Role of Circadian Transcription Factor Rev-Erb in Metabolism and Tissue Fibrosis
Abstract
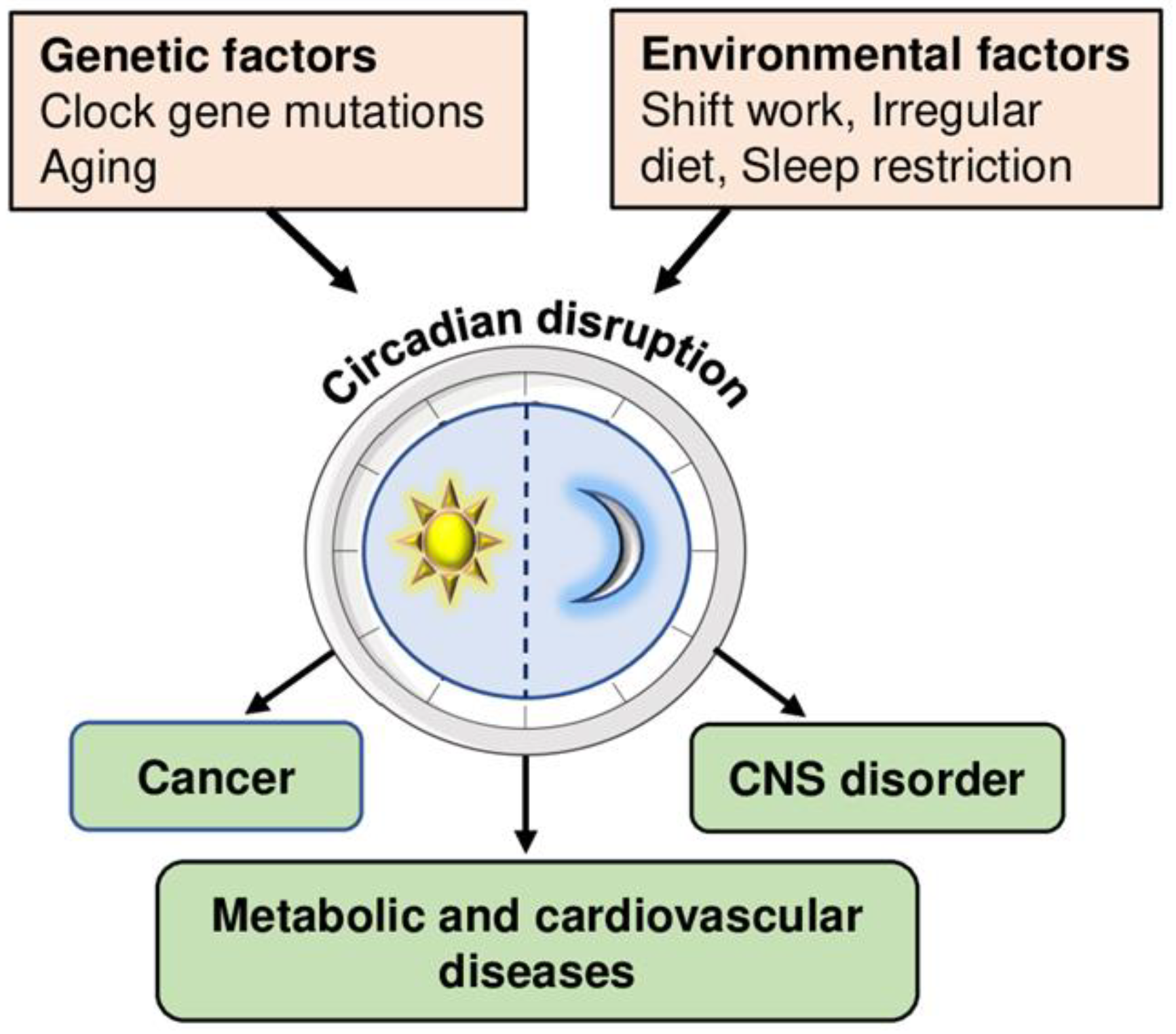
1. Introduction
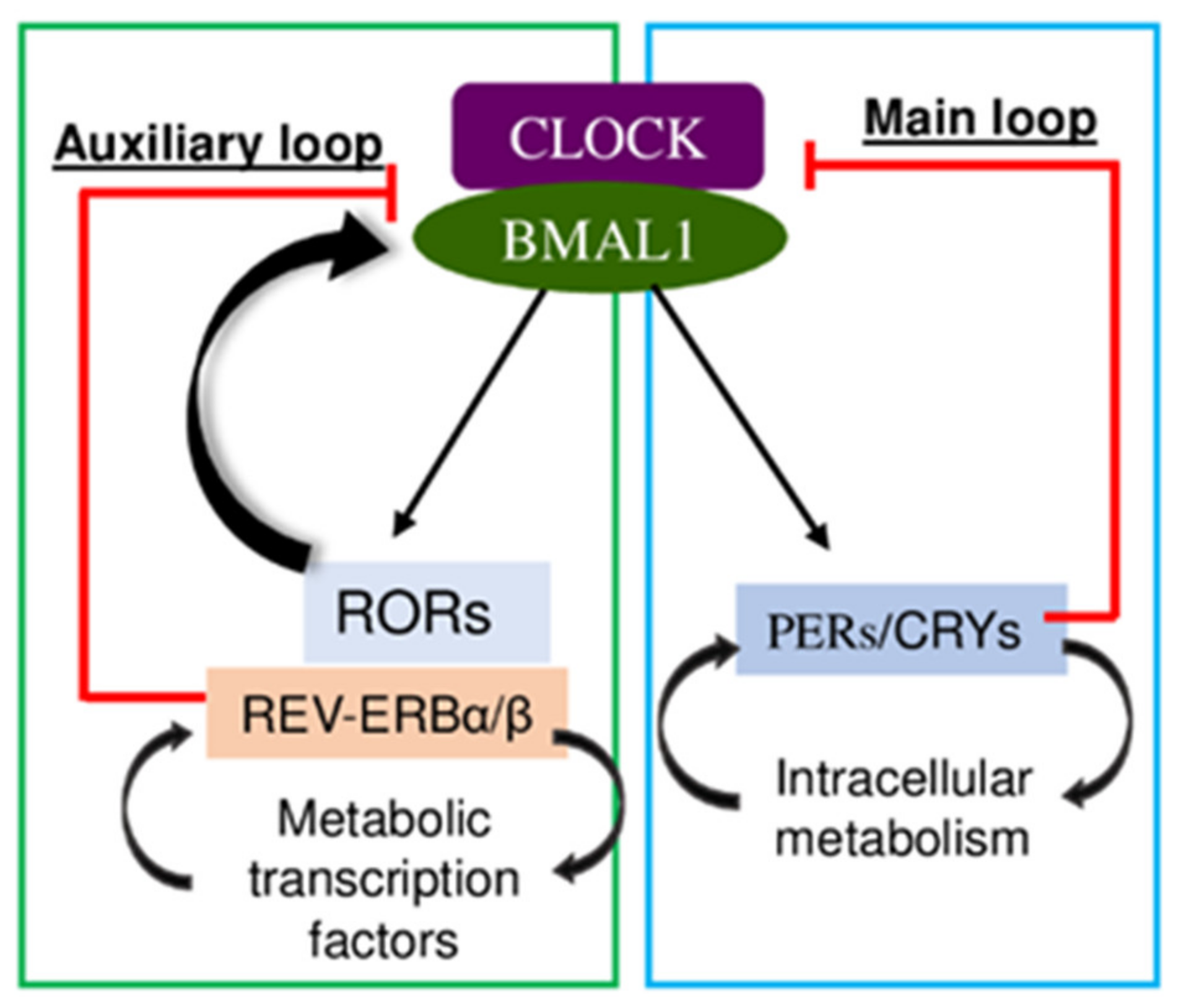
2. Circadian Rhythm
Circadian Repressor Rev-Erb
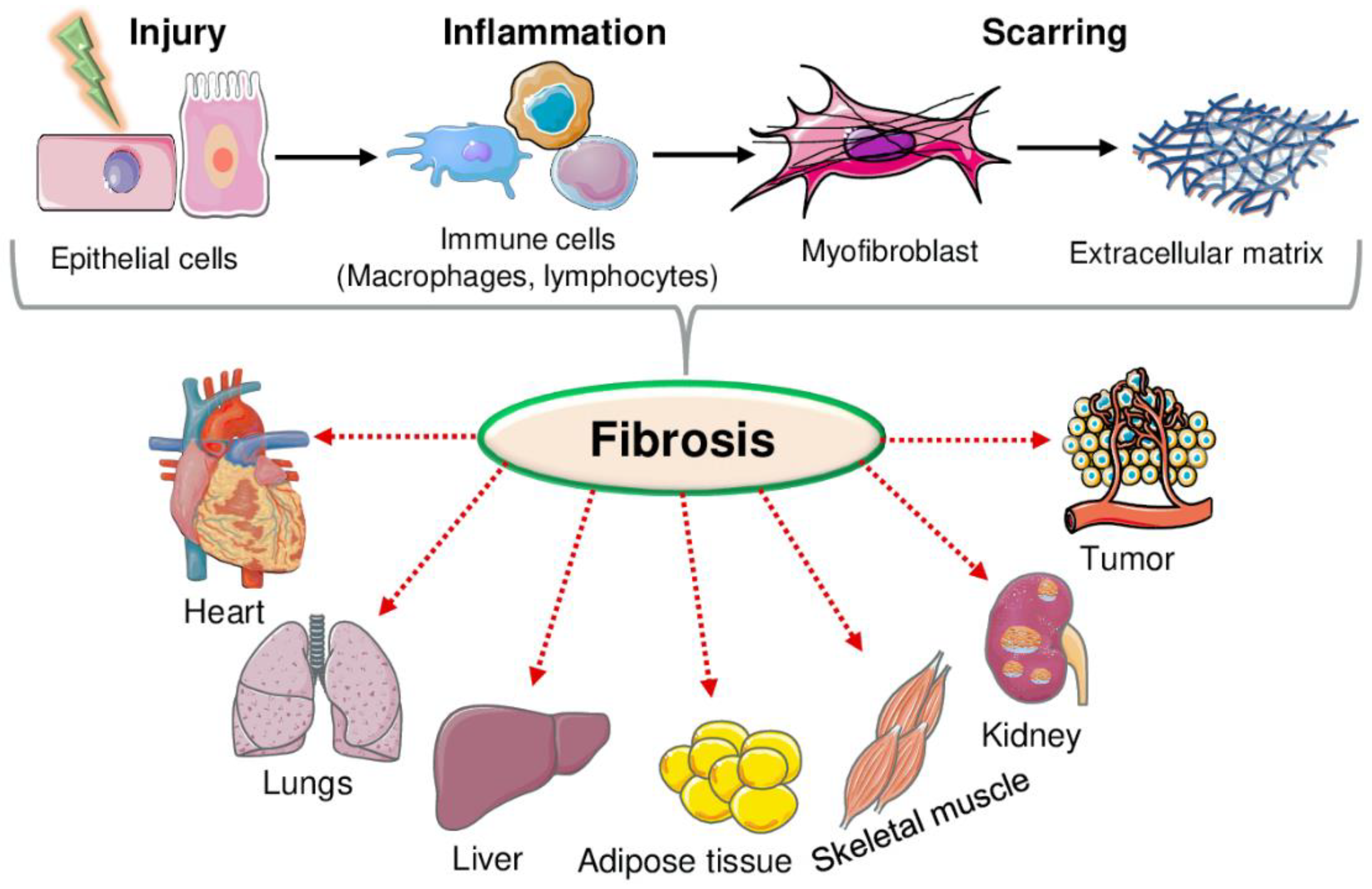
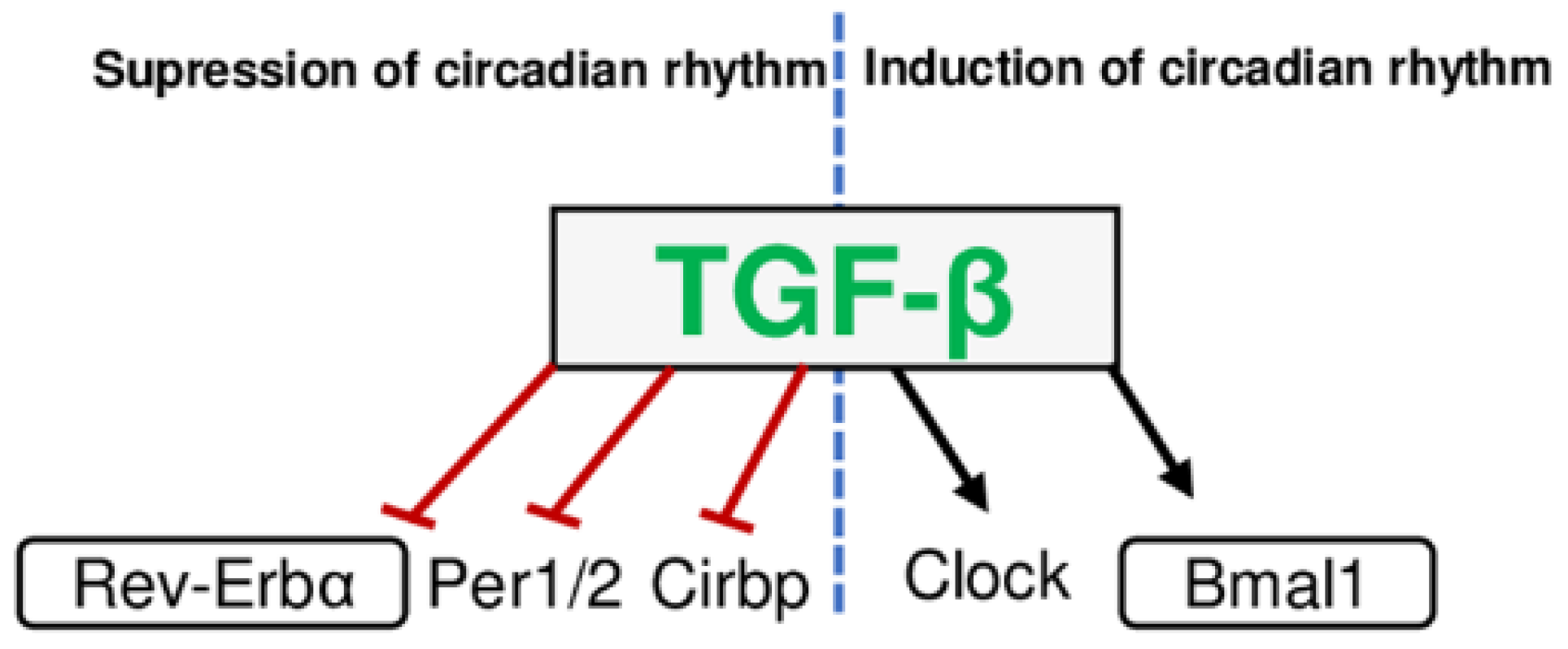
3. Fibrotic Mediators
4. Effect of Rev-Erb in Tissue Fibrosis
4.1. Rev-Erb in Heart Fibrosis
4.2. Rev-Erb in Lung Fibrosis
4.3. Rev-Erb in Liver Fibrosis
4.4. Rev-Erb in Adipose Tissue Fibrosis
4.5. Rev-Erb in Skeletal Muscle Fibrosis
4.6. Rev-Erb in Kidney Fibrosis
5. Summary and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Franceschi, C.; Campisi, J. Chronic Inflammation (Inflammaging) and Its Potential Contribution to Age-Associated Diseases. J. Gerontol. A. Biol. Sci. Med. Sci. 2014, 69 (Suppl. S1), S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From Mechanisms to Medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Buechler, C.; Krautbauer, S.; Eisinger, K. Adipose Tissue Fibrosis. World J. Diabetes 2015, 6, 548. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the Extracellular Matrix in Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of Fibrosis: Therapeutic Translation for Fibrotic Disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Manou, D.; Karamanos, N.K. The Extracellular Matrix as a Multitasking Player in Disease. FEBS J. 2019, 286, 2830–2869. [Google Scholar] [CrossRef] [PubMed]
- Klingberg, F.; Hinz, B.; White, E.S. The Myofibroblast Matrix: Implications for Tissue Repair and Fibrosis. J. Pathol. 2013, 229, 298. [Google Scholar] [CrossRef]
- Wynn, T.A. Cellular and Molecular Mechanisms of Fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The Extracellular Matrix at a Glance. J. Cell Sci. 2010, 123, 4195. [Google Scholar] [CrossRef]
- Hinz, B.; Phan, S.H.; Thannickal, V.J.; Galli, A.; Bochaton-Piallat, M.L.; Gabbiani, G. The Myofibroblast: One Function, Multiple Origins. Am. J. Pathol. 2007, 170, 1807–1816. [Google Scholar] [CrossRef]
- Darby, I.A.; Laverdet, B.; Bonté, F.; Desmoulière, A. Fibroblasts and Myofibroblasts in Wound Healing. Clin. Cosmet. Investig. Dermatol. 2014, 7, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Petrov, V.V.; Fagard, R.H.; Lijnen, P.J. Stimulation of Collagen Production by Transforming Growth Factor-Beta1 during Differentiation of Cardiac Fibroblasts to Myofibroblasts. Hypertension 2002, 39, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Gibb, A.A.; Lazaropoulos, M.P.; Elrod, J.W. Myofibroblasts and Fibrosis: Mitochondrial and Metabolic Control of Cellular Differentiation. Circ. Res. 2020, 127, 427–447. [Google Scholar] [CrossRef] [PubMed]
- Hinz, B. Formation and Function of the Myofibroblast during Tissue Repair. J. Investig. Dermatol. 2007, 127, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.; Duffy, H.S. Fibroblasts and Myofibroblasts: What Are We Talking About? J. Cardiovasc. Pharmacol. 2011, 57, 376. [Google Scholar] [CrossRef]
- Fontani, F.; Domazetovic, V.; Marcucci, T.; Vincenzini, M.T.; Iantomasi, T. Tumor Necrosis Factor-Alpha Up-Regulates ICAM-1 Expression and Release in Intestinal Myofibroblasts by Redox-Dependent and -Independent Mechanisms. J. Cell. Biochem. 2016, 117, 370–381. [Google Scholar] [CrossRef]
- Domazetovic, V.; Bonanomi, A.G.; Stio, M.; Vincenzini, M.T.; Iantomasi, T. Resveratrol Decreases TNFα-Induced ICAM-1 Expression and Release by Sirt-1-Independent Mechanism in Intestinal Myofibroblasts. Exp. Cell Res. 2019, 382, 111479. [Google Scholar] [CrossRef]
- Hinz, B.; Phan, S.H.; Thannickal, V.J.; Prunotto, M.; Desmoulire, A.; Varga, J.; De Wever, O.; Mareel, M.; Gabbiani, G. Recent Developments in Myofibroblast Biology: Paradigms for Connective Tissue Remodeling. Am. J. Pathol. 2012, 180, 1340–1355. [Google Scholar] [CrossRef]
- Kida, Y.; Duffield, J.S. Pivotal Role of Pericytes in Kidney Fibrosis. Clin. Exp. Pharmacol. Physiol. 2011, 38, 467–473. [Google Scholar] [CrossRef]
- Di Carlo, S.E.; Peduto, L. The Perivascular Origin of Pathological Fibroblasts. J. Clin. Investig. 2018, 128, 54–63. [Google Scholar] [CrossRef]
- Lombardi, A.A.; Gibb, A.A.; Arif, E.; Kolmetzky, D.W.; Tomar, D.; Luongo, T.S.; Jadiya, P.; Murray, E.K.; Lorkiewicz, P.K.; Hajnóczky, G.; et al. Mitochondrial Calcium Exchange Links Metabolism with the Epigenome to Control Cellular Differentiation. Nat. Commun. 2019, 10, 4509. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Rivera, S.; Monclus, E.A.; Synenki, L.; Zirk, A.; Eisenbart, J.; Feghali-Bostwick, C.; Mutlu, G.M.; Budinger, G.R.S.; Chandel, N.S. Mitochondrial Reactive Oxygen Species Regulate Transforming Growth Factor-β Signaling. J. Biol. Chem. 2013, 288, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Tomasek, J.J.; Gabbiani, G.; Hinz, B.; Chaponnier, C.; Brown, R.A. Myofibroblasts and Mechano-Regulation of Connective Tissue Remodelling. Nat. Rev. Mol. Cell Biol. 2002, 3, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Stempien-Otero, A.; Kim, D.H.; Davis, J. Molecular Networks Underlying Myofibroblast Fate and Fibrosis. J. Mol. Cell. Cardiol. 2016, 97, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Huang, L.; Zhao, J.; Chen, S.; Liu, J.; Li, G. The Circadian Clock and Inflammation: A New Insight. Clin. Chim. Acta. 2021, 512, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, J.C. Molecular Bases for Circadian Clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef]
- Mitsui, A.; Kumazawa, S.; Takahashi, A.; Ikemoto, H.; Cao, S.; Arai, T. Strategy by Which Nitrogen-Fixing Unicellular Cyanobacteria Grow Photoautotrophically. Nature 1986, 323, 720–722. [Google Scholar] [CrossRef]
- Vollmers, C.; Gill, S.; DiTacchio, L.; Pulivarthy, S.R.; Le, H.D.; Panda, S. Time of Feeding and the Intrinsic Circadian Clock Drive Rhythms in Hepatic Gene Expression. Proc. Natl. Acad. Sci. USA 2009, 106, 21453–21458. [Google Scholar] [CrossRef]
- Bass, J.; Takahashi, J.S. Circadian Integration of Metabolism and Energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef]
- Vetter, C.; Dashti, H.S.; Lane, J.M.; Anderson, S.G.; Schernhammer, E.S.; Rutter, M.K.; Saxena, R.; Scheer, F.A.J.L. Night Shift Work, Genetic Risk, and Type 2 Diabetes in the UK Biobank. Diabetes Care 2018, 41, 762–769. [Google Scholar] [CrossRef]
