.; Cihlar, T.; Feldmann, H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA 2020, 117, 6771–6776. [Google Scholar] [CrossRef]
- Wu, J.; Wang, H.; Liu, Q.; Li, R.; Gao, Y.; Fang, X.; Zhong, Y.; Wang, M.; Wang, Q.; Rao, Z.; et al. Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex. Cell Rep. 2021, 37, 109882. [Google Scholar] [CrossRef] [PubMed]
- Malin, J.J.; Suárez, I.; Priesner, V.; Fätkenheuer, G.; Rybniker, J. Remdesivir against COVID-19 and other viral diseases. Clin Microbiol. Rev. 2020, 34, e00162-20. [Google Scholar] [CrossRef]
- Sesmero, E.; Brown, J.A.; Thorpe, I.F. Molecular simulations to delineate functional conformational transitions in the HCV polymerase. J. Comput. Chem. 2017, 38, 1125–1137. [Google Scholar] [CrossRef]
- Venkataraman, S.; Prasad, B.V.L.S.; Selvarajan, R. RNA-dependent RNA polymerases: Insights from structure, function and evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Selisko, B.; Papageorgiou, N.; Ferron, F.; Canard, B. Structural and functional basis of the fidelity of nucleotide selection by flavivirus RNA-dependent RNA polymerases. Viruses 2018, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Sesmero, E.; Thorpe, I.F. Using the hepatitis C virus RNA-dependent RNA polymerase as a model to understand viral polymerase structure, function and dynamics. Viruses 2015, 7, 3974–3994. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ye, H.-Q.; Zhang, Q.-Y.; Lu, G.; Zhang, B.; Gong, P. A conformation-based intra-molecular initiation factor identified in the flavivirus RNA-dependent RNA polymerase. PLoS Pathog. 2020, 16, e1008484. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- De Clercq, E. Remdesivir: Quo vadis? Biochem. Pharmacol. 2021, 193, 114800. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The architecture of SARS-CoV-2 transcriptome. Cell 2020, 181, 914.e10–921.e10. [Google Scholar] [CrossRef]
- Bhatt, P.R.; Scaiola, A.; Loughran, G.; Leibundgut, M.; Kratzel, A.; Meurs, R.; Dreos, R.; O’Connor, K.M.; McMillan, A.; Bode, J.W.; et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 2021, 372, 1306–1313. [Google Scholar] [CrossRef]
- Wang, D.; Jiang, A.; Feng, J.; Li, G.; Guo, D.; Sajid, M.; Wu, K.; Zhang, Q.; Ponty, Y.; Will, S.; et al. The SARS-CoV-2 subgenome landscape and its novel regulatory features. Mol. Cell 2021, 81, 2135.e5–2147.e5. [Google Scholar] [CrossRef]
- Eriani, G.; Martin, F. Viral and cellular translation during SARS-CoV-2 infection. FEBS Open Bio. 2022, 12, 1584–1601. [Google Scholar] [CrossRef]
- Bouvet, M.; Imbert, I.; Subissi, L.; Gluais, L.; Canard, B.; Decroly, E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA 2012, 109, 9372–9377. [Google Scholar] [CrossRef] [PubMed]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019, 10, 2342. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef]
- Ferron, F.; Subissi, L.; Silveira De Morais, A.T.; Le, N.T.T.; Sevajol, M.; Gluais, L.; Decroly, E.; Vonrhein, C.; Bricogne, G.; Canard, B.; et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. USA 2018, 115, E162–E171. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell 2020, 182, 417.e413–428.e413. [Google Scholar] [CrossRef]
- Peng, Q.; Peng, R.; Yuan, B.; Zhao, J.; Wang, M.; Wang, X.; Wang, Q.; Sun, Y.; Fan, Z.; Qi, J.; et al. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020, 31, 107774. [Google Scholar] [CrossRef]
