Docetaxel Resistance in Castration-Resistant Prostate Cancer: Transcriptomic Determinants and the Effect of Inhibiting Wnt/β-Catenin Signaling by XAV939
Abstract
:1. Introduction
2. Results
2.1. Analysis of the Biological Pathways Associated with Docetaxel Resistance
2.2. Transcriptome Analysis of Plasma Exosomes in CRPC Patients during Docetaxel Therapy
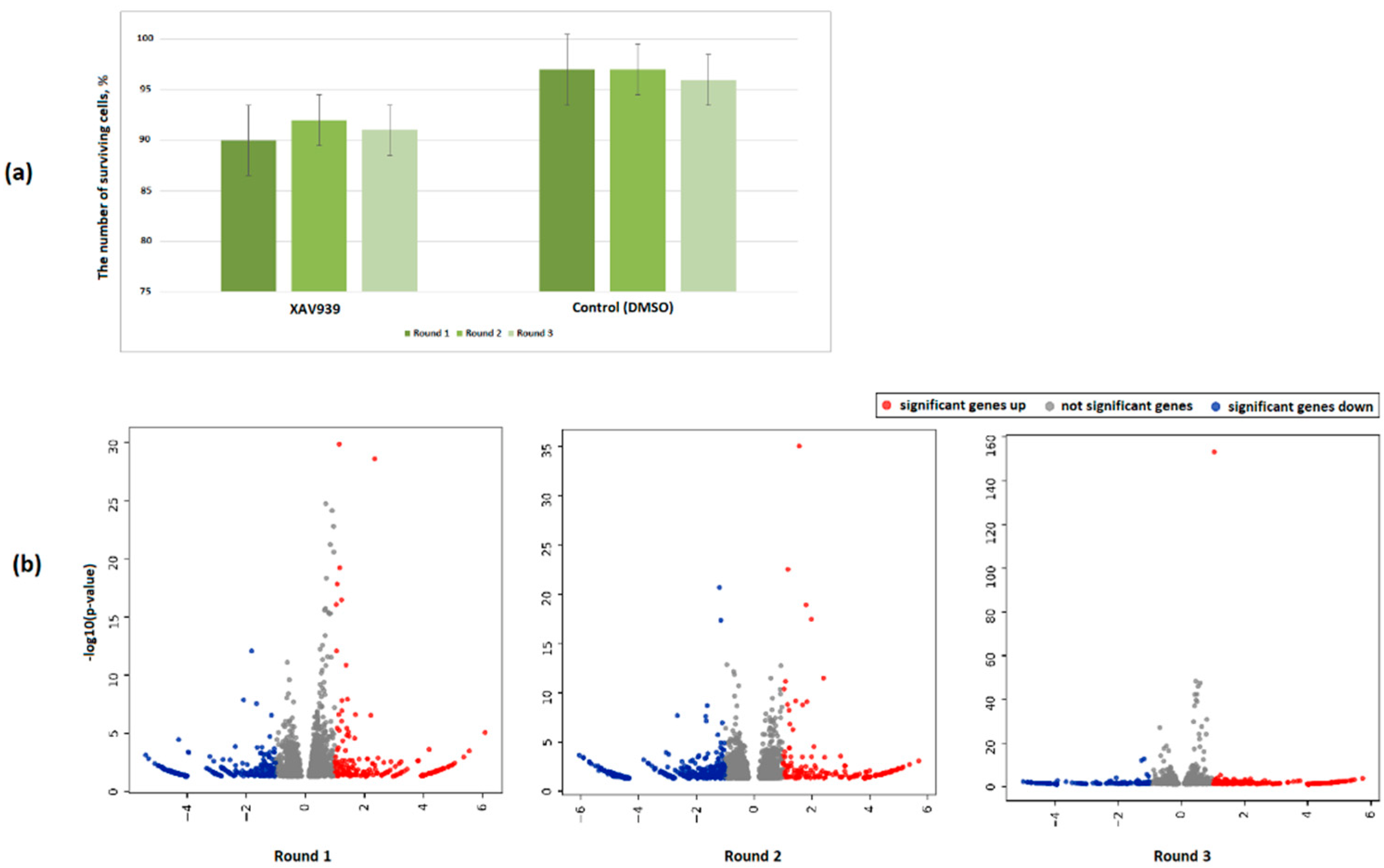
2.3. Features of the PC3 Cells Transcriptome Profile after Treatment with XAV939
2.4. Effect of the XAV939+Docetaxel Combination on the PC3 Cells Transcriptome
2.5. Identification of Promising lncRNAs Associated with Docetaxel Resistance
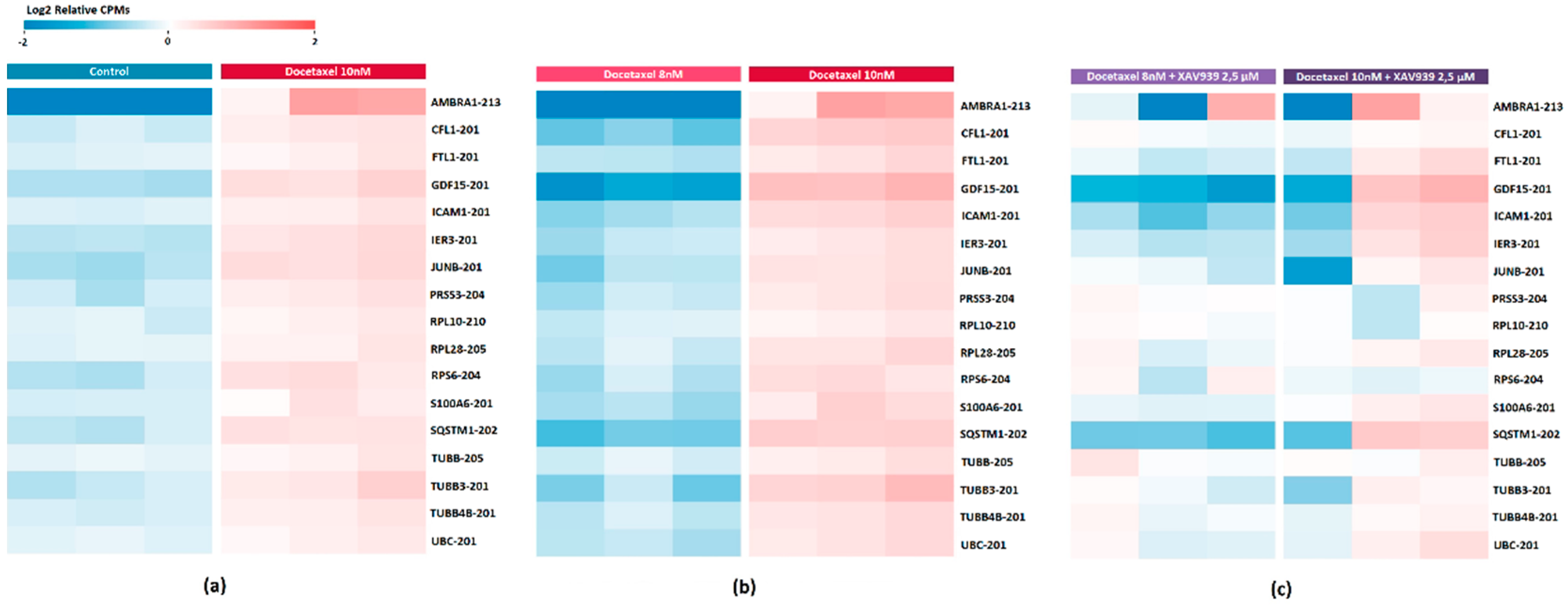
2.6. DE Transcripts as Potential Markers of Response to Docetaxel Based on PC3 Cell Data
2.7. Evaluation of the Effect of XAV939+Docetaxel Combination on PC3 Cells Based on the Identified DE Transcripts
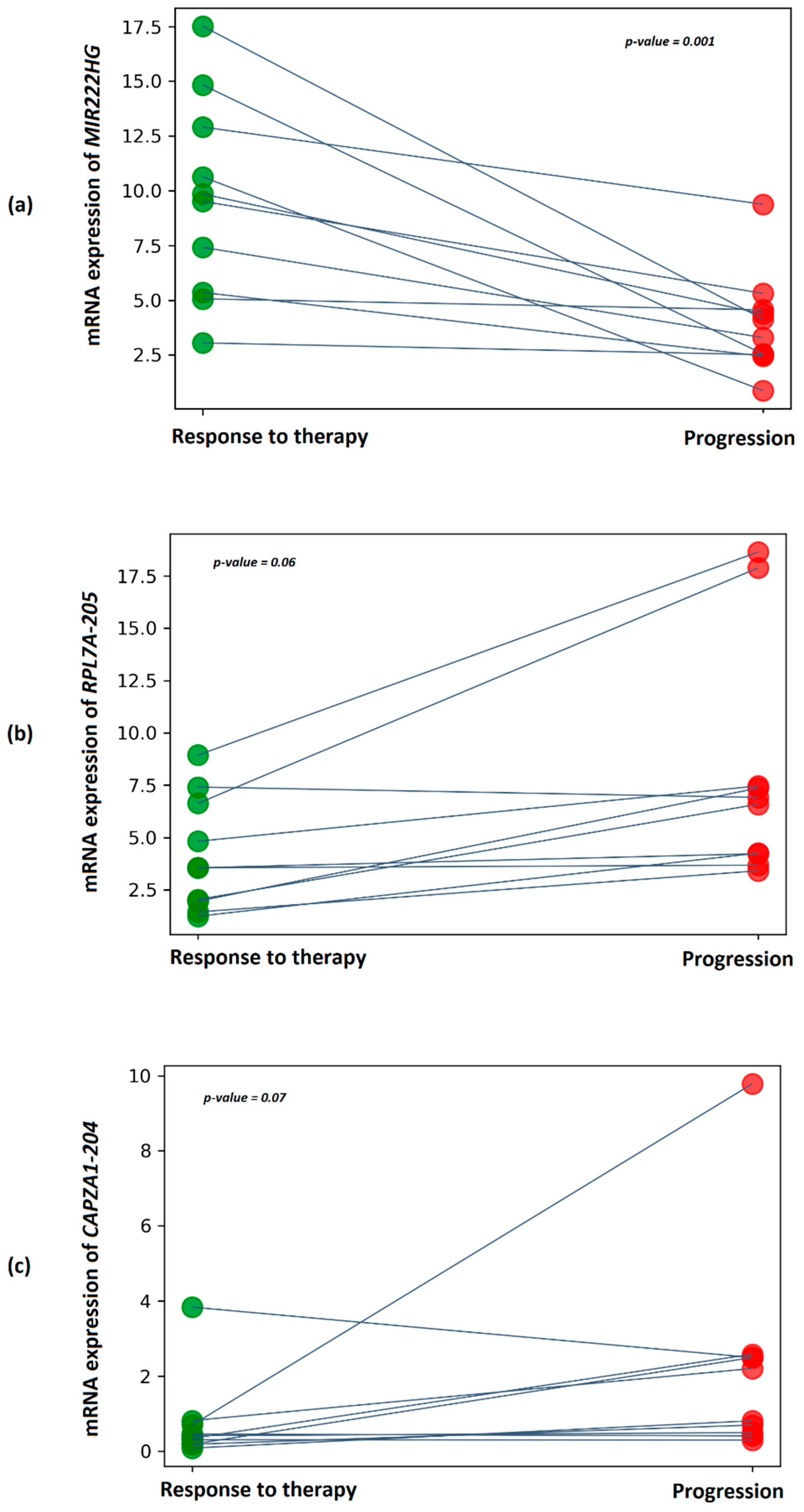
2.8. DE Transcripts as Potential Markers of Docetaxel Response Based on the Exosome Plasma Samples
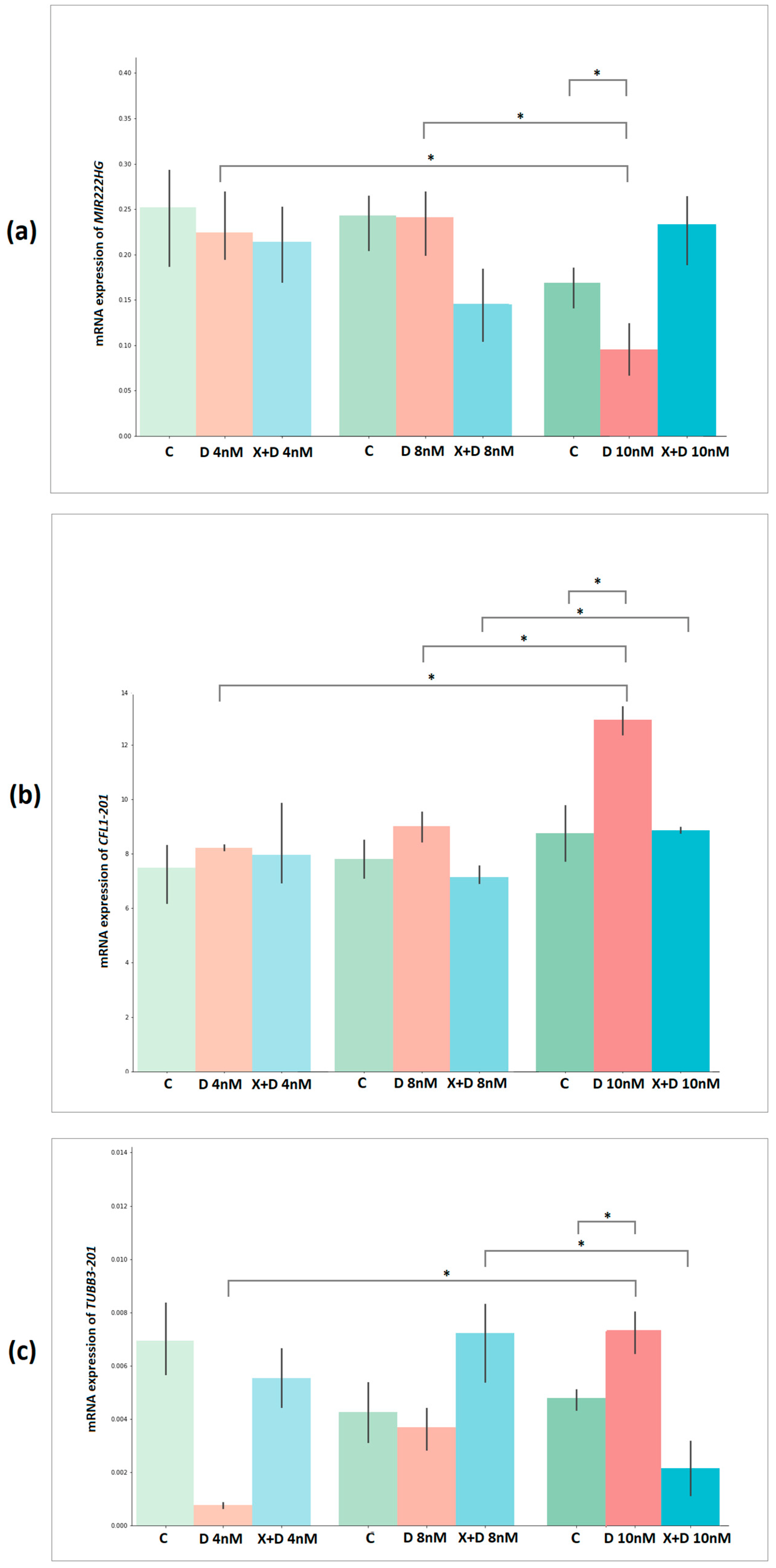
2.9. Validation of Docetaxel Resistance Potential Markers Expression by Quantitative PCR
3. Discussion
4. Materials and Methods
4.1. PC3 Cells
4.2. Treatments of PC3 Cells
4.3. Isolation of Total RNA from Cells
4.4. Plasma of Patients with CRPC Treated with Docetaxel
4.5. Isolation of Total Exosomal RNA from Blood Plasma Samples
4.6. Library Preparation and High Throughput Sequencing
4.7. Bioinformatics Analysis
4.8. Quantitative PCR (qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pernar, C.H.; Ebot, E.M.; Wilson, K.M.; Mucci, L.A. The Epidemiology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a030361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirby, M.; Hirst, C.; Crawford, E.D. Characterising the castration-resistant prostate cancer population: A systematic review. Int. J. Clin. Pract. 2011, 65, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Attard, G.; Parker, C.; Eeles, R.A.; Schroder, F.; Tomlins, S.A.; Tannock, I.; Drake, C.G.; de Bono, J.S. Prostate cancer. Lancet 2016, 387, 70–82. [Google Scholar] [CrossRef]
- Van der Toom, E.E.; Axelrod, H.D.; de la Rosette, J.J.; de Reijke, T.M.; Pienta, K.J.; Valkenburg, K.C. Prostate-specific markers to identify rare prostate cancer cells in liquid biopsies. Nat. Rev. Urol. 2019, 16, 7–22. [Google Scholar] [CrossRef]
- Rosenberg, E.E.; Gerashchenko, G.V.; Hryshchenko, N.V.; Mevs, L.V.; Nekrasov, K.A.; Lytvynenko, R.A.; Vitruk, Y.V.; Gryzodub, O.P.; Stakhovsky, E.A.; Kashuba, V.I. Expression of cancer-associated genes in prostate tumors. Exp. Oncol. 2017, 39, 131–137. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Bryzgunova, O.E.; Laktionov, P.P. miRNAs and radiotherapy response in prostate cancer. Andrology 2020, 9, 529–545. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exo-some-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef]
- Ji, J.; Chen, R.; Zhao, L.; Xu, Y.; Cao, Z.; Xu, H.; Chen, X.; Shi, X.; Zhu, Y.; Lyu, J.; et al. Circulating exosomal mRNA profiling identifies novel signatures for the detection of prostate cancer. Mol. Cancer 2021, 20, 58. [Google Scholar] [CrossRef]
- Safikhani, Z.; Smirnov, P.; Thu, K.L.; Silvester, J.; El-Hachem, N.; Quevedo, R.; Lupien, M.; Mak, T.W.; Cescon, D.; Haibe-Kains, B. Author Correction: Gene isoforms as expression-based biomarkers predictive of drug response in vitro. Nat. Commun. 2018, 9, 166. [Google Scholar] [CrossRef]