- Kim, H.J.; Choi, S.; Kim, K.; Park, H.; Kim, K.H.; Park, S.M. Association between Misalignment of Circadian Rhythm and Obesity in Korean Men: Sixth Korea National Health and Nutrition Examination Survey. Chronobiol. Int. 2020, 37, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Man, K.; Loudon, A.; Chawla, A. Immunity around the Clock. Science 2016, 354, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Dibner, C.; Schibler, U. Circadian Timing of Metabolism in Animal Models and Humans. J. Intern. Med. 2015, 277, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Tahara, Y.; Aoyama, S.; Shibata, S. The Mammalian Circadian Clock and Its Entrainment by Stress and Exercise. J. Physiol. Sci. 2017, 67, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.M.; Carter, A.M.; Grant, P.J. Association between Polymorphisms in the Clock Gene, Obesity and the Metabolic Syndrome in Man. Int. J. Obes. 2008, 32, 658–662. [Google Scholar] [CrossRef] [PubMed]
- Woon, P.Y.; Kaisaki, P.J.; Bragança, J.; Bihoreau, M.T.; Levy, J.C.; Farrall, M.; Gauguier, D. Aryl Hydrocarbon Receptor Nuclear Translocator-like (BMAL1) Is Associated with Susceptibility to Hypertension and Type 2 Diabetes. Proc. Natl. Acad. Sci. USA 2007, 104, 14412–14417. [Google Scholar] [CrossRef] [PubMed]
- Ruben, M.D.; Wu, G.; Smith, D.F.; Schmidt, R.E.; Francey, L.J.; Lee, Y.Y.; Anafi, R.C.; Hogenesch, J.B. A Database of Tissue-Specific Rhythmically Expressed Human Genes Has Potential Applications in Circadian Medicine. Sci. Transl. Med. 2018, 10, eaat8806. [Google Scholar] [CrossRef]
- Chaix, A.; Zarrinpar, A.; Panda, S. The Circadian Coordination of Cell Biology. J. Cell Biol. 2016, 215, 15–25. [Google Scholar] [CrossRef]
- Peek, C.B.; Levine, D.C.; Cedernaes, J.; Taguchi, A.; Kobayashi, Y.; Tsai, S.J.; Bonar, N.A.; McNulty, M.R.; Ramsey, K.M.; Bass, J. Circadian Clock Interaction with HIF1α Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab. 2017, 25, 86–92. [Google Scholar] [CrossRef]
- Liu, C.; Li, S.; Liu, T.; Borjigin, J.; Lin, J.D. Transcriptional Coactivator PGC-1alpha Integrates the Mammalian Clock and Energy Metabolism. Nature 2007, 447, 477–481. [Google Scholar] [CrossRef]
- Jee, H.U.; Yang, S.; Yamazaki, S.; Kang, H.; Viollet, B.; Foretz, M.; Chung, J.H. Activation of 5’-AMP-Activated Kinase with Diabetes Drug Metformin Induces Casein Kinase Iepsilon (CKIepsilon)-Dependent Degradation of Clock Protein MPer2. J. Biol. Chem. 2007, 282, 20794–20798. [Google Scholar] [CrossRef]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian Control of the NAD+ Salvage Pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Coogan, A.N.; Wyse, C.A. Neuroimmunology of the Circadian Clock. Brain Res. 2008, 1232, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Shimba, S.; Tezuka, M. Characterization of the Molecular Clock in Mouse Peritoneal Macrophages. Biol. Pharm. Bull. 2007, 30, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Hua, F.; Diandong, H.; Hong, Y. Effects of Sleep and Sleep Deprivation on Immunoglobulins and Complement in Humans. Brain. Behav. Immun. 2007, 21, 308–310. [Google Scholar] [CrossRef]
- Pekovic-Vaughan, V.; Gibbs, J.; Yoshitane, H.; Yang, N.; Pathiranage, D.; Guo, B.; Sagami, A.; Taguchi, K.; Bechtold, D.; Loudon, A.; et al. The Circadian Clock Regulates Rhythmic Activation of the NRF2/Glutathione-Mediated Antioxidant Defense Pathway to Modulate Pulmonary Fibrosis. Genes Dev. 2014, 28, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yang, G.; Jia, Z.; Zhang, H.; Aoyagi, T.; Soodvilai, S.; Symons, J.D.; Schnermann, J.B.; Gonzalez, F.J.; Litwin, S.E.; et al. Vascular PPARgamma Controls Circadian Variation in Blood Pressure and Heart Rate through Bmal1. Cell Metab. 2008, 8, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-ΚB, Inflammation, and Metabolic Disease. Cell Metab. 2011, 13, 11–22. [Google Scholar] [CrossRef]
- Kojetin, D.J.; Burris, T.P. REV-ERB and ROR Nuclear Receptors as Drug Targets. Nat. Rev. Drug Discov. 2014, 13, 197–216. [Google Scholar] [CrossRef]
- Amir, M.; Chaudhari, S.; Wang, R.; Campbell, S.; Mosure, S.A.; Chopp, L.B.; Lu, Q.; Shang, J.; Pelletier, O.B.; He, Y.; et al. REV-ERBα Regulates T H 17 Cell Development and Autoimmunity. Cell Rep. 2018, 25, 3733–3749.e8. [Google Scholar] [CrossRef]
- Burke, L.; Downes, M.; Carozzi, A.; Giguère, V.; Muscat, G.E.O. Transcriptional Repression by the Orphan Steroid Receptor RVR/Rev-Erb Beta Is Dependent on the Signature Motif and Helix 5 in the E Region: Functional Evidence for a Biological Role of RVR in Myogenesis. Nucleic Acids Res. 1996, 24, 3481–3489. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Lazar, M.A. The Orphan Nuclear Receptor Rev-Erbalpha Recruits the N-CoR/Histone Deacetylase 3 Corepressor to Regulate the Circadian Bmal1 Gene. Mol. Endocrinol. 2005, 19, 1452–1459. [Google Scholar] [CrossRef] [PubMed]
- Preitner, N.; Damiola, F.; Luis-Lopez-Molina; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The Orphan Nuclear Receptor REV-ERBalpha Controls Circadian Transcription within the Positive Limb of the Mammalian Circadian Oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, B.; Emmett, M.J.; Damle, M.; Sun, Z.; Feng, D.; Armour, S.M.; Remsberg, J.R.; Jager, J.; Soccio, R.E.; et al. GENE REGULATION. Discrete Functions of Nuclear Receptor Rev-Erbα Couple Metabolism to the Clock. Science 2015, 348, 1488–1492. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Liu, T.; Sun, Z.; Bugge, A.; Mullican, S.E.; Alenghat, T.; Liu, X.S.; Lazar, M.A. A Circadian Rhythm Orchestrated by Histone Deacetylase 3 Controls Hepatic Lipid Metabolism. Science 2011, 331, 1315–1319. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wu, N.; Curtin, J.C.; Qatanani, M.; Szwergold, N.R.; Reid, R.A.; Waitt, G.M.; Parks, D.J.; Pearce, K.H.; Wisely, G.B.; et al. Rev-Erbalpha, a Heme Sensor That Coordinates Metabolic and Circadian Pathways. Science 2007, 318, 1786–1789. [Google Scholar] [CrossRef]
- Gerhart-Hines, Z.; Lazar, M.A. Rev-Erbα and the Circadian Transcriptional Regulation of Metabolism. Diabetes. Obes. Metab. 2015, 17 (Suppl. S1), 12–16. [Google Scholar] [CrossRef]
