Reprograming of Gene Expression of Key Inflammatory Signaling Pathways in Human Peripheral Blood Mononuclear Cells by Soybean Lectin and Resveratrol
Abstract
1. Introduction
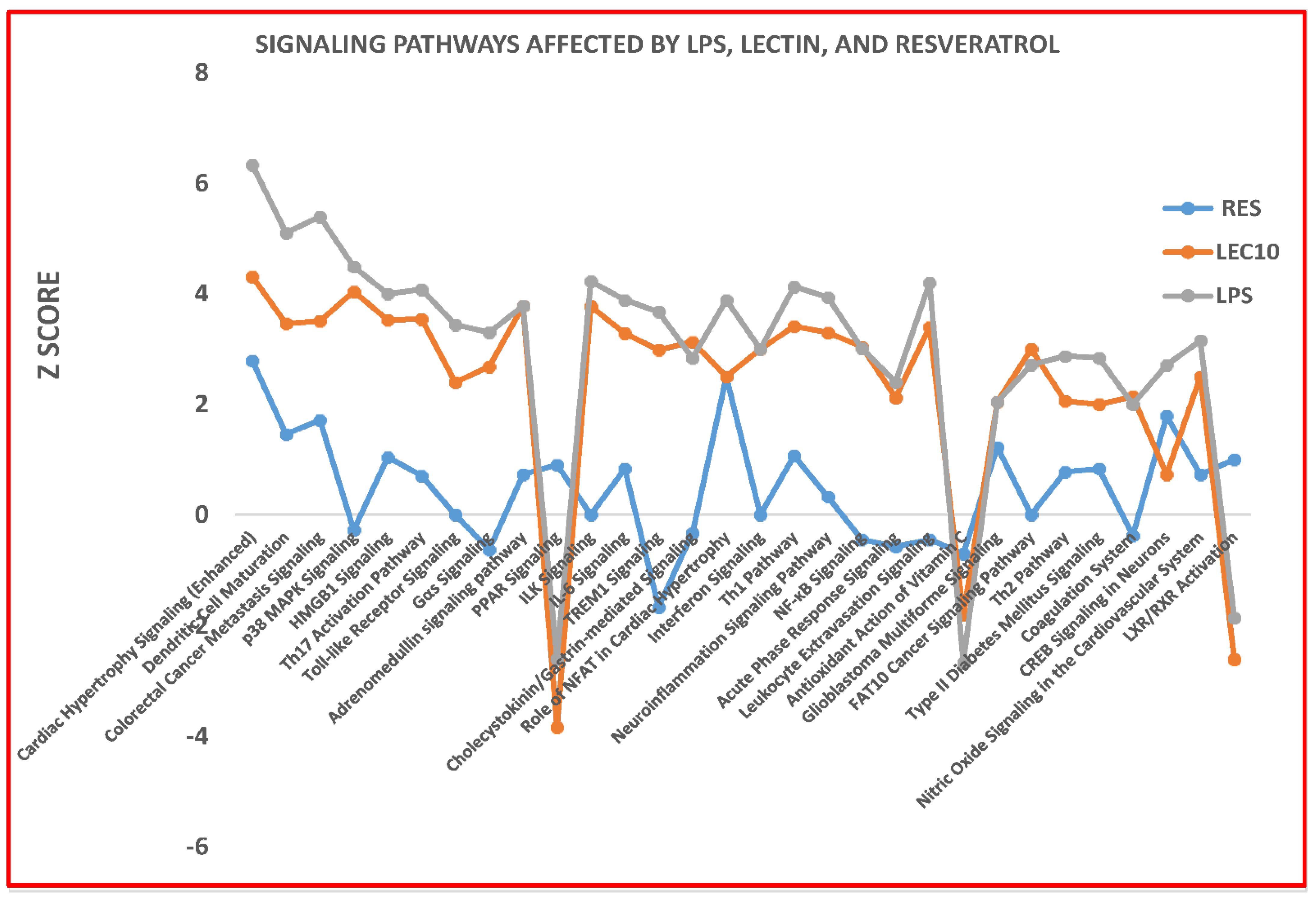
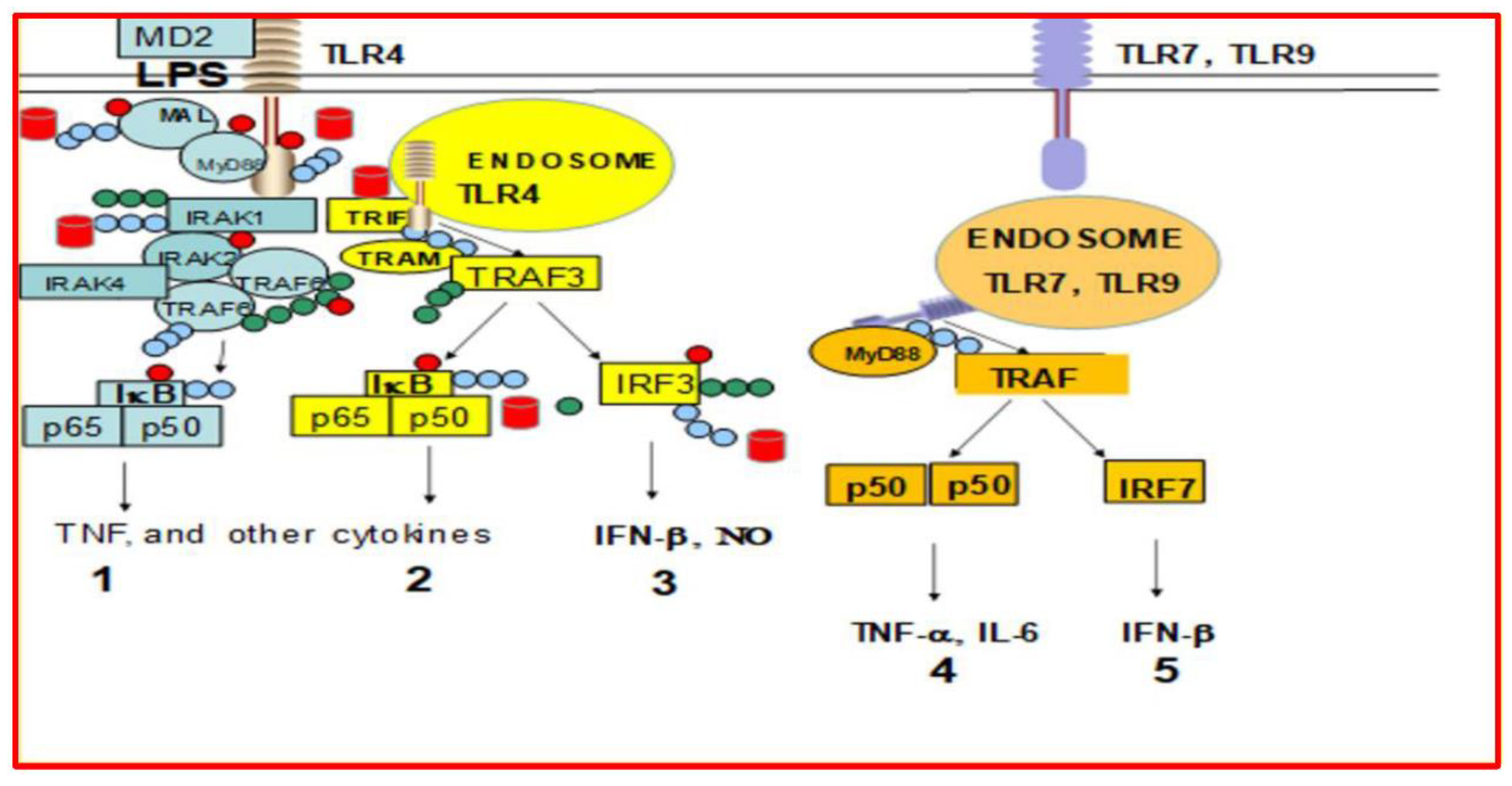
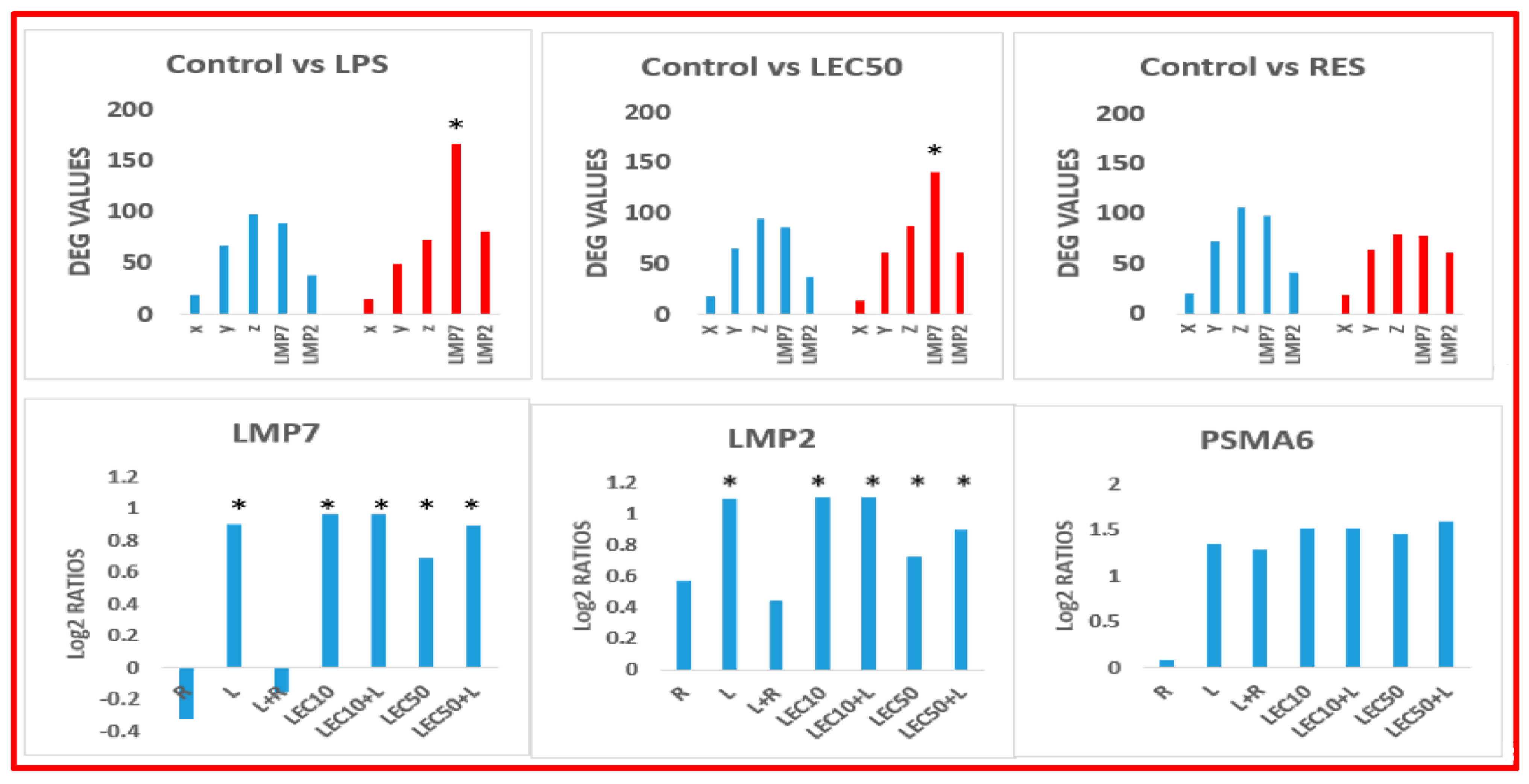
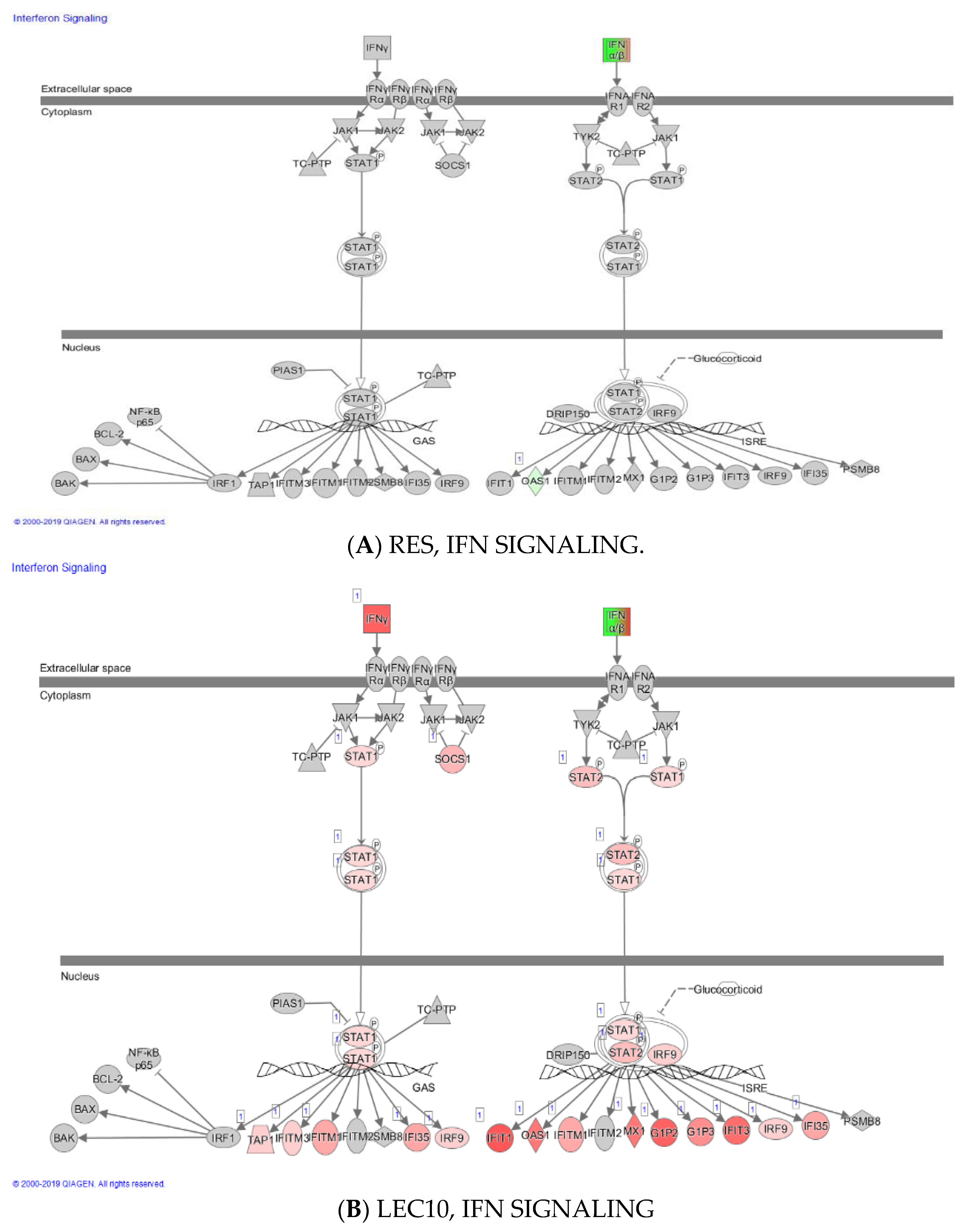
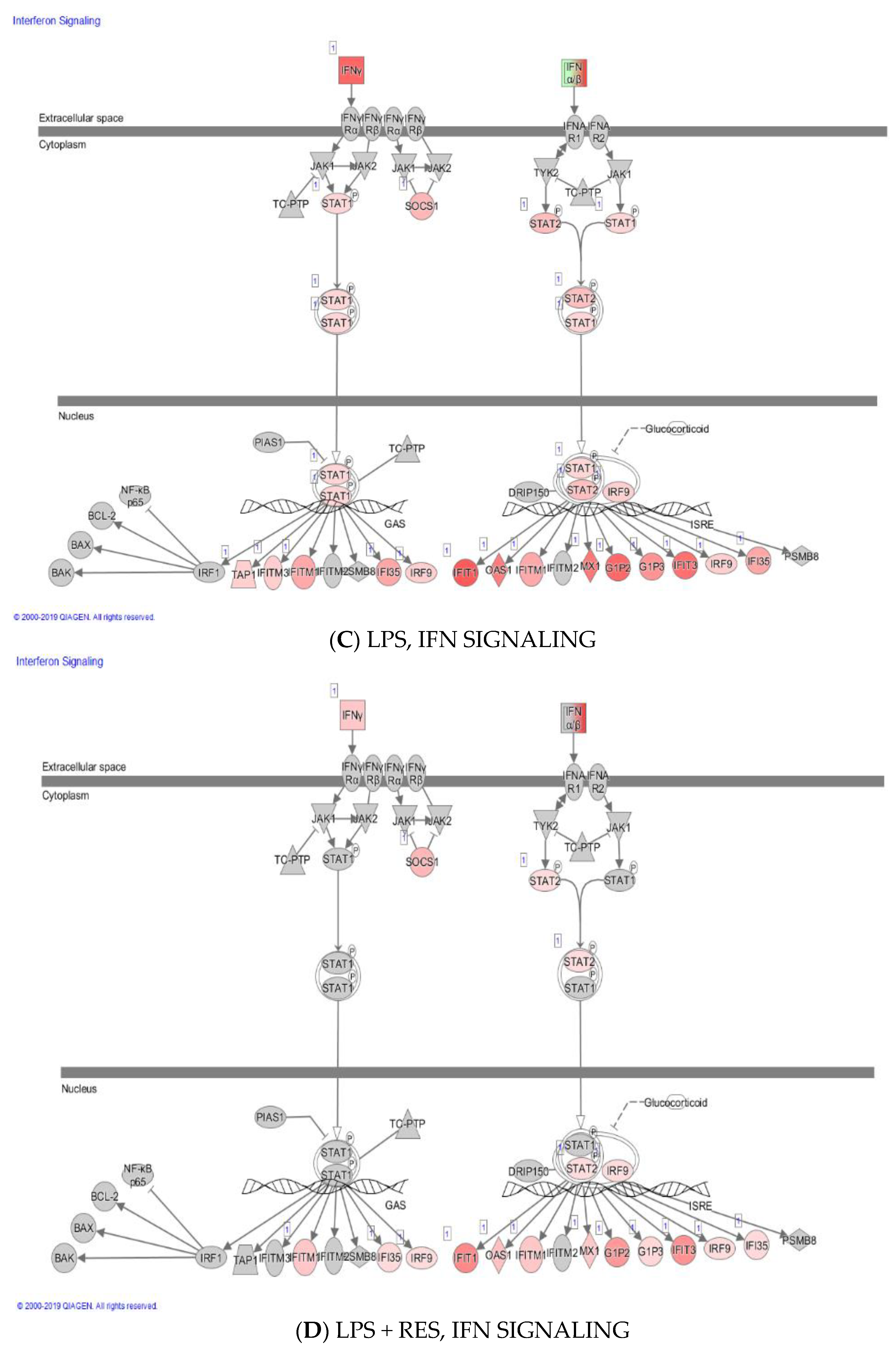
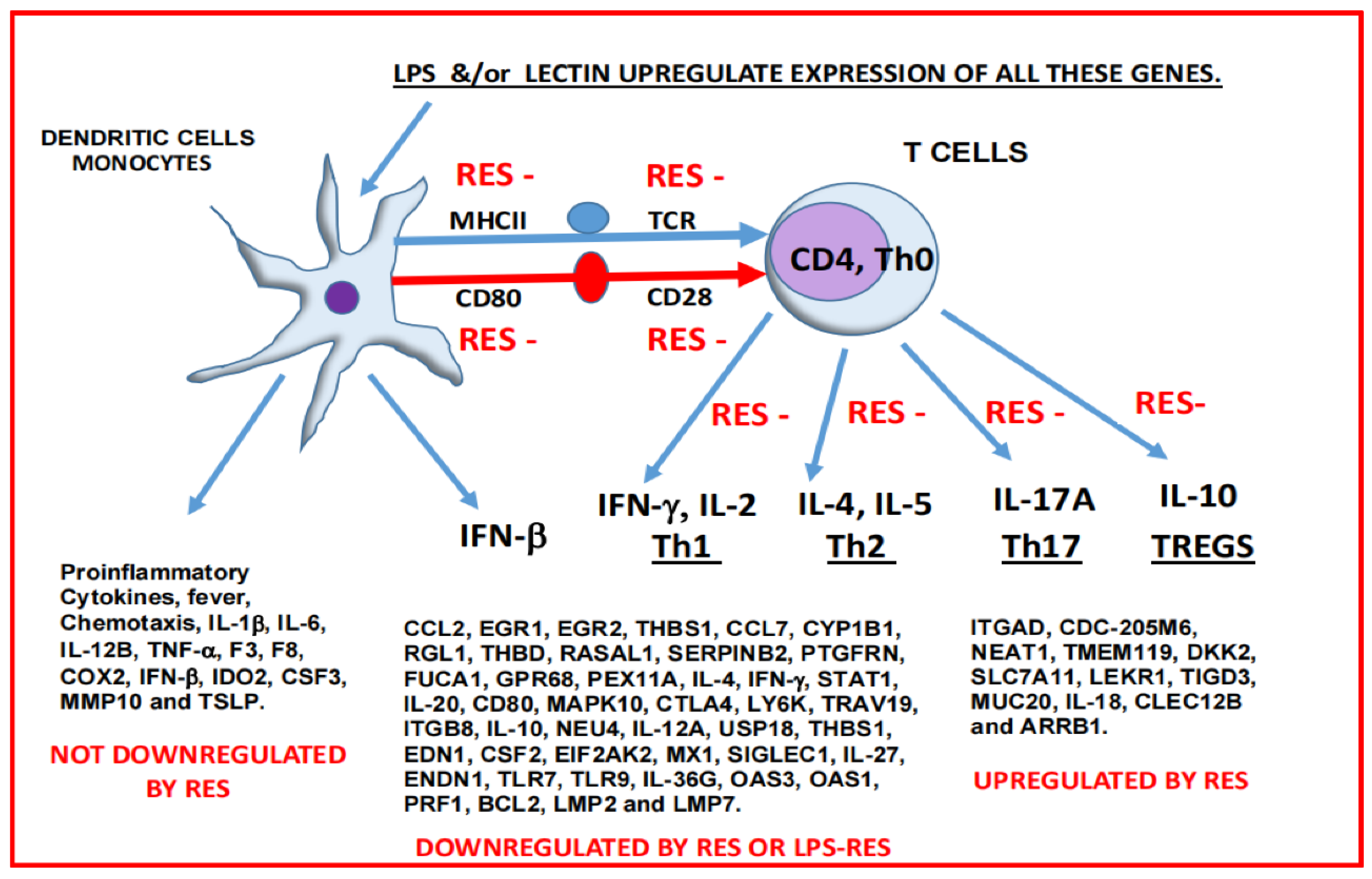
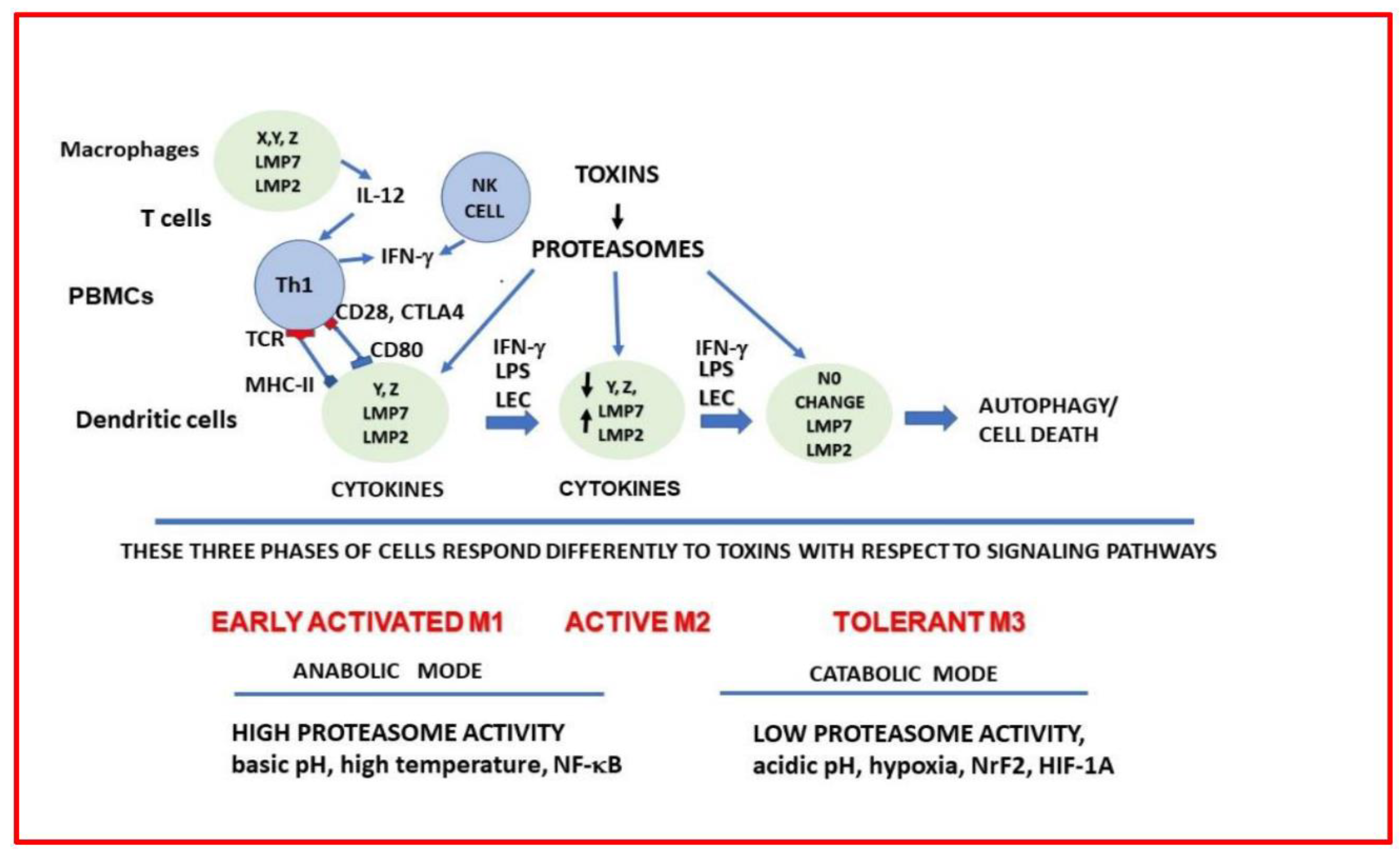
2. Results
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Detection of Cell Viability and Isolation of Total Cellular RNA
4.3. Experiments Prior to RNAseq Analysis
4.4. Sample Preparation for RNAseq Analysis
4.5. Data Analysis and Network and Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Angus, D.C.; van der Poll, T. Severe sepsis and septic shock. N. Engl. J. Med. 2013, 369, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). J. Am. Med. Assoc. 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Dallan, C.; Romano, F.; Siebert, J.; Politi, S.; Lacroix, L.; Sahyoun, C. Septic shock presentation in adolescents with COVID-19. Lancet Child Adolesc. Health 2020, 4, e21–e23. [Google Scholar] [CrossRef]
- Beltrán-García, J.; Osca-Verdegal, R.; Pallardó, F.V.; Ferreres, J.; Rodríguez, M.; Mulet, S.; Ferrando-Sánchez, C.; Carbonell, N.; García-Giménez, J.L. Sepsis, and coronavirus disease 2019: Common features and anti-inflammatory therapeutic approaches. Crit. Care Med. 2020, 48, 1841–1844. [Google Scholar] [CrossRef] [PubMed]
- Fink, M.P.; Warren, H.S. Strategies to improve drug development for sepsis. Nat. Rev. Drug Discov. 2014, 13, 741. [Google Scholar] [CrossRef]
- López-Collazo, E.; Cavaillon, J.M.; Biswas, S.K. Macrophages in sepsis progression. In Macrophages: Biology and Role in the Pathology of Diseases; Springer: New York, NY, USA, 2014; pp. 315–338. [Google Scholar]
- Rivers, E.P.; Jaehne, A.K.; Nguyen, H.B.; Papamatheakis, D.G.; Singer, D.; Yang, J.J.; Brown, S.; Klausner, H. Early biomarker activity in severe sepsis and septic shock and a contemporary review of immunotherapy trials: Not a time to give up, but to give it earlier. Shock 2013, 39, 127–137. [Google Scholar] [CrossRef]
- Vincent, J.L. Steroids in sepsis: Another swing of the pendulum in our clinical trials. Crit. Care 2008, 12, 141. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Immunosuppression in sepsis: A novel understanding of the disorder and a new therapeutic approach. Lancet Infect. Dis. 2013, 13, 260–268. [Google Scholar] [CrossRef]
- Qureshi, N.; Perera, P.Y.; Shen, J.; Zhang, G.; Lenschat, A.; Splitter, G.; Morrison, D.C.; Vogel, S.N. The proteasome as a lipopolysaccharide-binding protein in macrophages: Differential effects of proteasome inhibition on lipopolysaccharide-induced signaling events. J. Immunol. 2003, 171, 1515–1525. [Google Scholar] [CrossRef]
- Shen, J.; Gao, J.J.; Zhang, G.; Tan, X.; Morrison, D.C.; Papasian, C.J.; Vogel, S.N.; Qureshi, N. Proteasome inhibitor, lactacystin blocks CpG DNA-and peptidoglycan induced inflammatory genes, cytokines and mitogen-activated protein kinases in macrophages. Shock 2006, 25, 594–599. [Google Scholar] [CrossRef]
- Shen, J.; Reis, J.; Morrison, D.C.; Papasian, C.; Raghavakaimal, S.; Kolbert, C.; Qureshi, A.A.; Vogel, S.N.; Qureshi, N. Key inflammatory signaling pathways are regulated by the proteasome. Shock 2006, 25, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, N.; Morrison, D.C.; Reis, J. Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 2087–2093. [Google Scholar] [CrossRef] [PubMed]