- Bylehn, F.; Menendez, C.A.; Perez-Lemus, G.R.; Alvarado, W.; de Pablo, J.J. Modeling the binding mechanism of Remdesivir, Favilavir, and Ribavirin to SARS-CoV-2 RNA-dependent RNA polymerase. ACS Cent. Sci. 2021, 7, 164–174. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Gordon, C.J.; Woolner, E.; Kocinkova, D.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020, 295, 16156–16165. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, R.; Shu, B.; Jing, X.; Ye, H.Q.; Gong, P. Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures. Nat. Commun. 2020, 11, 2605. [Google Scholar] [CrossRef] [PubMed]
- Riccio, A.A.; Sullivan, E.D.; Copeland, W.C. Activation of the SARS-CoV-2 NSP14 3′-5′ exoribonuclease by NSP10 and response to antiviral inhibitors. J. Biol. Chem. 2022, 298, 101518. [Google Scholar] [CrossRef] [PubMed]
- Jockusch, S.; Tao, C.; Li, X.; Chien, M.; Kumar, S.; Morozova, I.; Kalachikov, S.; Russo, J.J.; Ju, J. Sofosbuvir-terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci. Rep. 2020, 10, 16577. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sacramento, C.Q.; Jockusch, S.; Chaves, O.A.; Tao, C.; Fintelman-Rodrigues, N.; Chien, M.; Temerozo, J.R.; Li, X.; Kumar, S.; et al. Combination of antiviral drugs inhibits SARS-CoV-2 polymerase and exonuclease and demonstrates COVID-19 therapeutic potential in viral cell culture. Commun. Biol. 2022, 5, 154. [Google Scholar] [CrossRef]
- Barik, S.; McLean, T.; Dupuy, L.C. Phosphorylation of Ser232 directly regulates the transcriptional activity of the P protein of human respiratory syncytial virus: Phosphorylation of Ser237 may play an accessory role. Virology 1995, 213, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Fearns, R.; Plemper, R.K. Polymerases of paramyxoviruses and pneumoviruses. Virus Res. 2017, 234, 87–102. [Google Scholar] [CrossRef]
- Beavis, A.C.; Tran, K.C.; Barrozo, E.R.; Phan, S.I.; Teng, M.N.; Biao, H. Respiratory syncytial virus phosphoprotein residue S156 plays a role in regulating genome transcription and replication. J. Virol. 2021, 95, e0120621. [Google Scholar] [CrossRef]
- Sacramento, C.Q.; De Melo, G.R.; De Freitas, C.S.; Rocha, N.; Hoelz, L.V.B.; Miranda, M.; Fintelman-Rodrigues, N.; Marttorelli, A.; Ferreira, A.C.; Barbosa-Lima, G.; et al. The clinically approved antiviral drug sofosbuvir inhibits Zika virus replication. Sci. Rep. 2017, 7, 40920. [Google Scholar] [CrossRef]
- Xu, H.T.; Colby-Germinario, S.P.; Hassounah, S.A.; Fogarty, C.; Osman, N.; Palanisamy, N.; Han, Y.; Oliveira, M.; Quan, Y.; Wainberg, M.A. Evaluation of Sofosbuvir (β-D-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine) as an inhibitor of Dengue virus replication. Sci. Rep. 2017, 7, 6345. [Google Scholar] [CrossRef]
- De Freitas, C.S.; Higa, L.M.; Sacramento, C.Q.; Ferreira, A.C.; Reis, P.A.; Delvecchio, R.; Monteiro, F.L.; Barbosa-Lima, G.; Westgarth, H.J.; Vieira, Y.R.; et al. Yellow fever virus is susceptible to sofosbuvir both in vitro and in vivo. PLoS Negl. Trop. Dis. 2019, 13, e0007072. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Jiang, W.R.; Robledo, N.; Leveque, V.; Ali, S.; Lara-Jaime, T.; Masjedizadeh, M.; Smith, D.B.; Cammack, N.; Klumpp, K.; et al. Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor beta-D-2′-Deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) and identification of a novel active 5′-triphosphate species. J. Biol. Chem. 2007, 282, 29812–29820. [Google Scholar] [CrossRef] [PubMed]