- Solt, L.A.; Wang, Y.; Banerjee, S.; Hughes, T.; Kojetin, D.J.; Lundasen, T.; Shin, Y.; Liu, J.; Cameron, M.D.; Noel, R.; et al. Regulation of Circadian Behaviour and Metabolism by Synthetic REV-ERB Agonists. Nature 2012, 485, 62–68. [Google Scholar] [CrossRef]
- Woldt, E.; Sebti, Y.; Solt, L.A.; Duhem, C.; Lancel, S.; Eeckhoute, J.; Hesselink, M.K.C.; Paquet, C.; Delhaye, S.; Shin, Y.; et al. Rev-Erb-α Modulates Skeletal Muscle Oxidative Capacity by Regulating Mitochondrial Biogenesis and Autophagy. Nat. Med. 2013, 19, 1039–1046. [Google Scholar] [CrossRef]
- Sulli, G.; Rommel, A.; Wang, X.; Kolar, M.J.; Puca, F.; Saghatelian, A.; Plikus, M.V.; Verma, I.M.; Panda, S. Pharmacological Activation of REV-ERBs Is Lethal in Cancer and Oncogene-Induced Senescence. Nature 2018, 553, 351–355. [Google Scholar] [CrossRef]
- Banerjee, S.; Wang, Y.; Solt, L.A.; Griffett, K.; Kazantzis, M.; Amador, A.; El-Gendy, B.M.; Huitron-Resendiz, S.; Roberts, A.J.; Shin, Y.; et al. Pharmacological Targeting of the Mammalian Clock Regulates Sleep Architecture and Emotional Behaviour. Nat. Commun. 2014, 5, 5759. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.E.; Blaikley, J.; Beesley, S.; Matthews, L.; Simpson, K.D.; Boyce, S.H.; Farrow, S.N.; Else, K.J.; Singh, D.; Ray, D.W.; et al. The Nuclear Receptor REV-ERBα Mediates Circadian Regulation of Innate Immunity through Selective Regulation of Inflammatory Cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Dierickx, P.; Emmett, M.J.; Jiang, C.; Uehara, K.; Liu, M.; Adlanmerini, M.; Lazar, M.A. SR9009 Has REV-ERB-Independent Effects on Cell Proliferation and Metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 12147–12152. [Google Scholar] [CrossRef] [PubMed]
- Rockey, D.C.; Bell, P.D.; Hill, J.A. Fibrosis—A Common Pathway to Organ Injury and Failure. N. Engl. J. Med. 2015, 372, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Ikeda, K.; Chen, Y.; Alba, D.L.; Stifler, D.; Shinoda, K.; Hosono, T.; Maretich, P.; Yang, Y.; Ishigaki, Y.; et al. Repression of Adipose Tissue Fibrosis through a PRDM16-GTF2IRD1 Complex Improves Systemic Glucose Homeostasis. Cell Metab. 2018, 27, 180–194.e6. [Google Scholar] [CrossRef]
- Gorelik, L.; Flavell, R.A. Transforming Growth Factor-Beta in T-Cell Biology. Nat. Rev. Immunol. 2002, 2, 46–53. [Google Scholar] [CrossRef]
- Munger, J.S.; Huang, X.; Kawakatsu, H.; Griffiths, M.J.D.; Dalton, S.L.; Wu, J.; Pittet, J.F.; Kaminski, N.; Garat, C.; Matthay, M.A.; et al. The Integrin Alpha v Beta 6 Binds and Activates Latent TGF Beta 1: A Mechanism for Regulating Pulmonary Inflammation and Fibrosis. Cell 1999, 96, 319–328. [Google Scholar] [CrossRef]
- Lopez, M.; Meier, D.; Müller, A.; Franken, P.; Fujita, J.; Fontana, A. Tumor Necrosis Factor and Transforming Growth Factor β Regulate Clock Genes by Controlling the Expression of the Cold Inducible RNA-Binding Protein (CIRBP). J. Biol. Chem. 2014, 289, 2736–2744. [Google Scholar] [CrossRef]
- Ertosun, M.G.; Kocak, G.; Ozes, O.N. The Regulation of Circadian Clock by Tumor Necrosis Factor Alpha. Cytokine Growth Factor Rev. 2019, 46, 10–16. [Google Scholar] [CrossRef]
- Gast, H.; Gordic, S.; Petrzilka, S.; Lopez, M.; Müller, A.; Gietl, A.; Hock, C.; Birchler, T.; Fontana, A. Transforming Growth Factor-Beta Inhibits the Expression of Clock Genes. Ann. N. Y. Acad. Sci. 2012, 1261, 79–87. [Google Scholar] [CrossRef]
- Kon, N.; Hirota, T.; Kawamoto, T.; Kato, Y.; Tsubota, T.; Fukada, Y. Activation of TGF-Beta/Activin Signalling Resets the Circadian Clock through Rapid Induction of Dec1 Transcripts. Nat. Cell Biol. 2008, 10, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Sato, H.; Jin, D.; Bhawal, U.K.; Wu, Y.; Noshiro, M.; Kawamoto, T.; Fujimoto, K.; Seino, H.; Morohashi, S.; et al. Smad3 and Snail Show Circadian Expression in Human Gingival Fibroblasts, Human Mesenchymal Stem Cell, and in Mouse Liver. Biochem. Biophys. Res. Commun. 2012, 419, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Roth, K.J.; Copple, B.L. Role of Hypoxia-Inducible Factors in the Development of Liver Fibrosis. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Imtiyaz, H.Z.; Williams, E.P.; Hickey, M.M.; Patel, S.A.; Durham, A.C.; Yuan, L.J.; Hammond, R.; Gimotty, P.A.; Keith, B.; Simon, M.C. Hypoxia-Inducible Factor 2alpha Regulates Macrophage Function in Mouse Models of Acute and Tumor Inflammation. J. Clin. Investig. 2010, 120, 2699–2714. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Liu, H.; Xu, L.; Li, Y.; Liu, X.; Shi, L.; Su, Y.; Qiu, X.; Zhang, X.; Yang, Y.; et al. Hypoxia-Inducible Factor-1α Perpetuates Synovial Fibroblast Interactions with T Cells and B Cells in Rheumatoid Arthritis. Eur. J. Immunol. 2016, 46, 742–751. [Google Scholar] [CrossRef]
- Manella, G.; Aviram, R.; Bolshette, N.; Muvkadi, S.; Golik, M.; Smith, D.F.; Asher, G. Hypoxia Induces a Time- And Tissue-Specific Response That Elicits Intertissue Circadian Clock Misalignment. Proc. Natl. Acad. Sci. USA 2020, 117, 779–786. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, D.; Liu, N.; Xiong, W.; Huang, H.; Li, Y.; Ma, Z.; Zhao, H.; Chen, P.; Qi, X.; et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017, 25, 73–85. [Google Scholar] [CrossRef]
- Kobayashi, M.; Morinibu, A.; Koyasu, S.; Goto, Y.; Hiraoka, M.; Harada, H. A Circadian Clock Gene, PER2, Activates HIF-1 as an Effector Molecule for Recruitment of HIF-1α to Promoter Regions of Its Downstream Genes. FEBS J. 2017, 284, 3804–3816. [Google Scholar] [CrossRef]
- Dimova, E.Y.; Jakupovic, M.; Kubaichuk, K.; Mennerich, D.; Chi, T.F.; Tamanini, F.; Oklejewicz, M.; Hänig, J.; Byts, N.; Mäkelä, K.A.; et al. The Circadian Clock Protein CRY1 Is a Negative Regulator of HIF-1α. iScience 2019, 13, 284–304. [Google Scholar] [CrossRef]
- Henderson, N.C.; Arnold, T.D.; Katamura, Y.; Giacomini, M.M.; Rodriguez, J.D.; McCarty, J.H.; Pellicoro, A.; Raschperger, E.; Betsholtz, C.; Ruminski, P.G.; et al. Targeting of Av Integrin Identifies a Core Molecular Pathway That Regulates Fibrosis in Several Organs. Nat. Med. 2013, 19, 1617–1624. [Google Scholar] [CrossRef]