- Hillebrand, A.C.; Pizzolato, L.S.; Neto, B.S.; Branchini, G.; Brum, I.S. Androgen receptor isoforms expression in benign prostatic hyperplasia and primary prostate cancer. PLoS ONE 2018, 13, e0200613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamurthy, N.; Kurzrock, R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2017, 62, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Koushyar, S.; Meniel, V.S.; Phesse, T.J.; Pearson, H.B. Exploring the Wnt Pathway as a Therapeutic Target for Prostate Cancer. Biomolecules 2022, 12, 309. [Google Scholar] [CrossRef]
- Yokoyama, N.N.; Shao, S.; Hoang, B.H.; Mercola, D.; Zi, X. Wnt signaling in castration-resistant prostate cancer: Implications for therapy. Am. J. Clin. Exp. Urol. 2014, 2, 27–44. [Google Scholar]
- Li, C.; Zheng, X.; Han, Y.; Lv, Y.; Lan, F.; Zhao, J. XAV939 inhibits the proliferation and migration of lung adenocarcinoma A549 cells through the WNT pathway. Oncol. Lett. 2018, 15, 8973–8982. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Shen, F.; Yang, L.; Zhang, L.; Guo, W.; Tian, J. Inhibitory effects of XAV939 on the proliferation of small-cell lung cancer H446 cells and Wnt/beta-catenin signaling pathway in vitro. Oncol. Lett. 2018, 16, 1953–1958. [Google Scholar] [PubMed] [Green Version]
- Emons, G.; Spitzner, M.; Reineke, S.; Moller, J.; Auslander, N.; Kramer, F.; Hu, Y.; Beissbarth, T.; Wolff, H.A.; Rave-Frank, M.; et al. Chemoradiotherapy Resistance in Colorectal Cancer Cells is Mediated by Wnt/beta-catenin Signaling. Mol. Cancer Res. 2017, 15, 1481–1490. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Gao, M.; Luo, G.; Han, X.; Bao, W.; Cheng, Y.; Tian, W.; Yan, M.; Yang, G.; An, J. Enhancement of Radiation Sensitivity in Lung Cancer Cells by a Novel Small Molecule Inhibitor That Targets the beta-Catenin/Tcf4 Interaction. PLoS ONE 2016, 11, e0152407. [Google Scholar]
- Stakheev, D.; Taborska, P.; Strizova, Z.; Podrazil, M.; Bartunkova, J.; Smrz, D. The WNT/beta-catenin signaling inhibitor XAV939 enhances the elimination of LNCaP and PC-3 prostate cancer cells by prostate cancer patient lymphocytes in vitro. Sci. Rep. 2019, 9, 4761. [Google Scholar] [CrossRef] [Green Version]
- Field, J.J.; Kanakkanthara, A.; Miller, J.H. Microtubule-targeting agents are clinically successful due to both mitotic and interphase impairment of microtubule function. Bioorg. Med. Chem. 2014, 22, 5050–5059. [Google Scholar] [CrossRef]
- Gligorov, J.; Lotz, J.P. Preclinical Pharmacology of the Taxanes: Implications of the Differences. Oncologist 2004, 9 (Suppl. 2), 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terry, S.; Ploussard, G.; Allory, Y.; Nicolaiew, N.; Boissiere-Michot, F.; Maille, P.; Kheuang, L.; Coppolani, E.; Ali, A.; Bibeau, F.; et al. Increased expression of class III beta-tubulin in castra-tion-resistant human prostate cancer. Br. J. Cancer 2009, 101, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Oprea-Lager, D.E.; Bijnsdorp, I.V.; Van Moorselaar, R.J.A.; Eertwegh, A.J.M.V.D.; Hoekstra, O.S.; Geldof, A.A. ABCC4 Decreases docetaxel and not cabazitaxel efficacy in prostate cancer cells in vitro. Anticancer Res. 2013, 33, 387–391. [Google Scholar] [PubMed]
- Singh, S.K.; Apata, T.; Gordetsky, J.B.; Singh, R. Docetaxel Combined with Thymoquinone Induces Apoptosis in Prostate Cancer Cells via Inhibition of the PI3K/AKT Signaling Pathway. Cancers 2019, 11, 1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Liu, X.; Li, J.; Ledet, E.; Alvarez, X.; Qi, Y.; Fu, X.; Sartor, O.; Dong, Y.; Zhang, H. Androgen receptor splice variants circumvent AR blockade by microtubule-targeting agents. Oncotarget 2015, 6, 23358–23371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maloney, S.M.; Hoover, C.A.; Morejon-Lasso, L.V.; Prosperi, J.R. Mechanisms of Taxane Resistance. Cancers 2020, 12, 3323. [Google Scholar] [CrossRef]