- Fung, A.; Jin, Z.; Dyatkina, N.; Wang, G.; Beigelman, L.; Deval, J. Efficiency of incorporation and chain termination determines the inhibition potency of 2′-modified nucleotide analogs against hepatitis C virus polymerase. Antimicrob. Agents Chemother. 2014, 58, 3636–3645. [Google Scholar] [CrossRef]
- Stuyver, L.J.; McBrayer, T.R.; Tharnish, P.M.; Clark, J.L.; Hollecher, L.; Lostia, S.; Nachman, T.; Grier, J.; Bennett, M.A.; Xie, M.-Y.; et al. Inhibition of hepatitis C replicon RNA synthesis by β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine: A specific inhibitor of hepatitis C virus replication. Antivir. Chem. Chemother. 2006, 17, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Murakami, E.; Niu, C.; Bao, H.; Steuer, H.M.; Whitaker, T.; Nachman, T.; Sofia, M.A.; Wang, P.; Otto, M.J.; Furman, P.A. The mechanism of action of β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine involves a second metabolic pathway leading to β-d-2′-deoxy-2′-fluoro-2′-C-methyluridine 5′-triphosphate, a potent inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 2008, 52, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.T.; Hassounah, S.A.; Colby-Germinario, S.P.; Oliveira, M.; Fogarty, C.; Quan, Y.; Han, Y.; Golubkov, O.; Ibanescu, I.; Brenner, B.; et al. Purification of Zika virus RNA-dependent RNA polymerase and its use to identify small-molecule Zika inhibitors. J. Antimicrob. Chemother. 2017, 72, 727–734. [Google Scholar] [CrossRef]
- Xu, S.; Doehle, B.; Rajyaguru, S.; Han, B.; Barauskas, O.; Feng, J.; Perry, J.; Dvory-Sobol, H.; Svarovskaia, E.S.; Miller, M.D.; et al. In vitro selection of resistance to sofosbuvir in HCV replicons of genotype-1 to -6. Antivir. Ther. 2017, 22, 587–597. [Google Scholar] [CrossRef]
- Shoun, A.A.; Abozahra, R.; Baraka, K.; Mehrez, M.; Abdelhamid, S.M. Identifying different mutation sites leading to resistance to the direct-acting antiviral (DAA) Sofosbuvir in hepatitis C virus patients from Egypt. Microorganisms 2022, 10, 679. [Google Scholar] [CrossRef]
- Younas, S.; Sumrin, A.; Hussain, N.; Bilal, M. Identification of NS5B resistance against SOFOSBUVIR in hepatitis C virus genotype 3a, naive and treated patients. J. Appl. Microbiol. 2022. Online ahead of print. [Google Scholar] [CrossRef]
- Mosley, R.T.; Edwards, T.E.; Murakami, E.; Lam, A.M.; Grice, R.L.; Du, J.; Sofia, M.J.; Furman, P.A.; Otto, M.J. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 2012, 86, 6503–6511. [Google Scholar] [CrossRef]
- Dutartre, H.; Bussetta, C.; Boretto, J.; Canard, B. General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob. Agents Chemother. 2006, 50, 4161–4169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Bell, E.; Yin, M.; Zhang, Y. EDock: Blind protein-ligand docking by replica-exchange Monte Carlo simulation. J. Cheminfor. 2020, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Appleby, T.C.; Perry, J.K.; Murakami, E.; Barauskas, O.; Feng, J.; Cho, A.; Fox, D.; Wetmore, D.R.; McGrath, M.E.; Ray, A.S.; et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 2015, 347, 771–775. [Google Scholar] [CrossRef]
- Bjork, J.A.; Wallace, K.B. Remdesivir: Molecular and functional measures of mitochondrial safety. Toxicol. Appl. Pharmacol. 2021, 433, 115783. [Google Scholar] [CrossRef]