- Hewlett, J.C.; Kropski, J.A.; Blackwell, T.S. Idiopathic Pulmonary Fibrosis: Epithelial-Mesenchymal Interactions and Emerging Therapeutic Targets. Matrix Biol. 2018, 71–72, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Vannella, K.M.; Ramalingam, T.R.; Borthwick, L.A.; Barron, L.; Hart, K.M.; Thompson, R.W.; Kindrachuk, K.N.; Cheever, A.W.; White, S.; Budelsky, A.L.; et al. Combinatorial Targeting of TSLP, IL-25, and IL-33 in Type 2 Cytokine-Driven Inflammation and Fibrosis. Sci. Transl. Med. 2016, 8, 337ra65. [Google Scholar] [CrossRef] [PubMed]
- Gieseck, R.L.; Wilson, M.S.; Wynn, T.A. Type 2 Immunity in Tissue Repair and Fibrosis. Nat. Rev. Immunol. 2018, 18, 62–76. [Google Scholar] [CrossRef]
- Park, M.J.; Moon, S.J.; Lee, E.J.; Jung, K.A.; Kim, E.K.; Kim, D.S.; Lee, J.H.; Kwok, S.K.; Min, J.K.; Park, S.H.; et al. IL-1-IL-17 Signaling Axis Contributes to Fibrosis and Inflammation in Two Different Murine Models of Systemic Sclerosis. Front. Immunol. 2018, 9, 1611. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Wang, H.; Zhang, L.; Yang, X.; Zhang, M.; Zhu, X.; Ji, X.; Wang, H. Role of Interleukin 17 in TGF-β Signaling-Mediated Renal Interstitial Fibrosis. Cytokine 2018, 106, 80–88. [Google Scholar] [CrossRef] [PubMed]
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 354 Diseases and Injuries for 195 Countries and Territories, 1990-2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- Groenewegen, A.; Rutten, F.H.; Mosterd, A.; Hoes, A.W. Epidemiology of Heart Failure. Eur. J. Heart Fail. 2020, 22, 1342–1356. [Google Scholar] [CrossRef]
- Kwak, H.-B. Aging, Exercise, and Extracellular Matrix in the Heart. J. Exerc. Rehabil. 2013, 9, 338–347. [Google Scholar] [CrossRef]
- Kong, P.; Christia, P.; Frangogiannis, N.G. The Pathogenesis of Cardiac Fibrosis. Cell. Mol. Life Sci. 2014, 71, 549–574. [Google Scholar] [CrossRef]
- Leibetseder, V.; Humpeler, S.; Svoboda, M.; Schmid, D.; Thalhammer, T.; Zuckermann, A.; Marktl, W.; Ekmekcioglu, C. Clock Genes Display Rhythmic Expression in Human Hearts. Chronobiol. Int. 2009, 26, 621–636. [Google Scholar] [CrossRef]
- Young, M.E.; Razeghi, P.; Taegtmeyer, H. Clock Genes in the Heart: Characterization and Attenuation with Hypertrophy. Circ. Res. 2001, 88, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Lefta, M.; Campbell, K.S.; Feng, H.Z.; Jin, J.P.; Esser, K.A. Development of Dilated Cardiomyopathy in Bmal1-Deficient Mice. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H475–H485. [Google Scholar] [CrossRef] [PubMed]
- Young, M.E.; Brewer, R.A.; Peliciari-Garcia, R.A.; Collins, H.E.; He, L.; Birky, T.L.; Peden, B.W.; Thompson, E.G.; Ammons, B.J.; Bray, M.S.; et al. Cardiomyocyte-Specific BMAL1 Plays Critical Roles in Metabolism, Signaling, and Maintenance of Contractile Function of the Heart. J. Biol. Rhythms 2014, 29, 257–276. [Google Scholar] [CrossRef] [PubMed]
- Alibhai, F.J.; LaMarre, J.; Reitz, C.J.; Tsimakouridze, E.V.; Kroetsch, J.T.; Bolz, S.S.; Shulman, A.; Steinberg, S.; Burris, T.P.; Oudit, G.Y.; et al. Disrupting the Key Circadian Regulator CLOCK Leads to Age-Dependent Cardiovascular Disease. J. Mol. Cell. Cardiol. 2017, 105, 24–37. [Google Scholar] [CrossRef]
- Wang, N.; Sun, Y.; Zhang, H.; Wang, B.; Chen, C.; Wang, Y.; Chen, J.; Tan, X.; Zhang, J.; Xia, F.; et al. Long-Term Night Shift Work Is Associated with the Risk of Atrial Fibrillation and Coronary Heart Disease. Eur. Heart J. 2021, 42, 4180–4188. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, R.; Tien, C.L.; Chan, R.E.; Sugi, K.; Fu, C.; Griffin, A.C.; Shen, Y.; Burris, T.P.; Liao, X.; et al. REV-ERBα Ameliorates Heart Failure through Transcription Repression. JCI Insight 2017, 2, e95177. [Google Scholar] [CrossRef]
- Stujanna, E.N.; Murakoshi, N.; Tajiri, K.; Xu, D.; Kimura, T.; Qin, R.; Feng, D.; Yonebayashi, S.; Ogura, Y.; Yamagami, F.; et al. Rev-Erb Agonist Improves Adverse Cardiac Remodeling and Survival in Myocardial Infarction through an Anti-Inflammatory Mechanism. PLoS ONE 2017, 12, e0189330. [Google Scholar] [CrossRef]
- Sitaula, S.; Billon, C.; Kamenecka, T.M.; Solt, L.A.; Burris, T.P. Suppression of Atherosclerosis by Synthetic REV-ERB Agonist. Biochem. Biophys. Res. Commun. 2015, 460, 566–571. [Google Scholar] [CrossRef]
- Cunningham, P.S.; Meijer, P.; Nazgiewicz, A.; Anderson, S.G.; Borthwick, L.A.; Bagnall, J.; Kitchen, G.B.; Lodyga, M.; Begley, N.; Venkateswaran, R.V.; et al. The Circadian Clock Protein REVERBα Inhibits Pulmonary Fibrosis Development. Proc. Natl. Acad. Sci. USA 2020, 117, 1139–1147. [Google Scholar] [CrossRef]
- Sundar, I.K.; Rashid, K.; Sellix, M.T.; Rahman, I. The Nuclear Receptor and Clock Gene REV-ERBα Regulates Cigarette Smoke-Induced Lung Inflammation. Biochem. Biophys. Res. Commun. 2017, 493, 1390–1395. [Google Scholar] [CrossRef]
- Yao, H.; Sundar, I.K.; Huang, Y.; Gerloff, J.; Sellix, M.T.; Sime, P.J.; Rahman, I. Disruption of Sirtuin 1-Mediated Control of Circadian Molecular Clock and Inflammation in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2015, 53, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Sundar, I.K.; Yao, H.; Sellix, M.T.; Rahman, I. Circadian Clock Function Is Disrupted by Environmental Tobacco/Cigarette Smoke, Leading to Lung Inflammation and Injury via a SIRT1-BMAL1 Pathway. FASEB J. 2014, 28, 176–194. [Google Scholar] [CrossRef] [PubMed]
- Raspé, E.; Duez, H.; Mansén, A.; Fontaine, C.; Fiévet, C.; Fruchart, J.C.; Vennström, B.; Staels, B. Identification of Rev-Erbα as a Physiological Repressor of ApoC-III Gene Transcription. J. Lipid Res. 2002, 43, 2172–2179. [Google Scholar] [CrossRef]
- Duez, H.; van der Veen, J.N.; Duhem, C.; Pourcet, B.; Touvier, T.; Fontaine, C.; Derudas, B.; Baugé, E.; Havinga, R.; Bloks, V.W.; et al. Regulation of Bile Acid Synthesis by the Nuclear Receptor Rev-Erbalpha. Gastroenterology 2008, 135, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Xiong, Y.; Trinh, T.M.; Xiao, Y.; Hu, W.; Jiang, C.; Dierickx, P.; Jang, C.; Rabinowitz, J.D.; Lazar, M.A. The Hepatocyte Clock and Feeding Control Chronophysiology of Multiple Liver Cell Types. Science 2020, 369, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Smalling, R.V.; Huang, Y.; Jiang, Y.; Kusumanchi, P.; Bogaert, W.; Wang, L.; Delker, D.A.; Skill, N.J.; Han, S.; et al. The Role of SHP/REV-ERBα/CYP4A Axis in the Pathogenesis of Alcohol-Associated Liver Disease. JCI Insight 2021, 6, e140687. [Google Scholar] [CrossRef]