- Silswal, N.; Reis, J.; Qureshi, A.A.; Papasian, C.; Qureshi, N. Of Mice and Men: Proteasome’s role in LPS-induced inflammation and tolerance. Shock 2017, 47, 445–454. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rockwell, C.E.; Monaco, J.J.; Qureshi, N. A critical role for the inducible proteasomal subunits LMP7 and MECL1 in cytokine production by activated murine splenocytes. Pharmacology 2012, 89, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.; Tan, X.; Yang, R.; Rockwell, C.E.; Papasian, C.J.; Vogel, S.N.; Morrison, D.C.; Qureshi, A.A.; Qureshi, N. A combination of proteasome inhibitors and antibiotics prevents lethality in a septic shock model. Innate Immun. 2008, 14, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, N.; Khan, D.A.; Zuberi, A.; Vernon, K.; Kaja, S.; Drees, B.M.; Qureshi, A.A.; van Way, C.W.; Morrison, D.C.; Silswal, N. Levels of proteasome subunit expression provide information about host’s immune system status. Intern. Med. Rev. 2017, 3, 1–22. [Google Scholar] [CrossRef]
- Gaczynska, M.; Goldberg, A.L.; Tanaka, K.; Hendil, K.B.; Rock, K.L. Proteasome subunits X and Y alter peptidase activities in opposite ways to the interferon-γ-induced subunits LMP2 and LMP7. J. Biol. Chem. 1996, 271, 17275–17280. [Google Scholar] [CrossRef]
- Robertson, H.; Hayes, J.D.; Sutherland, C. A partnership with the proteasome; the destructive nature of GSK3. Biochem. Pharmacol. 2017, 147, 77–92. [Google Scholar] [CrossRef]
- Rockwell, C.E.; Morrison, D.C.; Qureshi, N. Lipid A-mediated tolerance and cancer therapy. In Lipid A in Cancer Therapy; Springer: New York, NY, USA, 2009; pp. 81–99. [Google Scholar]
- Rockwell, C.E.; Qureshi, N. Differential effects of lactacystin on cytokine production in activated Jurkat cells and murine splenocytes. Cytokine 2010, 51, 12–17. [Google Scholar] [CrossRef]
- Kraaij, M.D.; Vereyken, E.J.; Leenen, P.J.; van den Bosch, T.P.; Rezaee, F.; Betjes, M.G.; Baan, C.C.; Rowshani, A.T. Human monocytes produce interferon-gamma upon stimulation with LPS. Cytokine 2014, 67, 7–12. [Google Scholar] [CrossRef]
- Fultz, M.J.; Barber, S.A.; Dieffenbach, C.W.; Vogel, S.N. Induction of IFN-γ in macrophages by lipopolysaccharide. Int. Immunol. 1993, 5, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Lundkvist, G.; Dons, L.; Kristensson, K.; Rottenberg, M.E. Interferon-γ mediates neuronal killing of intracellular bacteria. Scand. J. Immunol. 2004, 60, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Ouadrhiri, Y.; Scorneaux, B.; Sibille, Y.; Tulkens, P.M. Mechanism of the intracellular killing and modulation of antibiotic susceptibility of Listeria monocytogenes in THP-1 macrophages activated by gamma interferon. Antimicrob. Agents Chemother. 1999, 43, 1242–1251. [Google Scholar] [CrossRef] [PubMed]
- Herbst, S.; Schaible, U.E.; Schneider, B.E. Interferon gamma activated macrophages kill mycobacteria by nitric oxide induced apoptosis. PLoS ONE 2011, 6, e19105. [Google Scholar] [CrossRef]
- Luzina, I.G.; Keegan, A.D.; Heller, N.M.; Rook, G.A.; Shea-Donohue, T.; Atamas, S.P. Regulation of inflammation by interleukin-4: A review of “alternatives”. J. Leukoc. Biol. 2012, 92, 753–764. [Google Scholar] [CrossRef]
- Chen, S.; Kammerl, I.E.; Vosyka, O.; Baumann, T.; Yu, Y.; Wu, Y.; Irmler, M.; Overkleeft, H.S.; Beckers, J.; Eickelberg, O.; et al. Immunoproteasome dysfunction augments alternative polarization of alveolar macrophages. Cell Death Differ. 2016, 23, 1026. [Google Scholar] [CrossRef]
- Gordon, S.; Taylor, P.R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005, 5, 953. [Google Scholar] [CrossRef]
- Tugal, D.; Liao, X.; Jain, M.K. Transcriptional control of macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1135–1144. [Google Scholar] [CrossRef]
- Brown, B.N.; Sicari, B.M.; Badylak, S.F. Rethinking regenerative medicine: A macrophage-centered approach. Front. Immunol. 2014, 5, 510. [Google Scholar] [CrossRef]
- Mills, C. M1 and M2 macrophages: Oracles of health and disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef]
- Kelly, B.; O’Neill, L.A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771. [Google Scholar] [CrossRef] [PubMed]
