A Heat Shock Transcription Factor TrHSFB2a of White Clover Negatively Regulates Drought, Heat and Salt Stress Tolerance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results
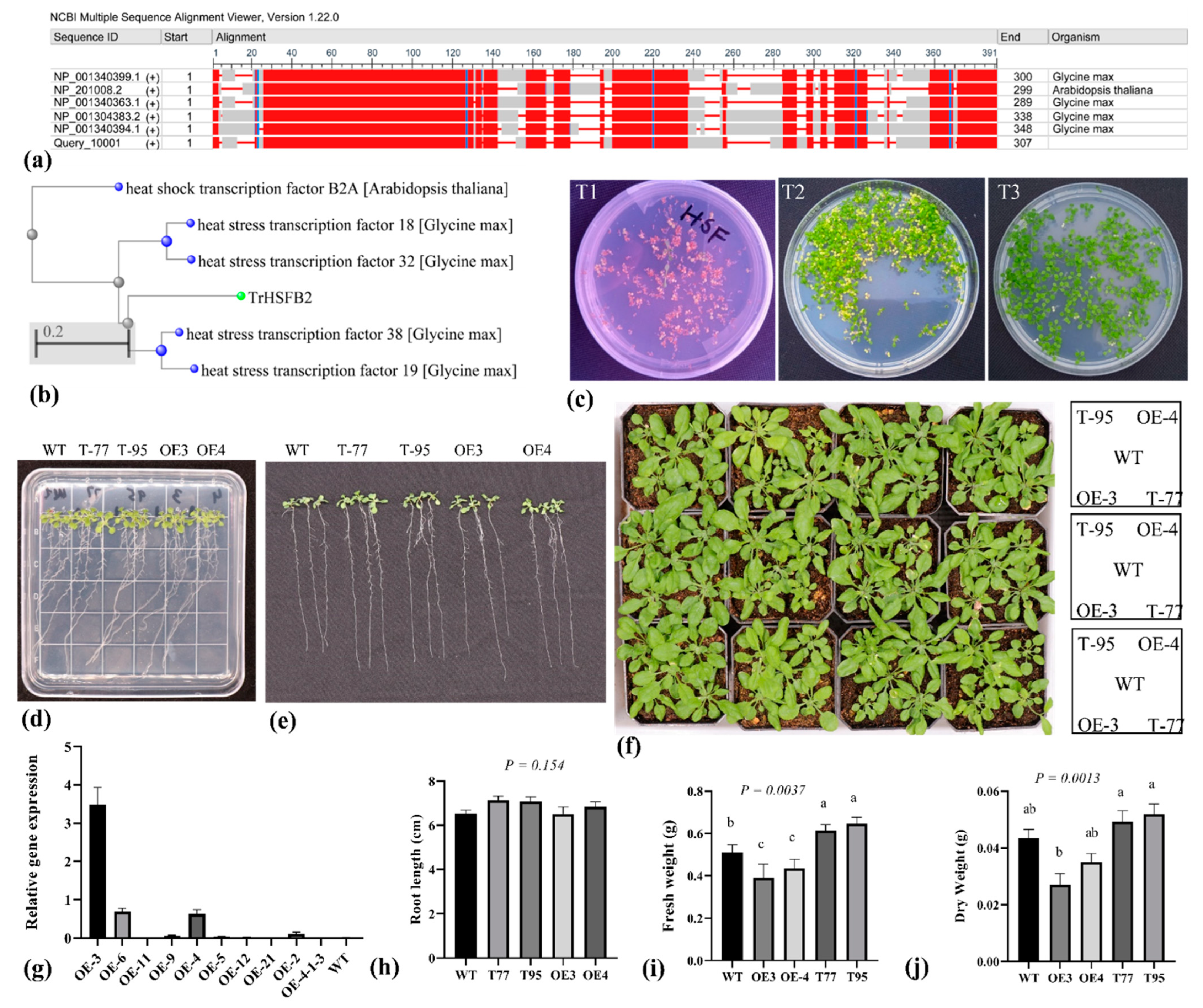
2.1. TrHSFB2a Was Predicated as One of the Class B HSF Proteins
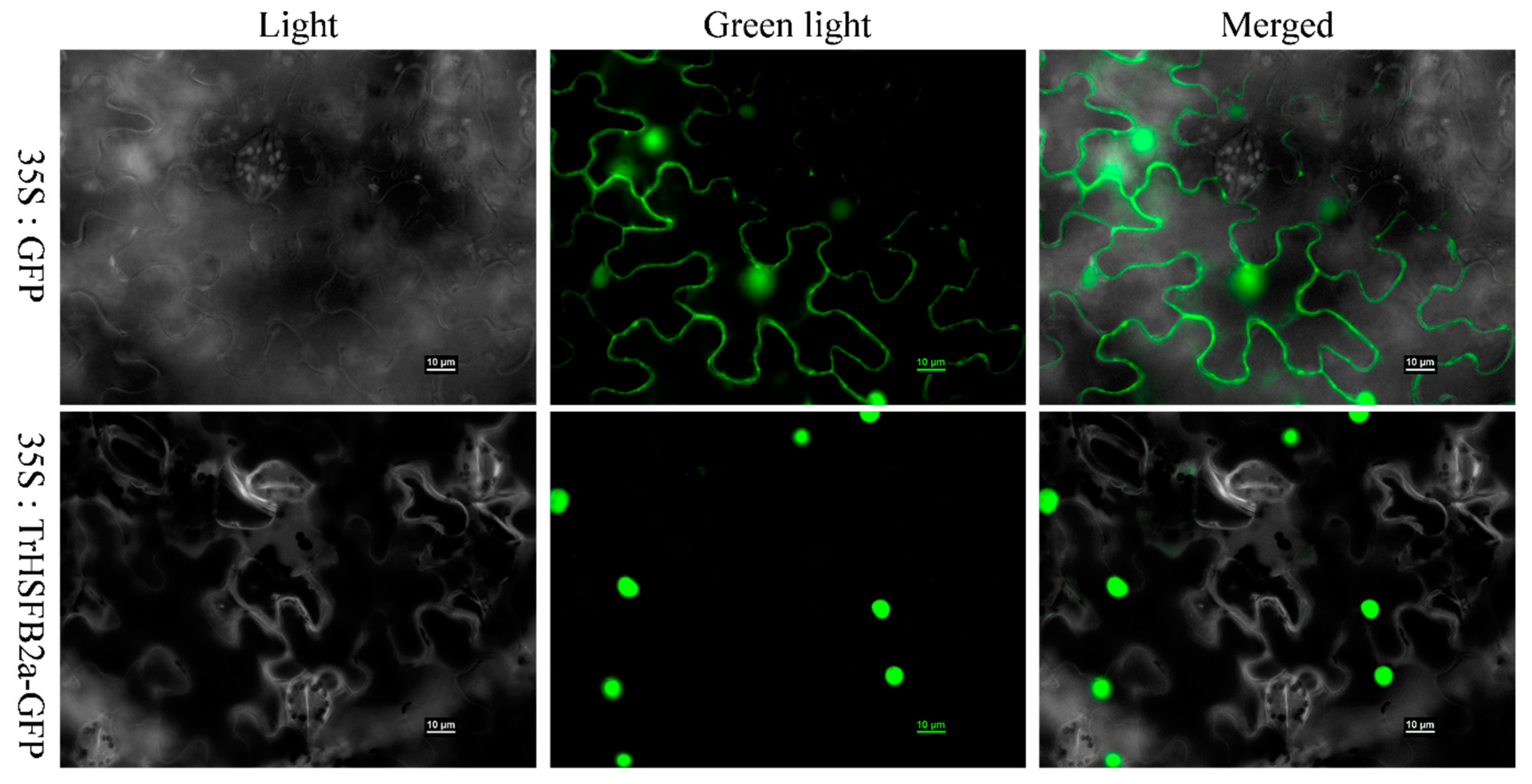
2.2. TrHSFB2a Is a Nuclear Protein
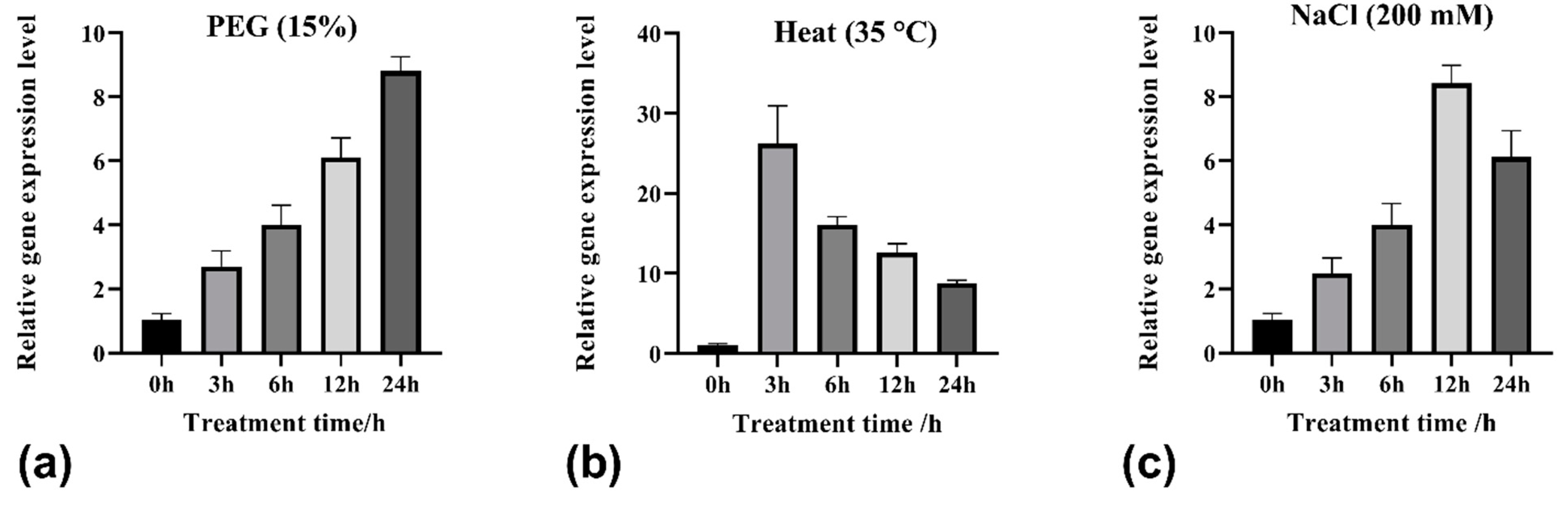
2.3. Relative Expression of TrHSFB2a in White Clover under Different Conditions
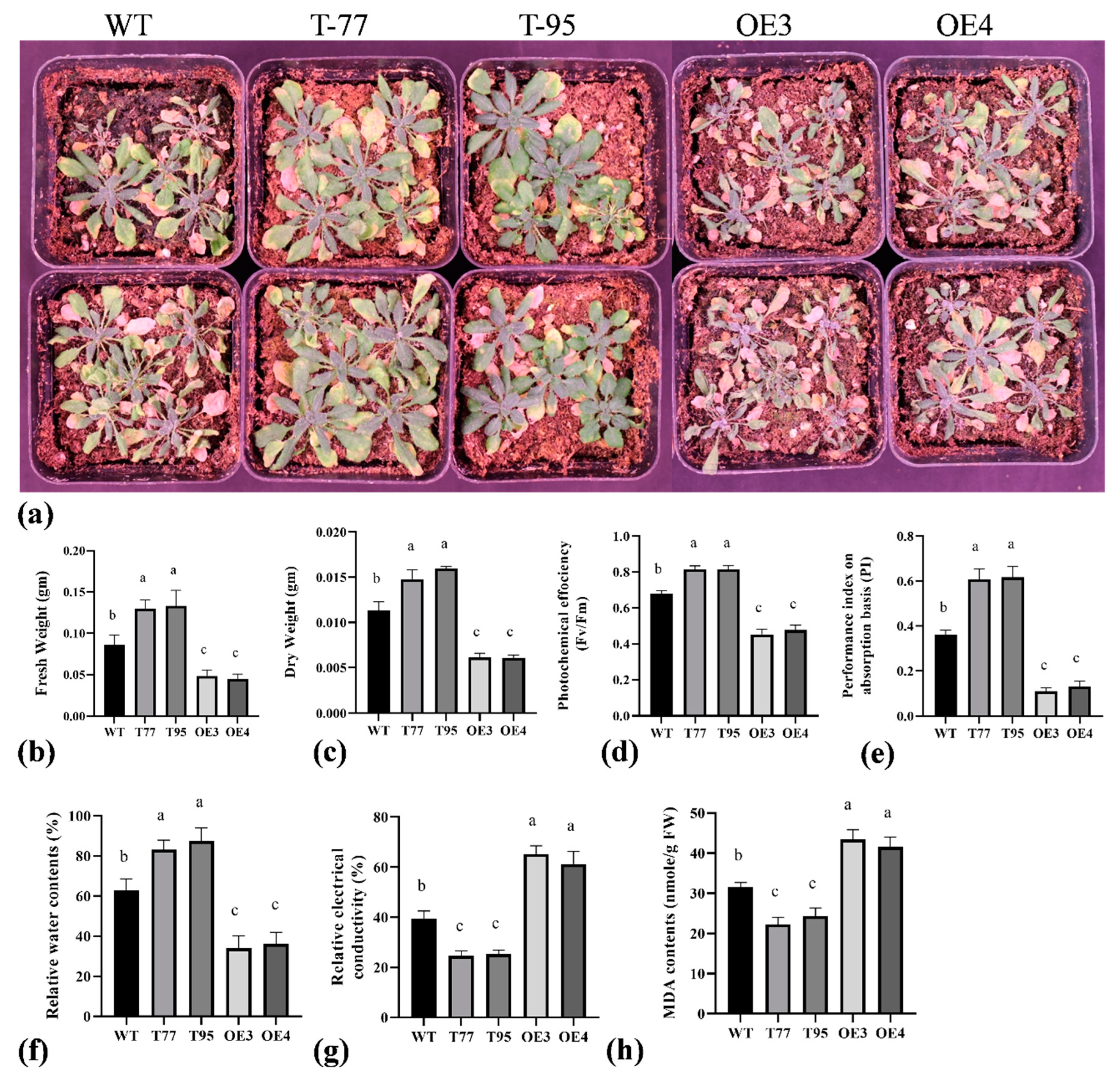
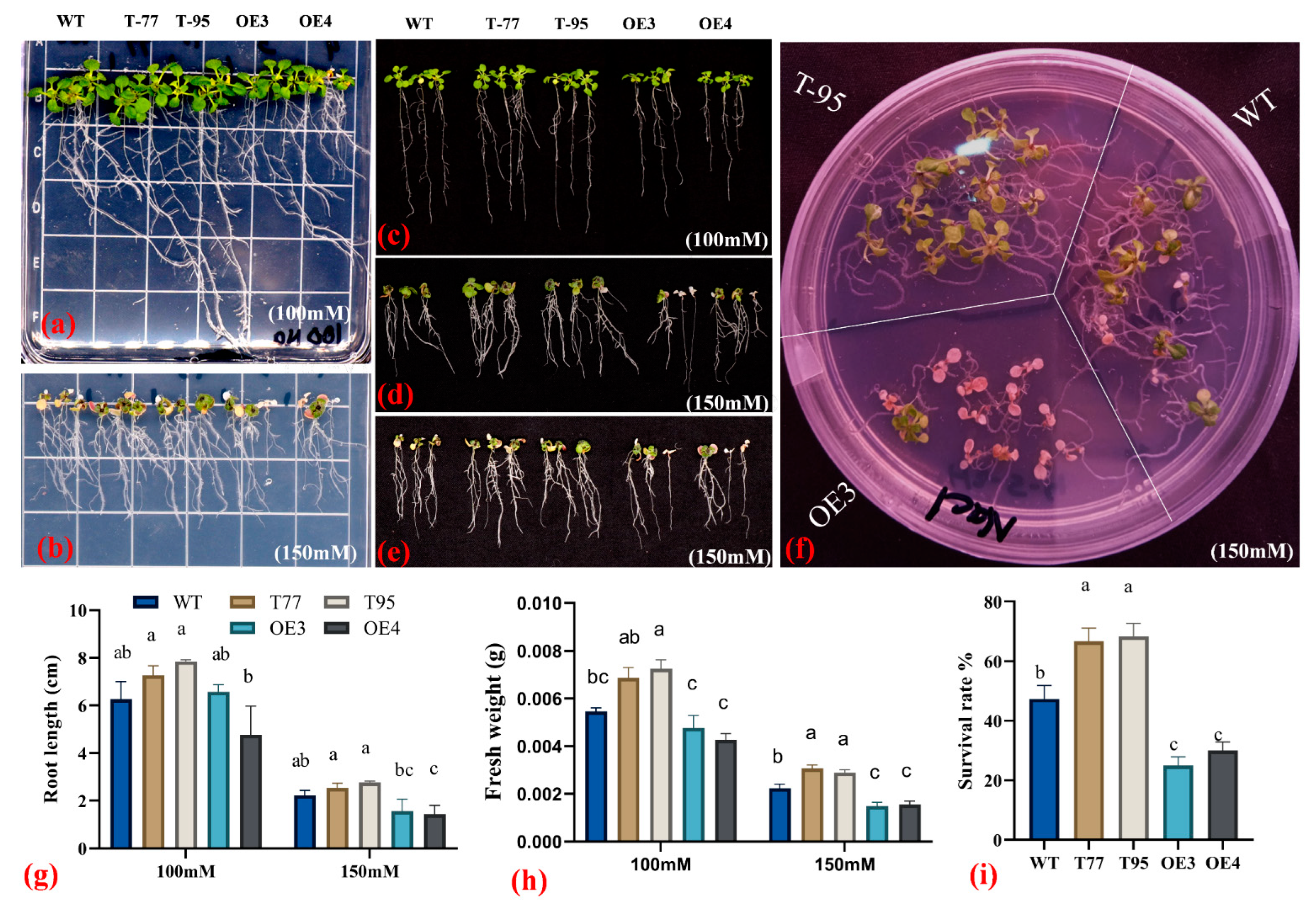
2.4. TrHSFB2a Negatively Regulates Drought Tolerance in Transgenic Overexpression Arabidopsis
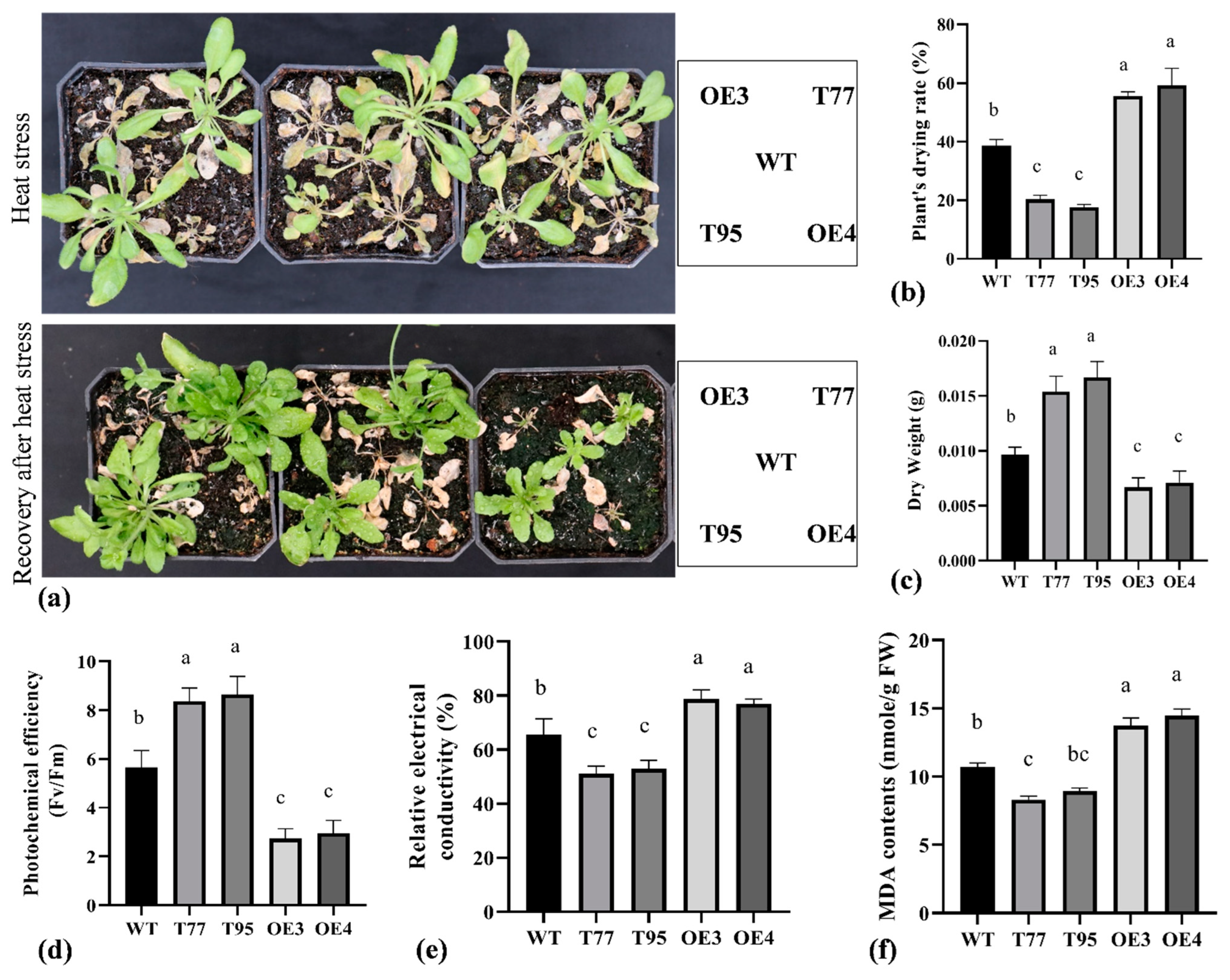
2.5. TrHSFB2a Negatively Regulates the Heat Tolerance in Arabidopsis
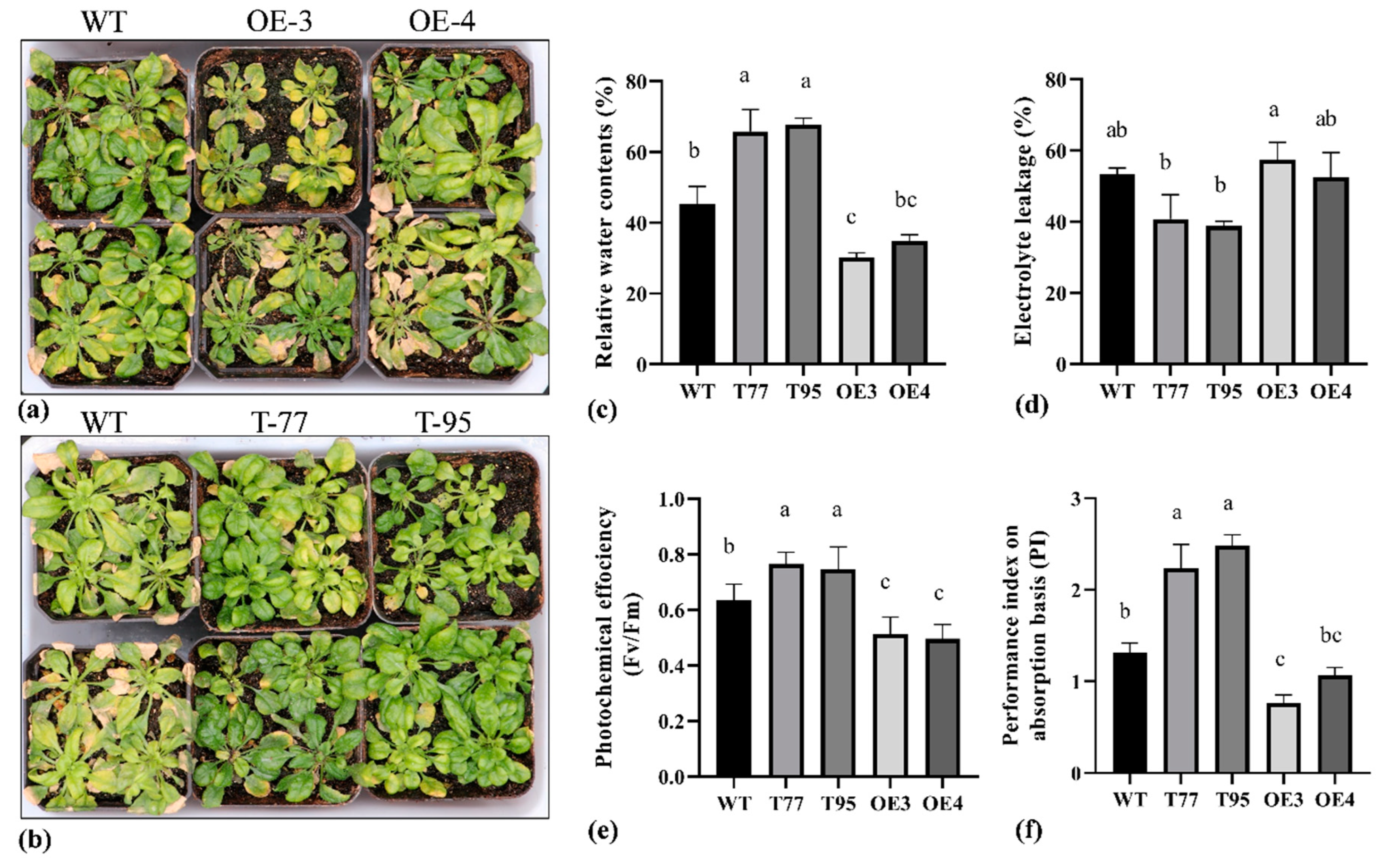
2.6. TrHSFB2a Negatively Regulates Salt Tolerance in Arabidopsis
3. Discussion
3.1. Abiotic Stress Treatments Induce TrHSFB2a Expression
3.2. TrHSFB2a Negatively Regulates Drought, Heat and Salt Stress in Arabidopsis
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. TrHSFB2 Gene Isolation from White Clover and Bioinformatics Analysis
4.3. Plasmid Construction, Genetic Transformation and Developing Homozygous Overexpression Lines of TrHSFB2a
4.4. Subcellular Localization
4.5. Relative Expression of TrHSFB2a
4.6. Drought, Salt and Heat Stress in Arabidopsis
4.7. Determination of Relative Water Contents (RWC)
4.8. Determination of Relative Electrical Conductivity (EC)
4.9. Determination of Chlorophyll Fluorescence Parameters
4.10. Determination of Malondialdehyde (MDA) Content