- Acharyya, S.; Oskarsson, T.; Vanharanta, S.; Malladi, S.; Kim, J.; Morris, P.G.; Manova-Todorova, K.; Leversha, M.; Hogg, N.; Seshan, V.E.; et al. A CXCL1 paracrine network links cancer chemoresistance and me-tastasis. Cell 2012, 150, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Mahon, K.; Lin, H.-M.; Castillo, L.; Lee, B.Y.; Lee-Ng, M.; Chatfield, M.; Chiam, K.; Breit, S.N.; Brown, D.A.; Molloy, M.; et al. Cytokine profiling of docetaxel-resistant castration-resistant prostate cancer. Br. J. Cancer 2015, 112, 1340–1348. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Yi, M.; Xiang, B. Novel Insights on Lipid Metabolism Alterations in Drug Resistance in Cancer. Front. Cell Dev. Biol. 2022, 10, 875318. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular Fatty Acid Metabolism and Cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Röhrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, F.; Amadori, D.; Carloni, S.; Brigliadori, G.; Tesei, A.; Ulivi, P.; Rosetti, M.; Vannini, I.; Arienti, C.; Zoli, W.; et al. Mitotic catastrophe and apoptosis induced by docetaxel in hormone-refractory prostate cancer cells. J. Cell. Physiol. 2008, 217, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Mediavilla-Varela, M.; Pacheco, F.J.; Almaguel, F.; Perez, J.; Sahakian, E.; Daniels, T.R.; Leoh, L.S.; Padilla, A.; Wall, N.R.; Lilly, M.B.; et al. Docetaxel-induced prostate cancer cell death involves concomitant activation of caspase and lysosomal pathways and is attenuated by LEDGF/p75. Mol. Cancer 2009, 8, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martens-Uzunova, E.S.; Böttcher, R.; Croce, C.M.; Jenster, G.; Visakorpi, T.; Calin, G.A. Long Noncoding RNA in Prostate, Bladder, and Kidney Cancer. Eur. Urol. 2013, 65, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.-P.; et al. RNA Exosome-Regulated Long Non-Coding RNA Transcription Controls Super-Enhancer Activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Du, S.Y.; Armenia, J.; Qu, F.; Fan, J.; Wang, X.; Fei, T.; Komura, K.; Liu, S.X.; Lee, G.M.; et al. Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer. Oncogenesis 2018, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Sun, L.; Wang, X.; Wang, N.; Xu, K.; Jiang, X.; Xu, S. A Five Immune-Related lncRNA Signature as a Prognostic Target for Glioblastoma. Front. Mol. Biosci. 2021, 8, 26. [Google Scholar] [CrossRef]
- Sekino, Y.; Han, X.; Kawaguchi, T.; Babasaki, T.; Goto, K.; Inoue, S.; Hayashi, T.; Teishima, J.; Shiota, M.; Yasui, W.; et al. TUBB3 Reverses Resistance to Docetaxel and Cabazitaxel in Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 3936. [Google Scholar] [CrossRef] [Green Version]
- Maahs, L.; Sanchez, B.E.; Gupta, N.; Van Harn, M.; Barrack, E.R.; Reddy, P.V.; Hwang, C. Class III beta-tubulin ex-pression as a predictor of docetaxel-resistance in metastatic castration-resistant prostate cancer. PLoS ONE 2019, 14, e0222510. [Google Scholar] [CrossRef]
- Mathew, R.; White, E. Autophagy in tumorigenesis and energy metabolism: Friend by day, foe by night. Curr. Opin. Genet. Dev. 2011, 21, 113–119. [Google Scholar] [CrossRef] [Green Version]
- White, E.; DiPaola, R.S. The Double-Edged Sword of Autophagy Modulation in Cancer. Clin. Cancer Res. 2009, 15, 5308–5316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falasca, L.; Torino, F.; Marconi, M.; Costantini, M.; Pompeo, V.; Sentinelli, S.; De Salvo, L.; Patrizio, M.; Padula, C.; Gallucci, M.; et al. AMBRA1 and SQSTM1 expression pattern in prostate cancer. Apoptosis 2015, 20, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, Z.; Guo, J.; Wang, L.; Liu, X. Ambra1 induces autophagy and desensitizes human prostate cancer cells to cisplatin. Biosci. Rep. 2019, 39, BSR20170770. [Google Scholar] [CrossRef] [Green Version]