- De Mejía, E.G.; Prisecaru, V.I. Lectins as bioactive plant proteins: A potential in cancer treatment. Crit. Rev. Food Sci. Nutr. 2005, 45, 425–445. [Google Scholar] [CrossRef] [PubMed]
- Maenz, D.D.; Irish, G.G.; Classen, H.L. Carbohydrate-binding and agglutinating lectins in raw and processed soybean meals. Anim. Feed. Sci. Technol. 1999, 76, 335–343. [Google Scholar] [CrossRef]
- Borra, M.T.; Smith, B.C.; Denu, J.M. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005, 280, 17187–17195. [Google Scholar] [CrossRef]
- Price, N.L.; Gomes, A.P.; Ling, A.J.; Duarte, F.V.; Martin-Montalvo, A.; North, B.J.; Agarwal, B.; Ye, L.; Ramadori, G.; Teodoro, J.S.; et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012, 15, 675–690. [Google Scholar] [CrossRef]
- Freitas, C.S.; Alves da Silva, G.; Perrone, D.; Vericimo, M.A.; Baião, D.S.; Pereira, P.R.; Paschoalin, V.M.F.; Del Aguila, E.M. Recovery of antimicrobials and bio accessible isoflavones and phenolics from soybean (Glycine max) meal by aqueous extraction. Molecules 2019, 24, 74. [Google Scholar] [CrossRef]
- Poltorak, A.; He, X.; Smirnova, L.; Liu, M.Y.; Huffel, C.V.; Du, X.; Birdwell, D.; Alejos, E.; Silva, M.; Galanos, C.; et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 1998, 597, 2085–2088. [Google Scholar] [CrossRef]
- Rajaiah, R.; Perkins, D.J.; Ireland, D.D.; Vogel, S.N. CD14 dependence of TLR4 endocytosis and TRIF signaling displays ligand specificity and is dissociable in endotoxin tolerance. Proc. Natl. Acad. Sci. USA 2015, 112, 8391–8396. [Google Scholar] [CrossRef]
- Wertz, I.E.; Dixit, V.M. Signaling to NF-κB: Regulation by ubiquitination. Cold Spring Harb. Perspect. Biol. 2010, 2, a003350. [Google Scholar] [CrossRef]
- Kaisho, T.; Akira, S. Regulation of dendritic cell function through Toll-like receptors. Curr. Mol. Med. 2003, 3, 759–771. [Google Scholar] [CrossRef]
- Ziegler-Heitbrock, L. The p50-homodimer mechanism in tolerance to LPS. J. Endotoxin Res. 2001, 7, 219–222. [Google Scholar] [CrossRef]
- Reis, J.; Hassan, F.; Guan, X.Q.; Shen, J.; Monaco, J.J.; Papasian, C.J.; Qureshi, A.A.; van Way, C.W.; Vogel, S.N.; Morrison, D.C.; et al. The immunoproteasomes regulate LPS-induced TRIF/TRAM signaling pathway in murine macrophages. Cell Biochem. Biophys. 2011, 60, 119–126. [Google Scholar] [CrossRef]
- Reis, J.; Guan, X.Q.; Kisselev, A.F.; Papasian, C.J.; Qureshi, A.A.; Morrison, D.C.; van Way, C.W.; Vogel, S.N.; Qureshi, N. LPS-induced formation of immunoproteasomes: TNF-α and nitric oxide production are regulated by altered composition of proteasome-active sites. Cell Biochem. Biophys. 2011, 60, 77–88. [Google Scholar] [CrossRef]
- Zong, Y.; Sun, L.; Liu, B.; Deng, Y.S.; Zhan, D.; Chen, Y.L.; He, Y.; Liu, J.; Zhang, Z.J.; Sun, J.; et al. Resveratrol inhibits LPS-induced MAPKs activation via activation of the phosphatidylinositol 3-kinase pathway in murine RAW 264.7 macrophage cells. PLoS ONE 2012, 7, e44107. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhang, L.; Kang, K.; Fei, D.; Gong, R.; Cao, Y.; Pan, S.; Zhao, M.; Zhao, M. Resveratrol ameliorates LPS-induced acute lung injury via NLRP3 inflammasome modulation. Biomed. Pharmacother. 2016, 84, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, T.L.; Koop, D.R. Effects of the wine polyphenolics quercetin and resveratrol on pro-inflammatory cytokine expression in RAW 264.7 macrophages. Biochem. Pharmacol. 1999, 57, 941–949. [Google Scholar] [CrossRef]
- Cianciulli, A.; Calvello, R.; Cavallo, P.; Dragone, T.; Carofiglio, V.; Panaro, M.A. Modulation of NF-kB activation by resveratrol in LPS treated human intestinal cells results in downregulation of PGE2 production and COX-2 expression. Toxicol. In Vitro 2012, 26, 1122–1128. [Google Scholar] [CrossRef]
- Birrell, M.A.; McCluskie, K.; Wong, S.; Donnelly, L.E.; Barnes, P.J.; Belvisi, M.G. Resveratrol, an extract of red wine, inhibits lipopolysaccharide induced airway neutrophilia and inflammatory mediators through an NF-kB-independent mechanism. FASEB J. 2005, 19, 840–841. [Google Scholar] [CrossRef]