- Xu, Y.; Barauskas, O.; Kim, C.; Babusis, D.; Murakami, E.; Kornyeyev, D.; Lee, G.; Stepan, G.; Perron, M.; Bannister, R.; et al. Off-target in vitro profiling demonstrates that remdesivir is a highly selective antiviral agent. Antimicrob. Agents Chemother. 2021, 65, e02237-20. [Google Scholar] [CrossRef]
- Ehteshami, M.; Zhou, L.; Amiralaei, S.; Shelton, J.R.; Cho, J.H.; Zhang, H.; Li, H.; Lu, X.; Ozturk, T.; Stanton, R.; et al. Nucleotide substrate specificity of anti-Hepatitis C virus nucleoside analogs for human mitochondrial RNA polymerase. Antimicrob. Agents Chemother. 2017, 61, e00492-17. [Google Scholar] [CrossRef] [PubMed]
- Stein, P.; Svoboda, P.; Anger, M.; Schultz, R.M. RNAi: Mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 2003, 9, 187–192. [Google Scholar] [CrossRef]
- Zhang, C.; Montgomery, T.A.; Fischer, S.E.; Garcia, S.M.; Riedel, C.G.; Fahlgren, N.; Sullivan, C.M.; Carrington, J.C.; Ruvkun, G. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr. Biol. 2012, 22, 881–890. [Google Scholar] [CrossRef]
- Hashemian, S.M.R.; Pourhanifeh, M.H.; Hamblin, M.R.; Shahrzad, M.K.; Mirzaei, H. RdRp inhibitors and COVID-19: Is molnupiravir a good option? Biomed. Pharmacother. 2022, 146, 112517. [Google Scholar] [CrossRef]
- Lu, G.; Zhang, X.; Zheng, W.; Sun, J.; Hua, L.; Xu, L.; Chu, X.-J.; Ding, S.; Xiong, W. Development of a simple in vitro assay to identify and evaluate nucleotide analogs against SARS-CoV-2 RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 2020, 65, e01508-20. [Google Scholar] [CrossRef]
- Antonov, L. Favipiravir tautomerism: A theoretical insight. Theor. Chem. Acc. 2020, 139, 145. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, C.; Pathirana, R.N.; Mahmood, N.; Devine, K.G.; Hay, A.J. Aryl phosphate derivatives of AZT retain activity against HIV1 in cell lines which are resistant to the action of AZT. Antivir. Res. 1992, 17, 311–321. [Google Scholar] [CrossRef]
- Mehellou, Y.; Rattan, H.S.; Balzarini, J. The ProTide prodrug technology: From the concept to the clinic. J. Med. Chem. 2018, 61, 2211–2226. [Google Scholar] [CrossRef] [PubMed]
- Slusarczyk, M.; Serpi, M.; Pertusati, F. Phosphoramidates and phosphonamidates (ProTides) with antiviral activity. Antivir. Chem. Chemother. 2018, 26, 2040206618775243. [Google Scholar] [CrossRef]
- Casida, J.E. Why prodrugs and propesticides succeed. Chem. Res. Toxicol. 2017, 30, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Traore, M.; Li, R.; Yuan, H.; He, M.; Wen, B.; Gao, W.; Jonsson, C.B.; Fitzpatrick, E.A.; Sun, D. Optimization of the prodrug moiety of Remdesivir to improve lung exposure/selectivity and enhance anti-SARS-CoV-2 activity. J. Med. Chem. 2022, 65, 12044–12054. [Google Scholar] [CrossRef]
- Sofia, M.J.; Bao, D.; Chang, W.; Du, J.; Nagarathnam, D.; Rachakonda, S.; Reddy, P.G.; Ross, B.S.; Wang, P.; Zhang, H.-R.; et al. Discovery of a β-D-2-deoxy-2-α-fluoro-2-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 2010, 53, 7202–7218. [Google Scholar] [CrossRef]
- Artursson, P. Epithelial transport of drugs in cell culture. I: A model for studying the passive diffusion of drugs over intestinal absorptive (Caco-2) cells. J. Pharm. Sci. 1990, 79, 476–482. [Google Scholar] [CrossRef]