- Patrikainen, L.; Porvari, K.; Kurkela, R.; Hirvikoski, P.; Soini, Y.; Vihko, P. Expression profiling of PC-3 cell line variants and comparison of MIC-1 transcript levels in benign and malignant prostate. Eur. J. Clin. Investig. 2007, 37, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Beer, T.M.; Higano, C.S.; True, L.D.; Vessella, R.; Lange, P.H.; Garzotto, M.; Nelson, P.S. Molecular alterations in prostate carcinomas that associate with in vivo exposure to chemotherapy: Identification of a cytoprotective mechanism involving growth differentiation factor 15. Clin. Cancer Res. 2007, 13, 5825–5833. [Google Scholar] [CrossRef] [Green Version]
- Hotulainen, P.; Paunola, E.; Vartiainen, M.K.; Lappalainen, P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell 2005, 16, 649–664. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Mouneimne, G.; Sidani, M.; Wyckoff, J.; Chen, X.; Makris, A.; Goswami, S.; Bresnick, A.R.; Condeelis, J.S. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 2006, 173, 395–404. [Google Scholar] [CrossRef] [Green Version]
- Castro, M.A.A.; Pizzol, F.D.; Zdanov, S.; Soares, M.; Müller, C.B.; Lopes, F.M.; Zanotto-Filho, A.; Fernandes, M.; Moreira, J.C.F.; Shacter, E.; et al. CFL1 expression levels as a prognostic and drug resistance marker in nonsmall cell lung cancer. Cancer 2010, 116, 3645–3655. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.I.; Fu, N.I.; Luo, X.U.; Li, X.Y.; Li, X.P. Overexpression of cofilin 1 in prostate cancer and the corresponding clinical implications. Oncol. Lett. 2015, 9, 2757–2761. [Google Scholar] [CrossRef] [Green Version]
- Pudova, E.A.; Kobelyatskaya, A.A.; Katunina, I.V.; Snezhkina, A.V.; Fedorova, M.S.; Guvatova, Z.G.; Nyushko, K.M.; Alekseev, B.Y.; Pavlov, V.S.; Savvateeva, M.V.; et al. Dynamic Profiling of Exosomal microRNAs in Blood Plasma of Patients with Castration-Resistant Prostate Cancer. Front. Biosci. (Schol. Ed.) 2022, 14, 15. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to ge-nomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, K.; Murphy, D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, e79. [Google Scholar] [CrossRef]
- Schnepp, P.M.; Shelley, G.; Dai, J.; Wakim, N.; Jiang, H.; Mizokami, A.; Keller, E.T. Single-Cell Transcriptomics Analysis Identifies Nuclear Protein 1 as a Regulator of Docetaxel Resistance in Prostate Cancer Cells. Mol. Cancer Res. 2020, 18, 1290–1301. [Google Scholar] [CrossRef]


| Pathway ID | Pathway Name | ES | NES | p-Value |
|---|---|---|---|---|
| Docetaxel 4 nM vs. Control 1 | ||||
| GO:0001819 | positive regulation of cytokine production | 0.51 | 1.45 | 1.00 × 10−2 |
| GO:0051896 | regulation of protein kinase B signaling | −0.64 | −1.37 | 2.00 × 10−2 |
| Docetaxel 8 nM vs. Control 2 | ||||
| GO:0071357 | cellular response to type I interferon | 0.63 | 1.60 | 1.00 × 10−2 |
| GO:0071346 | cellular response to interferon-gamma | 0.65 | 1.60 | 1.00 × 10−2 |
| GO:0006954 | inflammatory response | 0.63 | 1.58 | 1.00 × 10−2 |
| GO:0140546 | defense response to symbiont | 0.59 | 1.58 | 1.00 × 10−2 |
| GO:0051607 | defense response to virus | 0.57 | 1.54 | 1.00 × 10−2 |
| GO:0071427 | mRNA-containing ribonucleoprotein complex export from nucleus | −0.37 | −1.25 | 1.00 × 10−2 |
| GO:0042254 | ribosome biogenesis | −0.41 | −1.33 | 1.00 × 10−2 |
| GO:0090630 | activation of GTPase activity | −0.48 | −1.39 | 1.00 × 10−2 |
| GO:0000070 | mitotic sister chromatid segregation | −0.39 | −1.39 | 1.00 × 10−2 |
| GO:0016072 | rRNA metabolic process | −0.41 | −1.41 | 1.00 × 10−2 |
| GO:0140014 | mitotic nuclear division | −0.48 | −1.43 | 1.00 × 10−2 |
| GO:0006260 | DNA replication | −0.41 | −1.45 | 1.00 × 10−2 |
| GO:0006364 | rRNA processing | −0.45 | −1.52 | 1.00 × 10−2 |
| GO:0048015 | phosphatidylinositol-mediated signaling | −0.67 | −1.64 | 1.00 × 10−2 |