- Cho, C.E.; Damle, S.S.; Wancewicz, E.V.; Mukhopadhyay, S.; Hart, C.E.; Mazur, C.; Swayze, E.E.; Kamme, F.A. modular analysis of microglia gene expression, insights into the aged phenotype. BMC Genom. 2019, 20, 164. [Google Scholar] [CrossRef]
- Holthoff, J.H.; Wang, Z.; Seely, K.A.; Gokden, N.; Mayeux, P.R. Resveratrol improves renal microcirculation, protects the tubular epithelium, and prolongs survival in a mouse model of sepsis-induced acute kidney injury. Kidney Int. 2012, 81, 370–378. [Google Scholar] [CrossRef]
- Qureshi, A.A.; Guan, X.Q.; Reis, J.C.; Papasian, C.J.; Jabre, S.; Morrison, D.C.; Qureshi, N. Inhibition of nitric oxide and inflammatory cytokines in LPS-stimulated murine macrophages by resveratrol, a potent proteasome inhibitor. Lipids Health Dis. 2012, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.A.; Tan, X.; Reis, J.C.; Badr, M.Z.; Papasian, C.J.; Morrison, D.C.; Qureshi, N. Suppression of nitric oxide induction and pro-inflammatory cytokines by novel proteasome inhibitors in various experimental models. Lipids Health Dis. 2011, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Silswal, N.; Reddy, N.S.; Qureshi, A.A.; Qureshi, N. Resveratrol downregulates biomarkers of sepsis via inhibition of proteasome’s proteases. Shock 2018, 50, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Liener, I. (Ed.) The Lectins: Properties, Functions, and Applications in Biology and Medicine; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Benjamin, C.F.; Figueiredo, R.C.; Henriques, M.G.; Barja-Fidalgo, C. Inflammatory and anti-inflammatory effects of soybean agglutinin. Braz. J. Med. Biol. Res. 1997, 30, 873–881. [Google Scholar] [CrossRef]
- Gong, T.; Wang, X.; Yang, Y.; Yan, Y.; Yu, C.; Zhou, R.; Jiang, W. Plant lectins activate the NLRP3 inflammasome to promote inflammatory disorders. J. Immunol. 2017, 198, 2082–2092. [Google Scholar] [CrossRef]
- Wang, K.; Liu, C.; Hou, Y.; Zhou, H.; Wang, X.; Mai, K.; He, G. Differential apoptotic and mitogenic effects of lectins in zebrafish. Front. Endocrinol. 2019, 10, 356. [Google Scholar] [CrossRef]
- Mishra, A.; Behura, A.; Mawatwal, S.; Kumar, A.; Naik, L.; Mohanty, S.S.; Manna, D.; Dokania, P.; Mishra, A.; Patra, S.K.; et al. Structure-function and application of plant lectins in disease biology and immunity. Food Chem. Toxicol. 2019, 134, 110827. [Google Scholar] [CrossRef]
- Van Buul, V.J.; Brouns, F.J. Health effects of wheat lectins: A review. J. Cereal Sci. 2014, 59, 112–117. [Google Scholar] [CrossRef]
- Schmidt, N.; Gonzalez, E.; Visekruna, A.; Kühl, A.A.; Loddenkemper, C.; Mollenkopf, H.; Kaufmann, S.H.; Steinhoff, U.; Joeris, T. Targeting the proteasome: Partial inhibition of the proteasome by bortezomib or deletion of the immunosubunit LMP7 attenuates experimental colitis. Gut 2010, 59, 896–906. [Google Scholar] [CrossRef]
- Verbrugge, S.E.; Scheper, R.J.; Lems, W.F.; de Gruijl, T.D.; Jansen, G. Proteasome inhibitors as experimental therapeutics of autoimmune diseases. Arthritis Res. Ther. 2015, 17, 17. [Google Scholar] [CrossRef]
- Bach, S.V.; Hegde, A.N. The proteasome and epigenetics: Zooming in on histone modifications. Biomol. Concepts 2016, 7, 215–227. [Google Scholar] [CrossRef]
- Fotheringham, S.; Epping, M.T.; Stimson, L.; Khan, O.; Wood, V.; Pezzella, F.; Bernards, R.; La Thangue, N.B. Genome-wide loss-of-function screen reveals an important role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer Cell 2009, 15, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Pallotta, M.T.; Orabona, C.; Volpi, C.; Vacca, C.; Belladonna, M.L.; Bianchi, R.; Servillo, G.; Brunacci, C.; Calvitti, M.; Bicciato, S. Mazza EM. Indoleamine 2, 3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 2011, 12, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Merlo, L.M.; DuHadaway, J.B.; Montgomery, J.D.; Peng, W.D.; Murray, P.J.; Prendergast, G.C.; Caton, A.J.; Muller, A.J.; Mandik-Nayak, L. Differential roles of IDO1 and IDO2 in T and B cell inflammatory immune responses. Front. Immunol. 2020, 11, 1861. [Google Scholar] [CrossRef]