- Hand, L.E.; Usan, P.; Cooper, G.J.S.; Xu, L.Y.; Ammori, B.; Cunningham, P.S.; Aghamohammadzadeh, R.; Soran, H.; Greenstein, A.; Loudon, A.S.I.; et al. Adiponectin Induces A20 Expression in Adipose Tissue to Confer Metabolic Benefit. Diabetes 2015, 64, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Delezie, J.; Dumont, S.; Dardente, H.; Oudart, H.; Gréchez-Cassiau, A.; Klosen, P.; Teboul, M.; Delaunay, F.; Pévet, P.; Challet, E. The Nuclear Receptor REV-ERBα Is Required for the Daily Balance of Carbohydrate and Lipid Metabolism. FASEB J. 2012, 26, 3321–3335. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.W.; Ditacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of Circadian Behaviour and Metabolism by REV-ERB-α and REV-ERB-β. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef]
- Mayeuf-Louchart, A.; Thorel, Q.; Delhaye, S.; Beauchamp, J.; Duhem, C.; Danckaert, A.; Lancel, S.; Pourcet, B.; Woldt, E.; Boulinguiez, A.; et al. Rev-Erb-α Regulates Atrophy-Related Genes to Control Skeletal Muscle Mass. Sci. Rep. 2017, 7, 14383. [Google Scholar] [CrossRef]
- Welch, R.D.; Guo, C.; Sengupta, M.; Carpenter, K.J.; Stephens, N.A.; Arnett, S.A.; Meyers, M.J.; Sparks, L.M.; Smith, S.R.; Zhang, J.; et al. Rev-Erb Co-Regulates Muscle Regeneration via Tethered Interaction with the NF-Y Cistrome. Mol. Metab. 2017, 6, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhang, T.; Wang, F.; Chen, X.; Xu, H.; Zhou, C.; Chen, M.; Yu, F.; Wang, S.; Yang, D.; et al. Targeted Inhibition of Rev-Erb-α/β Limits Ferroptosis to Ameliorate Folic Acid-Induced Acute Kidney Injury. Br. J. Pharmacol. 2021, 178, 328–345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, S.; Zhou, Y.; Su, W.; Ruan, X.; Wang, B.; Zheng, F.; Warner, M.; Gustafsson, J.Å.; Guan, Y. Ablation of Cytochrome P450 Omega-Hydroxylase 4A14 Gene Attenuates Hepatic Steatosis and Fibrosis. Proc. Natl. Acad. Sci. USA 2017, 114, 3181–3185. [Google Scholar] [CrossRef] [PubMed]
- Pariollaud, M.; Gibbs, J.E.; Hopwood, T.W.; Brown, S.; Begley, N.; Vonslow, R.; Poolman, T.; Guo, B.; Saer, B.; Jones, D.H.; et al. Circadian Clock Component REV-ERBα Controls Homeostatic Regulation of Pulmonary Inflammation. J. Clin. Investig. 2018, 128, 2281–2296. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sundar, I.K.; Lucas, J.H.; Muthumalage, T.; Rahman, I. Molecular Clock REV-ERBα Regulates Cigarette Smoke-Induced Pulmonary Inflammation and Epithelial-Mesenchymal Transition. JCI Insight 2021, 6, e145200. [Google Scholar] [CrossRef]
- Sitaula, S.; Zhang, J.; Ruiz, F.; Burris, T.P. Rev-Erb Regulation of Cholesterologenesis. Biochem. Pharmacol. 2017, 131, 68–77. [Google Scholar] [CrossRef]
- Thomes, P.G.; Brandon-Warner, E.; Li, T.; Donohue, T.M.; Schrum, L.W. Rev-Erb Agonist and TGF-β Similarly Affect Autophagy but Differentially Regulate Hepatic Stellate Cell Fibrogenic Phenotype. Int. J. Biochem. Cell Biol. 2016, 81, 137–147. [Google Scholar] [CrossRef]
- Griffett, K.; Bedia-Diaz, G.; Elgendy, B.; Burris, T.P. REV-ERB Agonism Improves Liver Pathology in a Mouse Model of NASH. PLoS ONE 2020, 15, e0236000. [Google Scholar] [CrossRef]
- Feng, G.; Li, X.P.; Niu, C.Y.; Liu, M.L.; Yan, Q.Q.; Fan, L.P.; Li, Y.; Zhang, K.L.; Gao, J.; Qian, M.R.; et al. Bioinformatics Analysis Reveals Novel Core Genes Associated with Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Gene 2020, 742, 144549. [Google Scholar] [CrossRef]
- González-Fernández, B.; Sánchez, D.I.; Crespo, I.; San-Miguel, B.; de Urbina, J.O.; González-Gallego, J.; Tuñón, M.J. Melatonin Attenuates Dysregulation of the Circadian Clock Pathway in Mice With CCl4-Induced Fibrosis and Human Hepatic Stellate Cells. Front. Pharmacol. 2018, 9, 556. [Google Scholar] [CrossRef]
- Li, T.; Eheim, A.L.; Klein, S.; Uschner, F.E.; Smith, A.C.; Brandon-Warner, E.; Ghosh, S.; Bonkovsky, H.L.; Trebicka, J.; Schrum, L.W. Novel Role of Nuclear Receptor Rev-Erbα in Hepatic Stellate Cell Activation: Potential Therapeutic Target for Liver Injury. Hepatology 2014, 59, 2383–2396. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Solt, L.A.; Wang, Y.; Rogers, P.M.; Bhattacharyya, G.; Kamenecka, T.M.; Stayrook, K.R.; Crumbley, C.; Floyd, Z.E.; Gimble, J.M.; et al. Regulation of Adipogenesis by Natural and Synthetic REV-ERB Ligands. Endocrinology 2010, 151, 3015–3025. [Google Scholar] [CrossRef] [PubMed]
- Welch, R.D.; Billon, C.; Valfort, A.C.; Burris, T.P.; Flaveny, C.A. Pharmacological Inhibition of REV-ERB Stimulates Differentiation, Inhibits Turnover and Reduces Fibrosis in Dystrophic Muscle. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hadden, H.; Soldin, S.J.; Massaro, D. Circadian Disruption Alters Mouse Lung Clock Gene Expression and Lung Mechanics. J. Appl. Physiol. 2012, 113, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.L.; Brenner, B.E.; Camargo, C.A. Circadian-Rhythm Differences among Emergency Department Patients with Chronic Obstructive Pulmonary Disease Exacerbation. Chronobiol. Int. 2007, 24, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Papagiannakopoulos, T.; Bauer, M.R.; Davidson, S.M.; Heimann, M.; Subbaraj, L.; Bhutkar, A.; Bartlebaugh, J.; Vander Heiden, M.G.; Jacks, T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016, 24, 324. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.; Ince, L.; Matthews, L.; Mei, J.; Bell, T.; Yang, N.; Saer, B.; Begley, N.; Poolman, T.; Pariollaud, M.; et al. An Epithelial Circadian Clock Controls Pulmonary Inflammation and Glucocorticoid Action. Nat. Med. 2014, 20, 919–926. [Google Scholar] [CrossRef]
- Dong, C.; Gongora, R.; Sosulski, M.L.; Luo, F.; Sanchez, C.G. Regulation of Transforming Growth Factor-Beta1 (TGF-Β1)-Induced pro-Fibrotic Activities by Circadian Clock Gene BMAL1. Respir. Res. 2016, 17, 4. [Google Scholar] [CrossRef]
- Kisseleva, T.; Brenner, D.A. Mechanisms of Fibrogenesis. Exp. Biol. Med. 2008, 233, 109–122. [Google Scholar] [CrossRef]
- Wallace, M.C.; Friedman, S.L.; Mann, D.A. Emerging and Disease-Specific Mechanisms of Hepatic Stellate Cell Activation. Semin. Liver Dis. 2015, 35, 107–118. [Google Scholar] [CrossRef]