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vaseva, I.; Akiscan, Y.; Demirevska, K.; Anders, I.; Feller, U. Drought stress tolerance of red and white clover–comparative analysis of some chaperonins and dehydrins. Sci. Hortic. 2011, 130, 653–659. [Google Scholar]
- Chaves, M.M.; Oliveira, M.M. Mechanisms underlying plant resilience to water deficits: Prospects for water-saving agriculture. J. Exp. Bot. 2004, 55, 2365–2384. [Google Scholar] [PubMed] [Green Version]
- Annicchiarico, P.; Piano, E. Indirect Selection for root development of white clover and implications for drought tolerance. J Agron. Crop Sci. 2004, 190, 28–34. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Andrási, N.; Pettkó-Szandtner, A.; Szabados, L. Diversity of plant heat shock factors: Regulation, interactions, and functions. J. Exp. Bot. 2021, 72, 1558–1575. [Google Scholar] [CrossRef]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. BBA-Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar]
- Scharf, K.-D.; Rose, S.; Zott, W.; Schöffl, F.; Nover, L.; Schöff, F. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990, 9, 4495–4501. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.-H.; Ma, X.; Luo, D.-X.; Gong, Z.-H.; Lu, M.-H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- Chung, E.; Kim, K.-M.; Lee, J.-H. Genome-wide analysis and molecular characterization of heat shock transcription factor family in Glycine max. J. Genet. Genom. 2013, 40, 127–135. [Google Scholar] [CrossRef]
- Lin, Y.-X.; Jiang, H.-Y.; Chu, Z.-X.; Tang, X.-L.; Zhu, S.-W.; Cheng, B.-J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.; Zhu, W.; Song, X.; Lin, X.; Cai, J.; Wang, M.; Yang, Q. Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Sci. 2016, 7, 490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Liu, G.; Duan, W.; Liu, T.; Huang, Z.; Ren, J.; Li, Y.; Hou, X. Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol. Genet. Genom. 2014, 289, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Liu, B.; Zhang, Y.; Li, G.; Guo, X. Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genom. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Li, G.; Fu, C.; Duan, S.; Hu, D.; Guo, X. Genome-wide identification, transcriptome analysis and alternative splicing events of Hsf family genes in maize. Sci. Rep. 2020, 10, 8073. [Google Scholar] [CrossRef]
- Xue, G.-P.; Sadat, S.; Drenth, J.; McIntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, Y.; Tao, X.; Wang, J.-Z.; Cheng, H.-Y.; Yang, H.; Ma, X.-R. Heat shock factor genes of tall fescue and perennial ryegrass in response to temperature stress by RNA-Seq analysis. Front. Plant Sci. 2016, 6, 1226. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, T.; Oberkofler, V.; Trindade, I.; Altmann, S.; Brzezinka, K.; Lämke, J.; Gorka, M.; Kappel, C.; Sokolowska, E.; Skirycz, A. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 2021, 12, 3426. [Google Scholar] [CrossRef]
- Yue, Z.; Wang, Y.; Zhang, N.; Zhang, B.; Niu, Y. Expression of the Amorphophallus albus heat stress transcription factor AaHsfA1 enhances tolerance to environmental stresses in Arabidopsis. Ind. Crop Prod. 2021, 174, 114231. [Google Scholar] [CrossRef]
- Fabri, J.H.T.M.; Rocha, M.C.; Fernandes, C.M.; Persinoti, G.F.; Ries, L.N.A.; Cunha, A.F.d.; Goldman, G.H.; Del Poeta, M.; Malavazi, I. The heat shock transcription factor HsfA is essential for thermotolerance and regulates cell wall integrity in Aspergillus fumigatus. Front. Microbiol. 2021, 12, 735. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.-D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperon. 2001, 6, 177. [Google Scholar] [CrossRef]