- Metz, R.; Smith, C.; DuHadaway, J.B.; Chandler, P.; Baban, B.; Merlo, L.M.; Pigott, E.; Keough, M.P.; Rust, S.; Mellor, A.L.; et al. IDO2 is critical for IDO1-mediated T-cell regulation and exerts a non-redundant function in inflammation. Int. Immunol. 2014, 26, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, N.; Takayama, K.; Mascagni, P.; Honovich, J.; Wong, R.O.; Cotter, R.J. Complete structural determination of lipopolysaccharide obtained from deep rough mutant of Escherichia coli. Purification by high performance liquid chromatography and direct analysis by plasma desorption mass spectrometry. J. Biol. Chem. 1988, 263, 11971–11976. [Google Scholar] [CrossRef]
- Qureshi, N.; Takayama, K.; Ribi, E. Purification and structural determination of nontoxic lipid A from the rough mutant of Salmonella typhimurium. J. Biol. Chem. 1982, 257, 11808–11815. [Google Scholar] [CrossRef]
- Qureshi, N.; Takayama, K.; Heller, D.; Fenselau, C. Position of ester groups in the lipid A backbone of lipopolysaccharides obtained from Salmonella typhimurium. J. Biol. Chem. 1983, 258, 12947–12951. [Google Scholar] [CrossRef]
- Ciechanover, A. Intracellular protein degradation: From a vague idea thru the lysosome and the ubiquitin–proteasome system and onto human diseases and drug targeting. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2012, 1824, 3–13. [Google Scholar] [CrossRef]
- Narayanan, S.; Cai, C.Y.; Assaraf, Y.G.; Guo, H.Q.; Cui, Q.; Wei, L.; Huang, J.J.; Ashby, C.R., Jr.; Chen, Z.S. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist. Updates 2020, 48, 100663. [Google Scholar] [CrossRef]

| RES | Control |
log2 Fold | p Value |
Gene Name | Gene Description |
|---|---|---|---|---|---|
| 4.635 | 107.400 | −4.532 | 2.31 × 10−20 | CCL2 | chemokine_(C-C_motif)_ligand_2 |
| 4.0240 | 77.572 | −4.266 | 1.96 × 10−18 | EGR2 | early_growth_response_2 |
| 136.81 | 2235.303 | −4.030 | 3.57 × 10−18 | THBS1 | thrombospondin_1 |
| 10.798 | 111.972 | −3.373 | 1.83 × 10−13 | CCL7 | chemokine_(C-C_motif)_ligand_7 |
| 121.84 | 1096.809 | −3.170 | 1.19 × 10−12 | CYP1B1 | cytochrome_P450_family_1_subfamily_B_polypeptide_1 |
| 1.273 | 14.142 | −3.466 | 1.68 × 10−11 | CYP1B1-AS1 | CYP1B1_antisense_RNA_1 |
| 5.755 | 37.271 | −2.693 | 3.39 × 10−9 | RGL1 | ral_guanine_nucleotide_dissociation_stimulator-like_1 |
| 15.739 | 96.447 | −2.614 | 3.48 × 10−9 | THBD | thrombomodulin |
| 2.699 | 16.375 | −2.597 | 5.70 × 10−8 | RASAL1 | RAS_protein_activator_like_1_(GAP1_like) |
| 64.894 | 292.372 | −2.171 | 4.32 × 10−7 | SERPINB2 | serpin_peptidase_inhibitor_clade_B_(ovalbumin)_member2 |
| 0.764 | 5.423 | −2.816 | 7.48 × 10−7 | PTGFRN | prostaglandin_F2_receptor_inhibitor |
| 42.635 | 182.580 | −2.098 | 1.07 × 10−6 | FUCA1 | fucosidase_alpha-L-_1_tissue |
| 10.085 | 44.555 | −2.142 | 1.22 × 10−6 | GPR68 | G_protein-coupled_receptor_68 |
| 0.050 | 1.754 | −4.938 | 1.30 × 10−06 | PEX11A | peroxisomal_biogenesis_factor_11_alpha |
| 8.251 | 36.420 | −2.141 | 1.36 × 10−6 | EGR1 | early_growth_response_1 |
| 1.782 | 9.410 | −2.395 | 1.36 × 10−6 | LY6K | lymphocyte_antigen_6_complex_locus_K |
| 0.458 | 3.828 | −3.044 | 1.39 × 10−6 | TRAV19 | T_cell_receptor_alpha_variable_19 |
| 2.597 | 12.813 | −2.299 | 1.58 × 10−6 | ITGB8 | integrin_beta_8 |
| 0.865 | 5.582 | −2.679 | 1.64 × 10−6 | IL10 | interleukin_10 |
| 13.498 | 55.933 | −2.050 | 2.61 × 10−6 | NEU4 | sialidase_4 |
| LPS + RES | LPS | log2 Fold | p Value | Gene Name | Gene Description |
|---|---|---|---|---|---|
| 0.0529 | 9.74207 | −7.3514 | 1.41 × 10−18 | CCL8 | chemokine_(C-C_motif)_ligand_8 |
| 27.447 | 476.382 | −4.1171 | 1.49 × 10−18 | CYP1B1 | cytochrome_P450_family_1_subfamily_B_polypeptide_1 |