- Molina-Jimenez, F.; Benedicto, I.; Dao Thi, V.L.; Gondar, V.; Lavillette, D.; Marin, J.J.; Briz, O.; Moreno-Otero, R.; Aldabe, R.; Baumert, T.F.; et al. Matrigel embedded 3D culture of Huh-7 cells as a hepatocyte-like polarized system to study hepatitis C virus cycle. Virology 2012, 425, 31–39. [Google Scholar] [CrossRef]
- Liu, S.; Chen, R.; Hagedorn, C.H. Direct visualization of hepatitis C virus-infected Huh7.5 cells with a high titre of infectious chimeric JFH1-EGFP reporter virus in three-dimensional Matrigel cell cultures. J. Gen. Virol. 2014, 95, 423–433. [Google Scholar] [CrossRef][Green Version]
- Ruedinger, F.; Lavrentieva, A.; Blume, C.; Pepelanova, I.; Scheper, T. Hydrogels for 3D mammalian cell culture: A starting guide for laboratory practice. Appl. Microbiol. Biotechnol. 2015, 99, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Dankó, T.; Petővári, G.; Raffay, R.; Sztankovics, D.; Moldvai, D.; Vetlényi, E.; Krencz, I.; Rókusz, A.; Sipos, K.; Visnovitz, T.; et al. Characterisation of 3D bioprinted human breast cancer model for in vitro drug and metabolic targeting. Int. J. Mol. Sci. 2022, 23, 7444. [Google Scholar] [CrossRef]
- Chan, M.C.W.; Chan, R.Y.W.; Yu, W.C.L.; Ho, C.C.C.; Chui, W.H.; Lo, C.K.; Yuen, K.M.; Guan, Y.I.; Nicholls, J.M.; Peiris, J.S.M. Influenza H5N1 virus infection of polarized human alveolar epithelial cells and lung microvascular endothelial cells. Respir. Res. 2009, 10, 102. [Google Scholar] [CrossRef]
- Villenave, R.; Shields, M.D.; Power, U.F. Respiratory syncytial virus interaction with human airway epithelium. Trends Microbiol. 2013, 21, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Bitko, V.; Garmon, N.E.; Cao, T.; Estrada, B.; Oakes, J.E.; Lausch, R.N.; Barik, S. Activation of cytokines and NF-kappa B in corneal epithelial cells infected by respiratory syncytial virus: Potential relevance in ocular inflammation and respiratory infection. BMC Microbiol. 2004, 4, 28. [Google Scholar] [CrossRef] [PubMed][Green Version]
- O’Brien, L.E.; Zegers, M.M.P.; Mostov, K.E. Opinion: Building epithelial architecture: Insights from three-dimensional culture models. Nat. Rev. Mol. Cell Biol. 2002, 3, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Louz, D.; Bergmans, H.E.; Loos, B.P.; Hoeben, R.C. Animal models in virus research: Their utility and limitations. Crit. Rev. Microbiol. 2013, 39, 325–361. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barik, S. Inhibition of Viral RNA-Dependent RNA Polymerases by Nucleoside Inhibitors: An Illustration of the Unity and Diversity of Mechanisms. Int. J. Mol. Sci. 2022, 23, 12649. https://doi.org/10.3390/ijms232012649
Barik S. Inhibition of Viral RNA-Dependent RNA Polymerases by Nucleoside Inhibitors: An Illustration of the Unity and Diversity of Mechanisms. International Journal of Molecular Sciences. 2022; 23(20):12649. https://doi.org/10.3390/ijms232012649
Chicago/Turabian StyleBarik, Sailen. 2022. "Inhibition of Viral RNA-Dependent RNA Polymerases by Nucleoside Inhibitors: An Illustration of the Unity and Diversity of Mechanisms" International Journal of Molecular Sciences 23, no. 20: 12649. https://doi.org/10.3390/ijms232012649
APA StyleBarik, S. (2022). Inhibition of Viral RNA-Dependent RNA Polymerases by Nucleoside Inhibitors: An Illustration of the Unity and Diversity of Mechanisms. International Journal of Molecular Sciences, 23(20), 12649. https://doi.org/10.3390/ijms232012649