| GO:0034470 | ncRNA processing | −0.46 | −1.78 | 1.00 × 10−2 |
| GO:0034470 | cytokine-mediated signaling pathway | 0.49 | 1.39 | 1.00 × 10−2 |
| GO:0060337 | type I interferon signaling pathway | 0.63 | 1.59 | 1.00 × 10−2 |
| GO:0034341 | response to interferon-gamma | 0.64 | 1.47 | 1.00 × 10−2 |
| GO:0045071 | negative regulation of viral genome replication | 0.54 | 1.37 | 2.00 × 10−2 |
| GO:0050708 | regulation of protein secretion | 0.60 | 1.46 | 4.00 × 10−2 |
| GO:0098662 | inorganic cation transmembrane transport | −0.48 | −1.43 | 4.00 × 10−2 |
| GO:0071345 | cellular response to cytokine stimulus | 0.46 | 1.31 | 4.00 × 10−2 |
| GO:0072594 | establishment of protein localization to organelle | −0.47 | −1.36 | 4.00 × 10−2 |
| Docetaxel 10 nM vs. Control 3 | ||||
| GO:0006954 | inflammatory response | 0.65 | 1.65 | 1.00 × 10−2 |
| GO:0060337 | type I interferon signaling pathway | 0.60 | 1.60 | 1.00 × 10−2 |
| GO:0050727 | regulation of inflammatory response | 0.53 | 1.49 | 1.00 × 10−2 |
| GO:0032675 | regulation of interleukin-6 production | 0.59 | 1.43 | 2.00 × 10−2 |
| GO:0048523 | negative regulation of cellular process | 0.45 | 1.39 | 2.00 × 10−2 |
| GO:0098662 | inorganic cation transmembrane transport | 0.63 | 1.58 | 2.00 × 10−2 |
| GO:0071357 | cellular response to type I interferon | 0.60 | 1.52 | 4.00 × 10−2 |
| GO:0060333 | interferon-gamma-mediated signaling pathway | 0.63 | 1.51 | 4.00 × 10−2 |
| GO:0001818 | negative regulation of cytokine production | 0.56 | 1.41 | 4.00 × 10−2 |
| GO:0070555 | response to interleukin-1 | 0.63 | 1.49 | 4.00 × 10−2 |
| GO:0019221 | cytokine-mediated signaling pathway | 0.44 | 1.40 | 4.00 × 10−2 |
| GO:0006915 | apoptotic process | 0.50 | 1.41 | 4.00 × 10−2 |
| GO:0071346 | cellular response to interferon-gamma | 0.62 | 1.61 | 4.00 × 10−2 |
| GO:0032101 | regulation of response to external stimulus | 0.57 | 1.45 | 5.00 × 10−2 |
| Docetaxel 8 nM vs. Docetaxel 10 nM | ||||
| GO:0071357 | cellular response to type I interferon | 0.69 | 3.3 | 1.00 × 10−2 |
| GO:0060337 | type I interferon signaling pathway | 0.69 | 3.2 | 1.00 × 10−2 |
| GO:0060333 | interferon-gamma-mediated signaling pathway | 0.56 | 2.39 | 1.38 × 10−3 |
| GO:0045824 | negative regulation of innate immune response | 0.57 | 2.32 | 4.50 × 10−3 |
| GO:0006811 | ion transport | 0.59 | 2.32 | 4.95 × 10−3 |
| GO:0071346 | cellular response to interferon-gamma | 0.46 | 2.2 | 1.55 × 10−2 |
| GO:0008203 | cholesterol metabolic process | 0.49 | 2.2 | 1.62 × 10−2 |
| GO:0034341 | response to interferon-gamma | 0.52 | 2.17 | 1.73 × 10−2 |
| GO:0071357 | cellular response to type I interferon | 0.69 | 3.3 | 1.00 × 10−2 |
| GO:0060337 | type I interferon signaling pathway | 0.69 | 3.2 | 1.00 × 10−2 |
| GO:0060333 | interferon-gamma-mediated signaling pathway | 0.56 | 2.39 | 1.38 × 10−3 |
| GO:0045824 | negative regulation of innate immune response | 0.57 | 2.32 | 4.50 × 10−3 |
| GO:0006811 | ion transport | 0.59 | 2.32 | 4.95 × 10−3 |
| GO:0071346 | cellular response to interferon-gamma | 0.46 | 2.2 | 1.55 × 10−2 |
| GO:0008203 | cholesterol metabolic process | 0.49 | 2.2 | 1.62 × 10−2 |
| GO:0034341 | response to interferon-gamma | 0.52 | 2.17 | 1.73 × 10−2 |
| GO:0048878 | chemical homeostasis | 0.54 | 2.13 | 2.09 × 10−2 |
| GO:0014070 | response to organic cyclic compound | 0.70 | 2.13 | 1.00 × 10−2 |
| GO:0019221 | cytokine-mediated signaling pathway | 0.32 | 2.12 | 2.26 × 10−2 |
| GO:0060338 | regulation of type I interferon-mediated signaling pathway | 0.56 | 2.1 | 2.54 × 10−2 |
| GO:0050727 | regulation of inflammatory response | 0.4 | 2.08 | 2.91 × 10−2 |
| GO:0034599 | cellular response to oxidative stress | 0.38 | 2.07 | 2.95 × 10−2 |