- von Koskull-Döring, P.; Scharf, K.-D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Fragkostefanakis, S.; Simm, S.; El-Shershaby, A.; Hu, Y.; Bublak, D.; Mesihovic, A.; Darm, K.; Mishra, S.K.; Tschiersch, B.; Theres, K. The repressor and co-activator HsfB1 regulates the major heat stress transcription factors in tomato. Plant Cell Environ. 2019, 42, 874–890. [Google Scholar] [CrossRef] [PubMed]
- Bharti, K.; von Koskull-Döring, P.; Bharti, S.; Kumar, P.; Tintschl-Körbitzer, A.; Treuter, E.; Nover, L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef] [Green Version]
- Jeong, G.; Jeon, M.; Shin, J.; Lee, I. Heat shock transcription factor B2b acts as a transcriptional repressor of VIN3, a gene induced by long-term cold for flowering. Sci. Rep. 2022, 12, 10963. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Busch, W.; Birke, H.; Kemmerling, B.; Nürnberger, T.; Schöffl, F. Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1. 2 expression and pathogen resistance in Arabidopsis. Mol. Plant 2009, 2, 152–165. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, Y.; Zhang, X.; Merewitz, E.; Peng, Y.; Ma, X.; Huang, L.; Yan, Y. Metabolic pathways regulated by chitosan contributing to drought resistance in white clover. J. Proteom. Res. 2017, 16, 3039–3052. [Google Scholar] [CrossRef]
- Li, Z.; Hou, J.; Zhang, Y.; Zeng, W.; Cheng, B.; Hassan, M.J.; Zhang, Y.; Pu, Q.; Peng, Y. Spermine regulates water balance associated with Ca2+-dependent aquaporin (TrTIP2-1, TrTIP2-2 and TrPIP2-7) expression in plants under water stress. Plant Cell Physiol. 2020, 61, 1576–1589. [Google Scholar] [CrossRef]