- Mederacke, I.; Hsu, C.C.; Troeger, J.S.; Huebener, P.; Mu, X.; Dapito, D.H.; Pradere, J.P.; Schwabe, R.F. Fate Tracing Reveals Hepatic Stellate Cells as Dominant Contributors to Liver Fibrosis Independent of Its Aetiology. Nat. Commun. 2013, 4, 2823. [Google Scholar] [CrossRef] [PubMed]
- Böker, K.H.W.; Pehle, B.; Steinmetz, C.; Breitenstein, K.; Bahr, M.; Lichtinghagen, R. Tissue Inhibitors of Metalloproteinases in Liver and Serum/Plasma in Chronic Active Hepatitis C and HCV-Induced Cirrhosis. Hepatogastroenterology 2000, 47, 812–819. [Google Scholar] [PubMed]
- Berardis, S.; Sattwika, P.D.; Najimi, M.; Sokal, E.M. Use of Mesenchymal Stem Cells to Treat Liver Fibrosis: Current Situation and Future Prospects. World J. Gastroenterol. 2015, 21, 742–758. [Google Scholar] [CrossRef] [PubMed]
- Sahar, S.; Sassone-Corsi, P. Regulation of Metabolism: The Circadian Clock Dictates the Time. Trends Endocrinol. Metab. 2012, 23, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and Metabolic Syndrome in Circadian Clock Mutant Mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Adamovich, Y.; Rousso-Noori, L.; Zwighaft, Z.; Neufeld-Cohen, A.; Golik, M.; Kraut-Cohen, J.; Wang, M.; Han, X.; Asher, G. Circadian Clocks and Feeding Time Regulate the Oscillations and Levels of Hepatic Triglycerides. Cell Metab. 2014, 19, 319–330. [Google Scholar] [CrossRef]
- Dallmann, R.; Viola, A.U.; Tarokh, L.; Cajochen, C.; Brown, S.A. The Human Circadian Metabolome. Proc. Natl. Acad. Sci. USA 2012, 109, 2625–2629. [Google Scholar] [CrossRef] [PubMed]
- Le Martelot, G.; Claudel, T.; Gatfield, D.; Schaad, O.; Kornmann, B.; Lo Sasso, G.; Moschetta, A.; Schibler, U. REV-ERBalpha Participates in Circadian SREBP Signaling and Bile Acid Homeostasis. PLoS Biol. 2009, 7, e1000181. [Google Scholar] [CrossRef]
- Zhou, D.; Wang, Y.; Chen, L.; Jia, L.; Yuan, J.; Sun, M.; Zhang, W.; Wang, P.; Zuo, J.; Xu, Z.; et al. Evolving Roles of Circadian Rhythms in Liver Homeostasis and Pathology. Oncotarget 2016, 7, 8625–8639. [Google Scholar] [CrossRef]
- Hardwick, J.P. Cytochrome P450 Omega Hydroxylase (CYP4) Function in Fatty Acid Metabolism and Metabolic Diseases. Biochem. Pharmacol. 2008, 75, 2263–2275. [Google Scholar] [CrossRef]
- Trevaskis, J.L.; Griffin, P.S.; Wittmer, C.; Neuschwander-Tetri, B.A.; Brunt, E.M.; Dolman, C.S.; Erickson, M.R.; Napora, J.; Parkes, D.G.; Roth, J.D. Glucagon-like Peptide-1 Receptor Agonism Improves Metabolic, Biochemical, and Histopathological Indices of Nonalcoholic Steatohepatitis in Mice. Am. J. Physiol. -Gastrointest. Liver Physiol. 2012, 302, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Elbel, E.E.; Lavine, J.E.; Downes, M.; Van Natta, M.; Yu, R.; Schwimmer, J.B.; Behling, C.; Brunt, E.M.; Tonascia, J.; Evans, R. Hepatic Nuclear Receptor Expression Associates with Features of Histology in Pediatric Nonalcoholic Fatty Liver Disease. Hepatol. Commun. 2018, 2, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, B.; Gogg, S.; Hedjazifar, S.; Jenndahl, L.; Hammarstedt, A.; Smith, U. Inflammation and Impaired Adipogenesis in Hypertrophic Obesity in Man. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E999–E1003. [Google Scholar] [CrossRef] [PubMed]
- Exley, M.A.; Hand, L.; O’Shea, D.; Lynch, L. Interplay between the Immune System and Adipose Tissue in Obesity. J. Endocrinol. 2014, 223, R41–R48. [Google Scholar] [CrossRef] [PubMed]
- Hosogai, N.; Fukuhara, A.; Oshima, K.; Miyata, Y.; Tanaka, S.; Segawa, K.; Furukawa, S.; Tochino, Y.; Komuro, R.; Matsuda, M.; et al. Adipose Tissue Hypoxia in Obesity and Its Impact on Adipocytokine Dysregulation. Diabetes 2007, 56, 901–911. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, J.W.; Osborne, O.; Oh, D.Y.; Sasik, R.; Schenk, S.; Chen, A.; Chung, H.; Murphy, A.; Watkins, S.M.; et al. Increased Adipocyte O2 Consumption Triggers HIF-1α, Causing Inflammation and Insulin Resistance in Obesity. Cell 2014, 157, 1339–1352. [Google Scholar] [CrossRef]
- Lolmède, K.; De Durand Saint Front, V.; Galitzky, J.; Lafontan, M.; Bouloumié, A. Effects of Hypoxia on the Expression of Proangiogenic Factors in Differentiated 3T3-F442A Adipocytes. Int. J. Obes. Relat. Metab. Disord. 2003, 27, 1187–1195. [Google Scholar] [CrossRef]
- Wueest, S.; Rapold, R.A.; Rytka, J.M.; Schoenle, E.J.; Konrad, D. Basal Lipolysis, Not the Degree of Insulin Resistance, Differentiates Large from Small Isolated Adipocytes in High-Fat Fed Mice. Diabetologia 2009, 52, 541–546. [Google Scholar] [CrossRef]
- Giordano, A.; Murano, I.; Mondini, E.; Perugini, J.; Smorlesi, A.; Severi, I.; Barazzoni, R.; Scherer, P.E.; Cinti, S. Obese Adipocytes Show Ultrastructural Features of Stressed Cells and Die of Pyroptosis. J. Lipid Res. 2013, 54, 2423–2436. [Google Scholar] [CrossRef]
- Vila, I.K.; Badin, P.M.; Marques, M.A.; Monbrun, L.; Lefort, C.; Mir, L.; Louche, K.; Bourlier, V.; Roussel, B.; Gui, P.; et al. Immune Cell Toll-like Receptor 4 Mediates the Development of Obesity- and Endotoxemia-Associated Adipose Tissue Fibrosis. Cell Rep. 2014, 7, 1116–1129. [Google Scholar] [CrossRef]
- Tanaka, M.; Ikeda, K.; Suganami, T.; Komiya, C.; Ochi, K.; Shirakawa, I.; Hamaguchi, M.; Nishimura, S.; Manabe, I.; Matsuda, T.; et al. Macrophage-Inducible C-Type Lectin Underlies Obesity-Induced Adipose Tissue Fibrosis. Nat. Commun. 2014, 5, 4982. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Lyon, C.J.; Minze, L.J.; Lin, J.; Zou, J.; Liu, J.Z.; Ren, Y.; Yin, Z.; Hamilton, D.J.; Reardon, P.R.; et al. Class II Major Histocompatibility Complex Plays an Essential Role in Obesity-Induced Adipose Inflammation. Cell Metab. 2013, 17, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Marcelin, G.; Silveira, A.L.M.; Martins, L.B.; Ferreira, A.V.M.; Clément, K. Deciphering the Cellular Interplays Underlying Obesity-Induced Adipose Tissue Fibrosis. J. Clin. Investig. 2019, 129, 4032–4040. [Google Scholar] [CrossRef] [PubMed]
- Marcelin, G.; Ferreira, A.; Liu, Y.; Atlan, M.; Aron-Wisnewsky, J.; Pelloux, V.; Botbol, Y.; Ambrosini, M.; Fradet, M.; Rouault, C.; et al. A PDGFRα-Mediated Switch toward CD9 High Adipocyte Progenitors Controls Obesity-Induced Adipose Tissue Fibrosis. Cell Metab. 2017, 25, 673–685. [Google Scholar] [CrossRef]