| GO:0006006 | glucose metabolic process | 0.48 | 2.03 | 3.56 × 10−2 |
| GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 0.49 | 2.03 | 3.66 × 10−2 |
| GO:2000377 | regulation of reactive oxygen species metabolic process | 0.47 | 2.02 | 3.66 × 10−2 |
| GO:0050728 | negative regulation of inflammatory response | 0.43 | 2.02 | 3.67 × 10−2 |
| GO:0010952 | positive regulation of peptidase activity | 0.55 | 2.01 | 3.67 × 10−2 |
| GO:0014070 | response to organic cyclic compound | 0.6 | 2.03 | 3.71 × 10−2 |
| GO:0055092 | sterol homeostasis | 0.51 | 2.01 | 3.71 × 10−2 |
| GO:0006695 | cholesterol biosynthetic process | 0.52 | 1.96 | 4.73 × 10−2 |
| GO:0019216 | regulation of lipid metabolic process | 0.42 | 1.97 | 4.83 × 10−2 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 0.5 | 1.96 | 4.88 × 10−2 |
| GO:0050728 | negative regulation of inflammatory response | 0.50 | 1.79 | 1.00 × 10−2 |
| GO:0034599 | cellular response to oxidative stress | 0.36 | 1.48 | 1.00 × 10−2 |
| Dataset | Parameters | GO:0014070 | GO:0050728 | GO:0034599 |
|---|---|---|---|---|
| PC3 | ES | 0.70 | 0.50 | 0.49 |
| NES | 2.14 | 1.79 | 1.47 | |
| p-value | 1.00 × 10−2 | 1.00 × 10−2 | 3.13 × 10−2 | |
| PC3-SC | ES | 0.58 | 0.44 | 0.38 |
| NES | 1.91 | 1.83 | 1.55 | |
| p-value | 1.00 × 10−2 | 1.00 × 10−2 | 2.30 × 10−2 | |
| DU145-SC | ES | −0.56 | 0.49 | 0.66 |
| NES | −1.87 | 1.65 | 2.55 | |
| p-value | 1.00 × 10−2 | 2.44 × 10−2 | 1.00 × 10−2 |
| Pathway ID | Pathway Name | ES | NES | FDR |
|---|---|---|---|---|
| GO:0045087 | innate immune response | 0.58 | 2.14 | 4.53 × 10−2 |
| GO:0010467 | gene expression | −0.34 | −1.79 | 4.01 × 10−2 |
| GO:0006396 | RNA processing | −0.5 | −1.88 | 1.59 × 10−2 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process nonsense-mediated decay | −0.48 | −2.02 | 6.09 × 10−3 |
| GO:0042254 | ribosome biogenesis | −0.45 | −2.04 | 4.43 × 10−3 |
| GO:0006334 | nucleosome assembly | −0.62 | −2.06 | 3.00 × 10−3 |
| GO:0045047 | protein targeting to ER | −0.49 | −2.09 | 2.74 × 10−3 |
| GO:0065004 | protein–DNA complex assembly | −0.52 | −2.11 | 2.45 × 10−3 |
| GO:0006281 | DNA repair | −0.41 | −2.12 | 2.11 × 10−3 |
| GO:0016072 | rRNA metabolic process | −0.54 | −2.14 | 2.28 × 10−3 |
| GO:0002181 | cytoplasmic translation | −0.51 | −2.16 | 1.87 × 10−3 |
| GO:0006613 | cotranslational protein targeting to membrane | −0.58 | −2.16 | 2.06 × 10−3 |
| GO:0034728 | nucleosome organization | −0.59 | −2.19 | 1.52 × 10−3 |
| GO:0031497 | chromatin assembly | −0.62 | −2.27 | 8.56 × 10−4 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | −0.61 | −2.29 | 9.79 × 10−4 |
| GO:0006364 | rRNA processing | −0.55 | −2.29 | 1.14 × 10−3 |
| GO:0022618 | ribonucleoprotein complex assembly | −0.61 | −2.3 | 1.37 × 10−3 |
| GO:0006397 | mRNA processing | −0.53 | −2.37 | 1.00 × 10−4 |
| GO:0000377 | RNA splicing via transesterification reactions with bulged adenosine | −0.57 | −2.43 | 1.00 × 10−4 |
| GO:0034470 | ncRNA processing | −0.55 | −2.46 | 1.00 × 10−4 |
| GO:0000398 | mRNA splicing via spliceosome | −0.57 | −2.46 | 1.00 × 10−4 |
| lncRNA | Data Set | Log2FC | FDR/p-Value QLF |
|---|---|---|---|
| AL157938.2 | PC3 | - | - |
| PC3-CS | −1.99 | 1.12 × 10−4 | |
| DU-145 SC | −5.08 | 1.23 × 10−9 | |
| Plasma exosomes | −3.49 | 2.15 × 10−2 | |
| LINC02582 | PC3 | - | - |
| PC3-CS | −1.2 | 5.07 × 10−10 | |
| DU-145 SC | −2.43 | 4.24 × 10−6 | |
| Plasma exosomes | −1.89 | 1.73 × 10−2 | |
| SNHG1 | PC3 | −0.61 | 9.97 × 10−10 |
| PC3-CS | −1.08 | 9.14 × 10−3 | |
| DU-145 SC | −1.45 | 2.21 × 10−5 | |
| Plasma exosomes | - | - | |
| KCNQ1OT1 | PC3 | −0.38 | 8.22 × 10−3 |
| PC3-CS | −2.05 | 1.05× 10−18 | |
| DU-145 SC | - | - | |
| Plasma exosomes | −1.83 | 4.30 × 10−2 | |
| MIR222HG | PC3 | −0.88 | 9.94 × 10−5 |