- Liu, H.-L.; Lee, Z.-X.; Chuang, T.-W.; Wu, H.-C. Effect of heat stress on oxidative damage and antioxidant defense system in white clover (Trifolium repens L.). Planta 2021, 254, 103. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999, 294(5), 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nature Methods. 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, L.; Cao, W.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Characterization and functional analysis of FaHsfC1b from Festuca arundinacea conferring heat tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Diouf, D.; Cissé, N. Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 2016, 7, 1522. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, R.; Schippers, J.H.; Welker, A.; Mieulet, D.; Guiderdoni, E.; Mueller-Roeber, B. Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants 2012, 2012, pls011. [Google Scholar] [CrossRef]
- Ma, H.; Wang, C.; Yang, B.; Cheng, H.; Wang, Z.; Mijiti, A.; Ren, C.; Qu, G.; Zhang, H.; Ma, L. CarHSFB2, a class B heat shock transcription factor, is involved in different developmental processes and various stress responses in chickpea (Cicer arietinum L.). Plant Mol. Biol. Rep. 2016, 34, 1–14. [Google Scholar] [CrossRef]
- Xiang, J.; Ran, J.; Zou, J.; Zhou, X.; Liu, A.; Zhang, X.; Peng, Y.; Tang, N.; Luo, G.; Chen, X. Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Rep. 2013, 32, 1795–1806. [Google Scholar] [CrossRef]