- Zvonic, S.; Ptitsyn, A.A.; Conrad, S.A.; Scott, L.K.; Floyd, Z.E.; Kilroy, G.; Wu, X.; Goh, B.C.; Mynatt, R.L.; Gimble, J.M. Characterization of Peripheral Circadian Clocks in Adipose Tissues. Diabetes 2006, 55, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zvonic, S.; Floyd, Z.E.; Kilroy, G.; Goh, B.C.; Hernandez, T.L.; Eckel, R.H.; Mynatt, R.L.; Gimble, J.M. Induction of Circadian Gene Expression in Human Subcutaneous Adipose-Derived Stem Cells. Obesity 2007, 15, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Fonken, L.K.; Workman, J.L.; Walton, J.C.; Weil, Z.M.; Morris, J.S.; Haim, A.; Nelson, R.J. Light at Night Increases Body Mass by Shifting the Time of Food Intake. Proc. Natl. Acad. Sci. USA 2010, 107, 18664–18669. [Google Scholar] [CrossRef]
- Ando, H.; Kumazaki, M.; Motosugi, Y.; Ushijima, K.; Maekawa, T.; Ishikawa, E.; Fujimura, A. Impairment of Peripheral Circadian Clocks Precedes Metabolic Abnormalities in Ob/Ob Mice. Endocrinology 2011, 152, 1347–1354. [Google Scholar] [CrossRef]
- Maury, E.; Navez, B.; Brichard, S.M. Circadian Clock Dysfunction in Human Omental Fat Links Obesity to Metabolic Inflammation. Nat. Commun. 2021, 12, 2388. [Google Scholar] [CrossRef]
- Huber, J.; Löffler, M.; Bilban, M.; Reimers, M.; Kadl, A.; Todoric, J.; Zeyda, M.; Geyeregger, R.; Schreiner, M.; Weichhart, T.; et al. Prevention of High-Fat Diet-Induced Adipose Tissue Remodeling in Obese Diabetic Mice by n-3 Polyunsaturated Fatty Acids. Int. J. Obes. 2007, 31, 1004–1013. [Google Scholar] [CrossRef]
- Divoux, A.; Tordjman, J.; Lacasa, D.; Veyrie, N.; Hugol, D.; Aissat, A.; Basdevant, A.; Guerre-Millo, M.; Poitou, C.; Zucker, J.D.; et al. Fibrosis in Human Adipose Tissue: Composition, Distribution, and Link with Lipid Metabolism and Fat Mass Loss. Diabetes 2010, 59, 2817–2825. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Muise, E.S.; Iyengar, P.; Wang, Z.V.; Chandalia, M.; Abate, N.; Zhang, B.B.; Bonaldo, P.; Chua, S.; Scherer, P.E. Metabolic Dysregulation and Adipose Tissue Fibrosis: Role of Collagen VI. Mol. Cell. Biol. 2009, 29, 1575–1591. [Google Scholar] [CrossRef] [PubMed]
- Spencer, M.; Yao-Borengasser, A.; Unal, R.; Rasouli, N.; Gurley, C.M.; Zhu, B.; Peterson, C.A.; Kern, P.A. Adipose Tissue Macrophages in Insulin-Resistant Subjects Are Associated with Collagen VI and Fibrosis and Demonstrate Alternative Activation. Am. J. Physiol. Endocrinol. Metab. 2010, 299. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Park, J.; Gupta, O.T.; Holland, W.L.; Auerbach, P.; Zhang, N.; Goncalves Marangoni, R.; Nicoloro, S.M.; Czech, M.P.; Varga, J.; et al. Endotrophin Triggers Adipose Tissue Fibrosis and Metabolic Dysfunction. Nat. Commun. 2014, 5, 3485. [Google Scholar] [CrossRef]
- Chaix, A.; Lin, T.; Le, H.D.; Chang, M.W.; Panda, S. Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab. 2019, 29, 303–319.e4. [Google Scholar] [CrossRef] [PubMed]
- Paschos, G.K.; Ibrahim, S.; Song, W.L.; Kunieda, T.; Grant, G.; Reyes, T.M.; Bradfield, C.A.; Vaughan, C.H.; Eiden, M.; Masoodi, M.; et al. Obesity in Mice with Adipocyte-Specific Deletion of Clock Component Arntl. Nat. Med. 2012, 18, 1768–1777. [Google Scholar] [CrossRef] [PubMed]
- Jager, J.; Wang, F.; Fang, B.; Lim, H.W.; Peed, L.C.; Steger, D.J.; Won, K.J.; Kharitonenkov, A.; Adams, A.C.; Lazar, M.A. The Nuclear Receptor Rev-Erbα Regulates Adipose Tissue-Specific FGF21 Signaling. J. Biol. Chem. 2016, 291, 10867–10875. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.L.; Pelekanou, C.E.; Barron, N.J.; Northeast, R.C.; Grudzien, M.; Adamson, A.D.; Downton, P.; Cornfield, T.; Cunningham, P.S.; Billaud, J.N.; et al. Adipocyte Nr1d1 Dictates Adipose Tissue Expansion during Obesity. Elife 2021, 10, e63324. [Google Scholar] [CrossRef]
- Pivovarova, O.; Gögebakan; Sucher, S.; Groth, J.; Murahovschi, V.; Kessler, K.; Osterhoff, M.; Rudovich, N.; Kramer, A.; Pfeiffer, A.F.H. Regulation of the Clock Gene Expression in Human Adipose Tissue by Weight Loss. Int. J. Obes. 2016, 40, 899–906. [Google Scholar] [CrossRef]
- Järvinen, T.A.H.; Järvinen, T.L.N.; Kääriäinen, M.; Kalimo, H.; Järvinen, M. Muscle Injuries: Biology and Treatment. Am. J. Sports Med. 2005, 33, 745–764. [Google Scholar] [CrossRef] [PubMed]
- Kjær, M. Role of Extracellular Matrix in Adaptation of Tendon and Skeletal Muscle to Mechanical Loading. Physiol. Rev. 2004, 84, 649–698. [Google Scholar] [CrossRef] [PubMed]
- Gillies, A.R.; Lieber, R.L. Structure and Function of the Skeletal Muscle Extracellular Matrix. Muscle Nerve 2011, 44, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Dyar, K.A.; Hubert, M.J.; Mir, A.A.; Ciciliot, S.; Lutter, D.; Greulich, F.; Quagliarini, F.; Kleinert, M.; Fischer, K.; Eichmann, T.O.; et al. Transcriptional Programming of Lipid and Amino Acid Metabolism by the Skeletal Muscle Circadian Clock. PLoS Biol. 2018, 16, e2005886. [Google Scholar] [CrossRef] [PubMed]
- Hodge, B.A.; Wen, Y.; Riley, L.A.; Zhang, X.; England, J.H.; Harfmann, B.D.; Schroder, E.A.; Esser, K.A. The Endogenous Molecular Clock Orchestrates the Temporal Separation of Substrate Metabolism in Skeletal Muscle. Skelet. Muscle 2015, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liang, J.; Chen, N. Do Not Neglect the Role of Circadian Rhythm in Muscle Atrophy. Ageing Res. Rev. 2020, 63, 101155. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.I.; Park, D.K.; Chung, J.W.; Kim, K.O.; Kwon, K.A.; Kim, Y.J. Circadian Rhythm Disruption Is Associated with an Increased Risk of Sarcopenia: A Nationwide Population-Based Study in Korea. Sci. Rep. 2019, 9, 12015. [Google Scholar] [CrossRef]
- Dyar, K.A.; Ciciliot, S.; Wright, L.E.; Biensø, R.S.; Tagliazucchi, G.M.; Patel, V.R.; Forcato, M.; Paz, M.I.P.; Gudiksen, A.; Solagna, F.; et al. Muscle Insulin Sensitivity and Glucose Metabolism Are Controlled by the Intrinsic Muscle Clock. Mol. Metab. 2013, 3, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Michael, M.D.; Previs, S.F.; Peroni, O.D.; Mauvais-Jarvis, F.; Neschen, S.; Kahn, B.B.; Kahn, C.R.; Shulman, G.I. Redistribution of Substrates to Adipose Tissue Promotes Obesity in Mice with Selective Insulin Resistance in Muscle. J. Clin. Investig. 2000, 105, 1791–1797. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Downes, M.; Yu, R.T.; Bookout, A.L.; He, W.; Straume, M.; Mangelsdorf, D.J.; Evans, R.M. Nuclear Receptor Expression Links the Circadian Clock to Metabolism. Cell 2006, 126, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Pircher, P.; Chomez, P.; Yu, F.; Vennström, B.; Larsson, L. Aberrant Expression of Myosin Isoforms in Skeletal Muscles from Mice Lacking the Rev-ErbAalpha Orphan Receptor Gene. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, R482–R490. [Google Scholar] [CrossRef]