| PC3-CS | −2.15 | 7.97 × 10−5 | |
| DU-145 SC | - | - | |
| Plasma exosomes | −3.57 | 9.22 × 10−3 |
| Transcript ID | Transcript Name | Transcript Type | Log2FC | QLF FDR |
|---|---|---|---|---|
| ENST00000429677 | PRSS3-204 | protein_coding | 0.74 | 2.54 × 10−3 |
| ENST00000331825 | FTL-201 | protein_coding | 0.8 | 1.83 × 10−6 |
| ENST00000264832 | ICAM1-201 | protein_coding | 1.04 | 5.92 × 10−6 |
| ENST00000558131 | RPL28-205 | protein_coding | 0.68 | 5.74 × 10−5 |
| ENST00000683756 | AMBRA1-213 | protein_coding | 4.86 | 2.19 × 10−2 |
| ENST00000252809 | GDF15-201 | protein_coding | 2.32 | 1.66 × 10−6 |
| ENST00000380394 | RPS6-204 | protein_coding | 0.82 | 3.04 × 10−3 |
| ENST00000259874 | IER3-201 | protein_coding | 0.73 | 8.69 × 10−4 |
| ENST00000458500 | RPL10-210 | protein_coding | 0.45 | 1.13 × 10−3 |
| ENST00000339647 | UBC-201 | protein_coding | 0.78 | 5.25 × 10−6 |
| ENST00000389805 | SQSTM1-202 | protein_coding | 1.45 | 1.57 × 10−7 |
| ENST00000302754 | JUNB-201 | protein_coding | 0.89 | 2.13 × 10−2 |
| ENST00000308162 | CFL1-201 | protein_coding | 1.42 | 4.91 × 10−9 |
| ENST00000340384 | TUBB4B-201 | protein_coding | 0.71 | 2.54 × 10−5 |
| ENST00000327892 | TUBB-205 | protein_coding | 0.51 | 3.48 × 10−2 |
| ENST00000368719 | S100A6-201 | protein_coding | 0.94 | 5.39 × 10−6 |
| ENST00000315491 | TUBB3-201 | protein_coding | 1.25 | 2.93 × 10−4 |
| Transcript Name | Protein Stable ID | UniProt Swiss-Prot ID | UniProt TrEMBL ID | Protein Length |
|---|---|---|---|---|
| PRSS3-204 | ENSP00000401828 | P35030 | - | 304 |
| FTL-201 | ENSP00000366525 | P02792 | A0A384MDR3 | 175 |
| ICAM1-201 | ENSP00000264832 | P05362 | A0A384MEK5 | 532 |
| RPL28-205 AMBRA1-213 | ENSP00000453285 ENSP00000508322 | - Q9C0C7 | H0YLP6 - | 89 1298 |
| GDF15-201 | ENSP00000252809 | Q99988 | - | 308 |
| RPS6-204 | ENSP00000369757 | P62753 | A2A3R6 | 249 |
| IER3-201 | ENSP00000259874 | P46695 | A0A1U9X7X2 | 156 |
| RPL10-210 | ENSP00000395025 | - | A6QRI9 | 181 |
| UBC-201 | ENSP00000344818 | P0CG48 | - | 685 |
| SQSTM1-202 | ENSP00000374455 | Q13501 | - | 440 |
| JUNB-201 | ENSP00000303315 | P17275 | Q5U079 | 347 |
| CFL1-201 | ENSP00000309629 | P23528 | V9HWI5 | 166 |
| TUBB4B-201 | ENSP00000341289 | P68371 | - | 445 |
| TUBB-205 | ENSP00000339001 | P07437 | Q5SU16 | 444 |
| S100A6-201 | ENSP00000357708 | P06703 | - | 90 |
| TUBB3-201 | ENSP00000320295 | Q13509 | - | 450 |
| Gene ID | Gene Name | PC3 Log2FC | QLF FDR | PC3-SC Log2FC | QLF FDR | DU145-SC Log2FC | DU145-SC QLF FDR |
|---|---|---|---|---|---|---|---|
| ENSG00000010438 | PRSS3 | 0.46 | 2.38 × 10−7 | 0.48 | 1.99 × 10−2 | 4.67 | 3.71 × 10−11 |
| ENSG00000087086 | FTL | 0.58 | 2.00 × 10−23 | 0.81 | 8.52 × 10−10 | - | - |
| ENSG00000150991 | UBC | 0.49 | 1.63 × 10−17 | 0.54 | 1.83 × 10−3 | - | - |
| ENSG00000161011 | SQSTM1 | 1.16 | 1.51 × 10−66 | 1.11 | 1.83 × 10−6 | - | - |
| ENSG00000172757 | CFL1 | 0.66 | 2.93 × 10−30 | 0.74 | 3.90 × 10−9 | - | - |
| Transcript ID | Transcript Name | DOX Log2FC | XAV939+DOX Log2FC |
|---|---|---|---|
| ENST00000429677 | PRSS3-204 | 0.74 | −0.1 |
| ENST00000331825 | FTL-201 | 0.8 | 0.39 |
| ENST00000264832 | ICAM1-201 | 1.04 | 0.9 |
| ENST00000558131 | RPL28-205 | 0.68 | 0.18 |
| ENST00000683756 | AMBRA1-213 | 4.86 | 0.08 |
| ENST00000252809 | GDF15-201 | 2.32 | 1.7 |
| ENST00000380394 | RPS6-204 | 0.82 | −0.12 |
| ENST00000259874 | IER3-201 | 0.73 | 0.54 |
| ENST00000458500 | RPL10-210 | 0.45 | −0.13 |
| ENST00000339647 | UBC-201 | 0.78 | 0.25 |
| ENST00000389805 | SQSTM1-202 | 1.45 | 1.14 |
| ENST00000302754 | JUNB-201 | 0.89 | −0.02 |
| ENST00000308162 | CFL1-201 | 1.42 | 0.05 |
| ENST00000340384 | TUBB4B-201 | 0.71 | 0.04 |
| ENST00000327892 | TUBB-205 | 0.51 | −0.01 |
| ENST00000368719 | S100A6-201 | 0.94 | 0.33 |