- Czarnecka-Verner, E.; Pan, S.; Salem, T.; Gurley, W.B. Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol. Biol. 2004, 56, 57–75. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Chidambaranathan, P.; Jagannadham, P.T.K.; Satheesh, V.; Kohli, D.; Basavarajappa, S.H.; Chellapilla, B.; Kumar, J.; Jain, P.K.; Srinivasan, R. Genome-wide analysis identifies chickpea (Cicer arietinum) heat stress transcription factors (Hsfs) responsive to heat stress at the pod development stage. J. Plant Res. 2018, 131, 525–542. [Google Scholar] [CrossRef] [PubMed]
- Arvin, M.; Donnelly, D. Screening potato cultivars and wild species to abiotic stresses using an electrolyte leakage bioassay. J. Agric. Sci. Technol.-Iran 2008, 10, 33–42. [Google Scholar]
- Mantri, N.; Patade, V.; Pang, E. Recent advances in rapid and sensitive screening for abiotic stress tolerance. In Improvement of Crops in the Era of Climatic Changes; Ahmed, P., Wani, M.R., Azooz, M.M., Tran, L.S.P., Eds.; Springer: New York, NY, USA, 2014; Volume 2, pp. 37–47. [Google Scholar]
- Tommasino, E.; Griffa, S.; Grunberg, K.; Ribotta, A.; Lopez Colomba, E.; Carloni, E.; Quiroga, M.; Luna, C.M. Malondialdehyde content as a potential biochemical indicator of tolerant Cenchrus ciliaris L. genotypes under heat stress treatment. Grass Forage Sci. 2012, 67, 456–459. [Google Scholar] [CrossRef]
- Luna, C.; Garcia-Seffino, L.; Arias, C.; Taleisnik, E. Oxidative stress indicators as selection tools for salt tolerance. Plant Breed. 2000, 119, 341–345. [Google Scholar] [CrossRef]
- Lü, X.-P.; Shao, K.-Z.; Xu, J.-Y.; Li, J.-L.; Ren, W.; Chen, J.; Zhao, L.-Y.; Zhao, Q.; Zhang, J.-L. A heat shock transcription factor gene (HaHSFA1) from a desert shrub, Haloxylon ammodendron, elevates salt tolerance in Arabidopsis thaliana. Environ. Exp. Bot. 2022, 201, 104954. [Google Scholar] [CrossRef]