| 7.7361 | 135.790 | −4.1325 | 4.13 × 10−18 | CCL2 | chemokine_(C-C_motif)_ligand_2 |
| 125.420 | 1442.97 | −3.5241 | 7.36 × 10−15 | RSAD2 | radical_S-adenosyl_methionine_domain_containing_2 |
| 4.715 | 61.064 | −3.6929 | 8.29 × 10−15 | RGL1 | ral_guanine_nucleotide_dissociation_stimulator-like_1 |
| 8.954 | 108.360 | −3.596 | 1.04 × 10−14 | CCL7 | chemokine_(C-C_motif)_ligand_7 |
| 62.895 | 689.129 | −3.453 | 2.18 × 10−14 | OAS3 | 2′-5′-oligoadenylate_synthetase_3_100kDa |
| 0.105 | 6.422 | −5.834 | 5.39 × 10−14 | IL19 | interleukin_19 |
| 57.067 | 593.613 | −3.378 | 6.55 × 10−14 | CMPK2 | cytidine_monophosphate_(UMP-CMP) kinase_2_mitochondrial |
| 5.0337 | 54.642 | −3.438 | 2.76 × 10−13 | CXCL11 | chemokine_(C-X-C_motif)_ligand_11 |
| 6.358 | 63.133 | −3.310 | 9.45 × 10−13 | IFNG | interferon_gamma |
| 2.331 | 25.960 | −3.473 | 1.33 × 10−12 | SIGLEC1 | sialic_acid_binding_Ig-like_lectin_1_sialoadhesin |
| 0.688 | 11.102 | −3.997 | 1.61 × 10−12 | GGT5 | gamma-glutamyltransferase_5 |
| 23.526 | 194.569 | −3.047 | 1.01 × 10−11 | NEU4 | sialidase_4 |
| 0.635 | 9.252 | −3.849 | 2.12 × 10−11 | IL10 | interleukin_10 |
| 1.006 | 12.136 | −3.582 | 2.65 × 10−11 | EDN1 | endothelin_1 |
| 6.093 | 50.397 | −3.046 | 3.61 × 10−11 | CSF2 | colony_stimulating_factor_2_(granulocyte-macrophage) |
| 17.538 | 134.484 | −2.938 | 5.35 × 10−11 | USP18 | ubiquitin_specific_peptidase_18 |
| 5.351 | 43.920 | −3.035 | 5.93 × 10−11 | EGR2 | early_growth_response_2 |
| 0.105 | 3.973 | −5.142 | 2.01 × 10−10 | LAMA2 | laminin_alpha_2 |
| 1023.76 | 6937.936 | −2.760 | 2.90 × 10−10 | THBS1 | thrombospondin_1 |
| 171.677 | 1129.917 | −2.718 | 5.22 × 10−10 | MX1 | myxovirus_(influenza_virus)_resistance_1_interferon-inducible_protein_p78_(mouse) |
| 237.487 | 1548.772 | −2.705 | 6.12 × 10−10 | HELZ2 | helicase_with_zinc_finger_2_transcriptional_coactivator |
| 68.035 | 395.343 | −2.538 | 5.47 × 10−9 | EIF2AK2 | eukaryotic_translation_initiation_factor_2-alpha_kinase_2 |
| 0.317 | 3.156 | −3.284 | 1.46 × 10−6 | GAPDHP14 | glyceraldehyde-3-phosphate_dehydrogenase_pseudogene_14 |
| 237.328 | 983.024 | −2.050 | 1.54 × 10−6 | OAS2 | 2′-5′-oligoadenylate_synthetase_2_69/71kDa |
| 4.821 | 21.606 | −2.162 | 2.29 × 10−6 | DAGLA | diacylglycerol_lipase_alpha |
| (A) | ||||||||
| LEC10 | Control1 | log2 Fold | p Value | Gene Name | Gene Description | |||
| 1105.120 | 2.180 | 8.980 | 1.96 × 10−51 | IL6 | interleukin_6_(interferon_beta_2) | |||
| 1116.645 | 2.824 | 8.623 | 2.62 × 10−49 | IL1A | interleukin_1_alpha | |||
| 2159.807 | 7.532 | 8.162 | 3.34 × 10−47 | CCL4 | chemokine_(C-C_motif)_ligand_4 | |||
| 226.943 | 0.446 | 8.969 | 5.46 × 10−45 | CCL3L1 | chemokine_(C-C_motif)_ligand_3-like_1 | |||
| 77.794 | 0 | 13.49 | 1.22 × 10−40 | CSF3 | colony_stimulating_factor_3_(granulocyte) | |||
| 1,9385.16 | 146.59 | 7.046 | 8.24 × 10−40 | IL1B | interleukin_1_beta | |||
| 199.06 | 0.941 | 7.713 | 2.26 × 10−39 | CCL20 | chemokine_(C-C_motif)_ligand_20 | |||
| 488.089 | 3.270 | 7.218 | 3.70 × 10−39 | F3 | coagulation_factor_III_(thromboplastin_tissue_factor) | |||
| 2886.299 | 31.916 | 6.498 | 1.50 × 10−35 | PTGS2, COX2 | Prostaglandin-endoperoxidesynthase_2_ (prostaglandin_ G/H_synthase_and_cyclooxygenase) | |||
| 178.034 | 1.784 | 6.635 | 1.10 × 10−33 | IRG1 | immunoresponsive_1_homolog_(mouse) | |||
| 128.475 | 1.139 | 6.808 | 2.52 × 10−33 | CCL3 | chemokine_(C-C_motif)_ligand_3 | |||
| 26.226 | 0.346 | 6.212 | 9.88 × 10−24 | CSF2 | colony_stimulating_factor_2 (granulocyte-macrophage) | |||
| 439.770 | 15.065 | 4.866 | 1.58 × 10−23 | TNF | tumor_necrosis_factor | |||
| 2759.83 | 109.77 | 4.651 | 1.99 × 10−22 | CXCL2 | chemokine_(C-X-C_motif)_ligand_2 | |||
| 2523.37 | 105.21 | 4.583 | 5.92 × 10−22 | CXCL3 | chemokine_(C-X-C_motif)_ligand_3 | |||
| 8.210 | 0.198 | 5.325 | 4.05 × 10−15 | IL36G | interleukin_36_gamma | |||