| ENST00000315491 | TUBB3-201 | 1.25 | 0 |
| Transcript ID | Transcript Name | Transcript Type | Log2FC | QLF p-Value | rs | rs p-Value |
|---|---|---|---|---|---|---|
| ENST00000468019 | RPL7A-205 | lncRNA | 4.86 | 2.19 × 10−3 | 0.53 | 1.59 × 10−2 |
| ENST00000598681 | NOP53-207 | lncRNA | 2.97 | 3.55 × 10−2 | 0.46 | 4.14 × 10−2 |
| ENST00000476936 | CAPZA1-204 | lncRNA | 3.19 | 2.37 × 10−2 | 0.64 | 2.57 × 10−3 |
| ENST00000508832 | MALAT1-201 | lncRNA | 2.84 | 3.39 × 10−2 | 0.48 | 3.18 × 10−2 |
| Groups | MIR222HG | p-Value | CFL1-201 | p-Value | TUBB3-201 | p-Value |
|---|---|---|---|---|---|---|
| DOX 4 nM vs. DOX 8 nM | −0.32 | 7.62 × 10−1 | −2.46 | 6.96 × 10−2 | −2.21 | 9.16 × 10−2 |
| DOX 4 nM vs. DOX 10 nM | 4.45 | 1.13 × 10−2 | −16.15 | 8.61 × 10−5 | −14.52 | 1.31 × 10−4 |
| DOX 8 nM vs. DOX 10 nM | 3.09 | 3.66 × 10−2 | −9.28 | 7.49 × 10−4 | −2.62 | 5.88 × 10−2 |
| Control 1 vs. DOX 4 nM | 0.46 | 6.69 × 10−1 | −1.12 | 3.27 × 10−1 | 1.83 | 1.41 × 10−1 |
| Control 2 vs. DOX 8 nM | 0.04 | 9.70 × 10−1 | −2.38 | 7.63 × 10−2 | 0.29 | 7.86 × 10−1 |
| Control 3 vs. DOX 10 nM | 3.46 | 2.58 × 10−2 | −6.38 | 3.09 × 10−3 | −5.06 | 7.16 × 10−3 |
| XAV939+DOX 8 nM vs. 10 nM | −0.91 | 4.13 × 10−1 | −8.36 | 1.12 × 10−3 | 4.68 | 9.45 × 10−3 |
| Patients | Age | Gleason Score | PSA at Diagnosis CRPC, ng/mL | Radionuclide Study of the Skeletal System |
|---|---|---|---|---|
| Pat1 | 66 | 9 (5 + 4) | 3000 | multiple bone metastasis |
| Pat2 | 68 | 9 (5 + 4) | 52 | multiple bone metastasis |
| Pat3 | 71 | 8 (4 + 4) | 1200 | bone metastasis |
| Pat4 | 68 | 8 (4 + 4) | 1900 | bone metastasis |
| Pat5 | 61 | 8 (4 + 4) | 23 | multiple bone metastasis |
| Pat6 | 68 | 8 (4 + 4) | 124 | bone metastasis |
| Pat7 | 70 | 8 (4 + 4) | 72 | bone metastasis |
| Pat8 | 73 | 8 (4 + 4) | 2950 | bone metastasis |
| Pat9 | 66 | 8 (4 + 4) | 334 | bone metastasis |
| Pat10 | 69 | 9 (4 + 5) | 37 | bone metastasis |
| Pat11 | 66 | - | 521 | bone metastasis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pudova, E.; Kobelyatskaya, A.; Katunina, I.; Snezhkina, A.; Nyushko, K.; Fedorova, M.; Pavlov, V.; Bulavkina, E.; Dalina, A.; Tkachev, S.; et al. Docetaxel Resistance in Castration-Resistant Prostate Cancer: Transcriptomic Determinants and the Effect of Inhibiting Wnt/β-Catenin Signaling by XAV939. Int. J. Mol. Sci. 2022, 23, 12837. https://doi.org/10.3390/ijms232112837
Pudova E, Kobelyatskaya A, Katunina I, Snezhkina A, Nyushko K, Fedorova M, Pavlov V, Bulavkina E, Dalina A, Tkachev S, et al. Docetaxel Resistance in Castration-Resistant Prostate Cancer: Transcriptomic Determinants and the Effect of Inhibiting Wnt/β-Catenin Signaling by XAV939. International Journal of Molecular Sciences. 2022; 23(21):12837. https://doi.org/10.3390/ijms232112837
Chicago/Turabian StylePudova, Elena, Anastasiya Kobelyatskaya, Irina Katunina, Anastasiya Snezhkina, Kirill Nyushko, Maria Fedorova, Vladislav Pavlov, Elizaveta Bulavkina, Alexandra Dalina, Sergey Tkachev, and et al. 2022. "Docetaxel Resistance in Castration-Resistant Prostate Cancer: Transcriptomic Determinants and the Effect of Inhibiting Wnt/β-Catenin Signaling by XAV939" International Journal of Molecular Sciences 23, no. 21: 12837. https://doi.org/10.3390/ijms232112837
APA StylePudova, E., Kobelyatskaya, A., Katunina, I., Snezhkina, A., Nyushko, K., Fedorova, M., Pavlov, V., Bulavkina, E., Dalina, A., Tkachev, S., Alekseev, B., Krasnov, G., Volodin, V., & Kudryavtseva, A. (2022). Docetaxel Resistance in Castration-Resistant Prostate Cancer: Transcriptomic Determinants and the Effect of Inhibiting Wnt/β-Catenin Signaling by XAV939. International Journal of Molecular Sciences, 23(21), 12837. https://doi.org/10.3390/ijms232112837