- Xia, H.; Zhou, Y.; Deng, H.; Lin, L.; Deng, Q.; Wang, J.; Lv, X.; Zhang, X.A.; Liang, D. Melatonin improves heat tolerance in Actinidia deliciosa via carotenoid biosynthesis and heat shock proteins expression. Physiol. Plant. 2021, 172, 1582–1593. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information, Basic Local Alignment Search Tool. Available online: http://www.ncbi.nlm.nih.gov/Blast (accessed on 12 April 2020).
- National Library of Medicine, National Center for Biotechnology Information. Trifolium Repens (assembly AgR_To_v5). Available online: https://www.ncbi.nlm.nih.gov/genome/13404 (accessed on 12 April 2020).
- Protein Identification and Analysis Tools on the ExPASy Server. Expasy, ProtParam Tool. Available online: https://web.expasy.org/protparam/ (accessed on 1 March 2022).
- Clustal Omega. Multiple Sequence Alignment. Available online: https://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 1 March 2022).
- PSORT: Protein Subcellular Localization Prediction Tool. Available online: https://www.genscript.com/psort.html (accessed on 10 December 2020).
- Search for Conserved Domains within a Protein or Coding Nucleotide Sequence, NCBI. Available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 1 March 2022).
- Open Reading Frame Finder, NIH, NCBI. Available online: https://www.ncbi.nlm.nih.gov/orffinder (accessed on 10 May 2020).
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jia, T.; Hou, J.; Iqbal, M.Z.; Zhang, Y.; Cheng, B.; Feng, H.; Li, Z.; Liu, L.; Zhou, J.; Feng, G. Overexpression of the white clover TrSAMDC1 gene enhanced salt and drought resistance in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 165, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Barrs, H.; Weatherley, P. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef] [Green Version]