| 3,0181.18 | 2646.306 | 3.511 | 8.38 × 10−15 | IL8 | interleukin_8 | |||
| 659.862 | 74.437 | 3.147 | 1.68 × 10−12 | G0S2 | G0/G1switch_2 | |||
| 758.803 | 87.323 | 3.119 | 2.47 × 10−12 | NLRP3 | NLR_family_pyrin_domain_containing_3 | |||
| 3.248 | 0.198 | 3.989 | 3.42 × 10−8 | GCKR | glucokinase_(hexokinase_4)_regulator | |||
| 17.130 | 4.113 | 2.056 | 8.44 × 10−6 | HDAC9 | histone_deacetylase_9 | |||
| 8.033 | 1.685 | 2.248 | 8.51 × 10−6 | GGT5 | gamma-glutamyltransferase_5 | |||
| 15.948 | 3.964 | 2.006 | 1.41 × 10−5 | RBKS | ribokinase | |||
| 2.421 | 0.297 | 2.997 | 1.87 × 10−5 | GPR141 | G_protein-coupled_receptor_141 | |||
| (B) | ||||||||
| LPS | LEC10 | log2 Fold | p Value | Gene Name | Gene Description | |||
| 66.397 | 6.369 | 3.380 | 3.99 × 10−13 | IFNG | Interferon-gamma | |||
| 131.870 | 16.719 | 2.979 | 3.19 × 10−11 | USP18 | ubiquitin_specific_peptidase_18 | |||
| 44.223 | 6.687 | 2.724 | 2.05 × 10−9 | CXCL11 | chemokine_(C-X-C_motif)_ligand_11 | |||
| 1395.944 | 265.063 | 2.396 | 2.88 × 10−8 | RSAD2 | radical_S-adenosyl_methionine_domain_containing_2 | |||
| 393.764 | 74.572 | 2.400 | 2.95 × 10−8 | IFI6 | interferon_alpha-inducible_protein_6 | |||
| 15.275 | 2.653 | 2.521 | 1.50 × 10−7 | CACNA1A | calcium_channel_voltage-dependent_P/Q_type_alpha_1A_subunit | |||
| 7.329 | 1.114 | 2.709 | 3.69 × 10−7 | FAM19A2 | family_with_sequence_similarity_19_(chemokine_(C-C_motif)-like)_member_A2 | |||
| 91.465 | 19.956 | 2.196 | 4.74 × 10−7 | LAMP3 | lysosomal-associated_membrane_protein_3 | |||
| 20.017 | 3.980 | 2.328 | 5.25 × 10−7 | BCL2L14 | BCL2-like_14_(apoptosis_facilitator) | |||
| 24.575 | 5.095 | 2.268 | 7.22 × 10−7 | SIGLEC1 | sialic_acid_binding_Ig-like_lectin_1_sialoadhesin | |||
| 1500.467 | 345.580 | 2.118 | 7.24 × 10−7 | ISG15 | ISG15_ubiquitin-like_modifier | |||
| 3.202 | 0.318 | 3.301 | 9.76 × 10−7 | CCNA1 | cyclin_A1 | |||
| (C) | ||||||||
| Symbol | Entrez Gene Name | LPS | LEC10 | LEC10 + LPS | LEC50 | LEC50 + LPS | ||
| IL6 | interleukin 6 | 9.67 | 9.724 | 9.724 | 9.917 | 9.865 | ||
| IL1A | interleukin 1 alpha | 8.788 | 8.816 | 8.816 | 9.167 | 8.843 | ||
| CCL20 | C-C motif chemokine ligand 20 | 8.674 | 8.764 | 8.764 | 9.231 | 8.888 | ||
| CSF3 | colony stimulating factor 3 | 14.018 | 14.054 | 14.054 | 14.629 | 14.428 | ||
| INHBA | inhibin subunit beta A | 7.711 | 7.668 | 7.668 | 7.583 | 7.587 | ||
| F3 | coagulation factor III, tissue factor | 7.579 | 7.674 | 7.674 | 8.117 | 7.857 | ||
| IL1B | interleukin 1 beta | 7.304 | 7.380 | 7.380 | 7.749 | 7.572 | ||
| ACOD1 | aconitate decarboxylase 1 | 7.546 | 7.539 | 7.539 | 7.252 | 7.811 | ||
| TNFAIP6 | TNF alpha induced protein 6 | 6.695 | 6.671 | 6.671 | 6.757 | 6.659 | ||
| CSF2 | colony stimulating factor 2 | 7.311 | 7.399 | 7.399 | 8.025 | 7.84 | ||
| CXCL11 | C-X-C motif chemokine ligand 11 | 6.662 | 6.324 | 6.324 | 5.338 | 6.432 | ||
| IL12B | interleukin 12B | 9.513 | 9.761 | 9.761 | 9.497 | 9.525 | ||
| IDO2 | indoleamine 2,3-dioxygenase 2 | 8.616 | 9.145 | 9.145 | 8.351 | 9.084 | ||
| IL36RN | interleukin 36 receptor antagonist | 8.515 | 7.942 | 7.942 | 8.142 | 8.774 | ||
| IFNB1 | interferon beta 1 | 8.443 | 8.194 | 8.194 | 8.534 | 8.851 | ||
| Symbol | RES | LPS | RES + LPS | LEC10 | Gene Description |
|---|---|---|---|---|---|
| IFNG | 0.190 | 4.913 | 1.586 | 4.953 | Interferon γ |
| IFN-l | Nc | 6.491 | - | 9.194 | Interferon lamda |
| IFNB1 | Nc | 8.443 | 7.453 | 8.194 | Interferon β |
| TNF-α | −0.043 | 5.790 | 5.392 | 5.986 | Tumor necrosis factor |
| IL12A | 2.130 | 6.216 | 4.646 | 6.080 | Interleukin 12 α |
| IL12B | Nc | 9.513 | 8.337 | 9.761 | Interleukin 12 β |
| IL27 | −0.636 | 3896 | 0.963 | 4.189 | Interleukin 27 |
| IL36RN | Nc | 8.515 | 4.204 | 7.942 | Interleukin 36 receptor antagonist |
| IL-36B | Nc | 5.984 | 3.280 | 5.846 | Interleukin 36 β |
| IL-20 | Nc | 5.984 | 5.164 | 6.833 | Interleukin 20 |
| IL6 | −0.095 | 9.670 | 10.139 | 9.724 | Interleukin 6 |