- Blum, A.; Ebercon, A. Cell membrane stability as a measure of drought and heat tolerance in wheat 1. Crop Sci. 1981, 21, 43–47. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iqbal, M.Z.; Jia, T.; Tang, T.; Anwar, M.; Ali, A.; Hassan, M.J.; Zhang, Y.; Tang, Q.; Peng, Y. A Heat Shock Transcription Factor TrHSFB2a of White Clover Negatively Regulates Drought, Heat and Salt Stress Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2022, 23, 12769. https://doi.org/10.3390/ijms232112769
Iqbal MZ, Jia T, Tang T, Anwar M, Ali A, Hassan MJ, Zhang Y, Tang Q, Peng Y. A Heat Shock Transcription Factor TrHSFB2a of White Clover Negatively Regulates Drought, Heat and Salt Stress Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2022; 23(21):12769. https://doi.org/10.3390/ijms232112769
Chicago/Turabian StyleIqbal, Muhammad Zafar, Tong Jia, Tao Tang, Muhammad Anwar, Asif Ali, Muhammad Jawad Hassan, Youzhi Zhang, Qilin Tang, and Yan Peng. 2022. "A Heat Shock Transcription Factor TrHSFB2a of White Clover Negatively Regulates Drought, Heat and Salt Stress Tolerance in Transgenic Arabidopsis" International Journal of Molecular Sciences 23, no. 21: 12769. https://doi.org/10.3390/ijms232112769
APA StyleIqbal, M. Z., Jia, T., Tang, T., Anwar, M., Ali, A., Hassan, M. J., Zhang, Y., Tang, Q., & Peng, Y. (2022). A Heat Shock Transcription Factor TrHSFB2a of White Clover Negatively Regulates Drought, Heat and Salt Stress Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences, 23(21), 12769. https://doi.org/10.3390/ijms232112769