| IL1B | −0.956 | 7.304 | 7.467 | 7.38 | Interleukin 1 β |
| IL1R1 | −0.904 | 1.184 | 0.095 | 1.170 | Interleukin 1 receptor 1 |
| TGFB2 | 1.636 | 3.349 | 1.609 | 3.377 | Transforming growth factor-β2 |
| MYD88 | −0.249 | 1.050 | −0.134 | 0.975 | Myeloid differentiation primary response 88 |
| RIPK1 | 0.128 | 1.031 | 0.485 | 1.012 | Receptor-interacting serine/threonine-protein kinase 1 |
| NLRP3 | 0.361 | 3.019 | 4.083 | 3.124 | NLR family pyrin domain containing 3 (NLRP3) |
| INSR | −0.424 | −1.683 | −1.424 | −2.114 | Insulin receptor |
| INSM1 | Nc | 6.173 | 3.28 | 6.064 | Insulinoma-associated protein 1 |
| F2RL1 | 0.175 | 1.867 | 1.32 | 1.842 | Thrombin receptor like 1,3 |
| LDLR | 0.300 | 1.095 | 0.406 | 1.105 | Lipoprotein receptor |
| CXCL10 | 0.873 | 4.645 | 2.060 | 4.560 | C-X-C motif chemokine ligand 10 |
| CXCL8 | −0.893 | 3.913 | 3.845 | 3.974 | chemokine (C-X-C motif) ligand 8 |
| CD80 | 0.936 | 3.12 | 1.541 | 3.183 | Cluster of differentiation 80 |
| CD40 | 0.383 | 1.284 | 1.495 | 1.188 | Cluster of differentiation 40 |
| CTLA4 | −0.119 | 1.171 | 0.451 | 1.181 | Cytotoxic T-lymphocyte-associated protein 4 |
| STAT1 | 0.090 | 1.356 | −0.206 | 1.249 | Signal transducer and activator of transcription 1 |
| HLA-DQB | 1.360 | 3.122 | 2.148 | 3.206 | HLA class II DQB |
| IRF7 | 0.178 | 2.492 | 1.519 | 2.556 | Interferon regulatory factor 7 |
| TLR7 | −0.116 | 3.156 | 1.854 | 3.089 | Toll like receptor 7 |
| NCOA7 | −0.477 | 2.355 | 0.015 | 2.330 | Nuclear receptor coactivator |
| PTGER3 | −1.025 | 2.269 | 0.233 | 2.560 | Prostaglandin EP3 receptor |
| SIGLEC1 | 0.490 | 3.694 | 0.204 | 3.582 | Sialo adhesin |
| 2 RES | 3LPS | 4LPS + RES | 5LEC10 | 6LEC10 + LPS | 7LEC50 | 8LEC50 + LPS |
|---|---|---|---|---|---|---|
| EGR1- | HDAC9 | NFKBIA | HDAC9 | HDAC9 | HDAC9 | HDAC9 |
| EGR2- | PARP12 | GADD45A | NFKBIA | PARP12 | NOTCH3- | PARP12 |
| NOTCH3- | EGR1 | GADD45A | NOTCH3- | NFKBIA | NOTCH3- | |
| NFKBIA | EGR2- | EHF | NFKBIA | NCOA7 | NFKBIA | |
| KLF5 | IRF5- | ETS2 | NCOA7 | GADD45A | KLF5 | |
| NCOA7 | EHF | E2F7 | GADD45A | EGR1 | NCOA7 | |
| GADD45A | ETS2 | MAFF | TRIM25 | TRIM5 | GADD45A | |
| TRIM25 | E2F7 | TRIM5 | EHF | EGR1 | ||
| TRIM5 | ZNF668- | EHF | PML | TRIM25 | ||
| EHF | HIC1 | PML | ETS2 | TRIM5 | ||
| PML | OLIG2- | ETS2 | E2F7 | TBX3- | ||
| ETS2 | E2F7 | MAFF | EHF | |||
| E2F7 | MAFF | IRF7 | PML | |||
| MAFF | IRF7 | OLIG2- | ETS2 | |||
| IRF7 | OLIG2- | E2F7 | ||||
| OLIG2- | MAFF | |||||
| IRF7 | ||||||
| OLIG2- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qureshi, N.; Desousa, J.; Siddiqui, A.Z.; Morrison, D.C.; Qureshi, A.A. Reprograming of Gene Expression of Key Inflammatory Signaling Pathways in Human Peripheral Blood Mononuclear Cells by Soybean Lectin and Resveratrol. Int. J. Mol. Sci. 2022, 23, 12946. https://doi.org/10.3390/ijms232112946
Qureshi N, Desousa J, Siddiqui AZ, Morrison DC, Qureshi AA. Reprograming of Gene Expression of Key Inflammatory Signaling Pathways in Human Peripheral Blood Mononuclear Cells by Soybean Lectin and Resveratrol. International Journal of Molecular Sciences. 2022; 23(21):12946. https://doi.org/10.3390/ijms232112946
Chicago/Turabian StyleQureshi, Nilofer, Julia Desousa, Adeela Z. Siddiqui, David C. Morrison, and Asaf A. Qureshi. 2022. "Reprograming of Gene Expression of Key Inflammatory Signaling Pathways in Human Peripheral Blood Mononuclear Cells by Soybean Lectin and Resveratrol" International Journal of Molecular Sciences 23, no. 21: 12946. https://doi.org/10.3390/ijms232112946
APA StyleQureshi, N., Desousa, J., Siddiqui, A. Z., Morrison, D. C., & Qureshi, A. A. (2022). Reprograming of Gene Expression of Key Inflammatory Signaling Pathways in Human Peripheral Blood Mononuclear Cells by Soybean Lectin and Resveratrol. International Journal of Molecular Sciences, 23(21), 12946. https://doi.org/10.3390/ijms232112946