- Ramakrishnan, S.N.; Lau, P.; Burke, L.J.; Muscat, G.E.O. Rev-Erbβ Regulates the Expression of Genes Involved in Lipid Absorption in Skeletal Muscle Cells: Evidence for Cross-Talk between Orphan Nuclear Receptors and Myokines. J. Biol. Chem. 2005, 280, 8651–8659. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin Is Recruited Selectively to Impaired Mitochondria and Promotes Their Autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Wang, H.; Lozano, R.; Davis, A.; Liang, X.; Zhou, M.; Vollset, S.E.; Abbasoglu Ozgoren, A.; Abdalla, S.; Abd-Allah, F.; et al. Global, Regional, and National Age-Sex Specific All-Cause and Cause-Specific Mortality for 240 Causes of Death, 1990–2013: A Systematic Analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385, 117–171. [Google Scholar] [CrossRef]
- Rhee, C.M.; Kovesdy, C.P. Epidemiology: Spotlight on CKD Deaths—Increasing Mortality Worldwide. Nat. Rev. Nephrol. 2015, 11, 199–200. [Google Scholar] [CrossRef] [PubMed]
- Jager, K.J.; Kovesdy, C.; Langham, R.; Rosenberg, M.; Jha, V.; Zoccali, C. A Single Number for Advocacy and Communication-Worldwide More than 850 Million Individuals Have Kidney Diseases. Kidney Int. 2019, 96, 1048–1050. [Google Scholar] [CrossRef] [PubMed]
- Duffield, J.S. Cellular and Molecular Mechanisms in Kidney Fibrosis. J. Clin. Investig. 2014, 124, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Takemura, T.; Tohda, M.; Akano, N.; Miyamoto, H.; Ooshima, A.; Maki, S. Glomerular Localization of Type III Collagen in Human Kidney Disease. Kidney Int. 1989, 35, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Nerlich, A.G.; Schleicher, E. Identification of Lymph and Blood Capillaries by Immunohistochemical Staining for Various Basement Membrane Components. Histochemistry 1991, 96, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Lebleu, V.S.; Taduri, G.; O’Connell, J.; Teng, Y.; Cooke, V.G.; Woda, C.; Sugimoto, H.; Kalluri, R. Origin and Function of Myofibroblasts in Kidney Fibrosis. Nat. Med. 2013, 19, 1047–1053. [Google Scholar] [CrossRef]
- Carew, R.M.; Wang, B.; Kantharidis, P. The Role of EMT in Renal Fibrosis. Cell Tissue Res. 2012, 347, 103–116. [Google Scholar] [CrossRef]
- Solocinski, K.; Gumz, M.L. The Circadian Clock in the Regulation of Renal Rhythms. J. Biol. Rhythms 2015, 30, 470–486. [Google Scholar] [CrossRef] [PubMed]
- Voogel, A.J.; Koopman, M.G.; Hart, A.A.M.; Van Montfrans, G.A.; Arisz, L. Circadian Rhythms in Systemic Hemodynamics and Renal Function in Healthy Subjects and Patients with Nephrotic Syndrome. Kidney Int. 2001, 59, 1873–1880. [Google Scholar] [CrossRef] [PubMed]
- Ansermet, C.; Centeno, G.; Nikolaeva, S.; Maillard, M.P.; Pradervand, S.; Firsov, D. The Intrinsic Circadian Clock in Podocytes Controls Glomerular Filtration Rate. Sci. Rep. 2019, 9, 16089. [Google Scholar] [CrossRef]
- Yoshida, Y.; Matsunaga, N.; Nakao, T.; Hamamura, K.; Kondo, H.; Ide, T.; Tsutsui, H.; Tsuruta, A.; Kurogi, M.; Nakaya, M.; et al. Alteration of Circadian Machinery in Monocytes Underlies Chronic Kidney Disease-Associated Cardiac Inflammation and Fibrosis. Nat. Commun. 2021, 12, 2783. [Google Scholar] [CrossRef] [PubMed]
- Egstrand, S.; Mace, M.L.; Olgaard, K.; Lewin, E. The Vascular Circadian Clock in Chronic Kidney Disease. Cells 2021, 10, 1769. [Google Scholar] [CrossRef] [PubMed]
- Zha, M.; Tian, T.; Xu, W.; Liu, S.; Jia, J.; Wang, L.; Yan, Q.; Li, N.; Yu, J.; Huang, L. The Circadian Clock Gene Bmal1 Facilitates Cisplatin-Induced Renal Injury and Hepatization. Cell Death Dis. 2020, 11, 446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, C.; Liang, Q.; Zheng, F.; Guan, Y.; Yang, G.; Chen, L. Postnatal Deletion of Bmal1 in Mice Protects against Obstructive Renal Fibrosis via Suppressing Gli2 Transcription. FASEB J. 2021, 35, e21530. [Google Scholar] [CrossRef] [PubMed]
- Kramann, R. Hedgehog Gli Signalling in Kidney Fibrosis. Nephrol. Dial. Transplant 2016, 31, 1989–1995. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.D.; Yeh, J.K.; Peng, M.T.; Shie, S.S.; Lin, S.L.; Yang, C.H.; Chen, T.H.; Hung, K.C.; Wang, C.C.; Hsieh, I.C.; et al. Circadian CLOCK Mediates Activation of Transforming Growth Factor-β Signaling and Renal Fibrosis through Cyclooxygenase 2. Am. J. Pathol. 2015, 185, 3152–3163. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, H.; Tahara, Y.; Whittaker, D.S.; Wang, H.B.; Yamaji, T.; Wakui, H.; Haraguchi, A.; Yamazaki, M.; Miyakawa, H.; Hama, K.; et al. The Circadian Clock Is Disrupted in Mice with Adenine-Induced Tubulointerstitial Nephropathy. Kidney Int. 2020, 97, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, N.; Ikeda, E.; Kakimoto, K.; Watanabe, M.; Shindo, N.; Tsuruta, A.; Ikeyama, H.; Hamamura, K.; Higashi, K.; Yamashita, T.; et al. Inhibition of G0/G1 Switch 2 Ameliorates Renal Inflammation in Chronic Kidney Disease. EBioMedicine 2016, 13, 262. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Ueno, T.; Tanaka, S.; Kobayashi, H.; Okamura, M.; Hemmi, S.; Fuke, Y.; Matsumoto, Y.; Abe, M.; Fukuda, N. Identification of Clock Genes Related to Hypertension in Kidney From Spontaneously Hypertensive Rats. Am. J. Hypertens. 2020, 33, 1136. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Brown, S.A.; Marcacci, L.; Tronche, F.; Kellendonk, C.; Reichardt, H.M.; Schutz, G.; Schibler, U. Resetting of Circadian Time in Peripheral Tissues by Glucocorticoid Signaling. Science 2000, 289, 2344–2347. [Google Scholar] [CrossRef] [PubMed]
 = decrease,
= decrease,  = increase).
= increase).
 = decrease,
= decrease,  = increase).
= increase).
| A | ||||
| Effector Organ | Rev-Erb | Functions | Reference | |
| Lung | Knockout mice |
| [99,100] | |
| Lower mRNA and protein expression |
| [101] | ||
| Reduced Rev-Erbα mRNA and protein expression |
| [102] | ||
| Liver | Knockout mice |
| [55,103,104] | |
| Knockout of Rev-Erbα/β mice fed with HFD |
| [105] | ||
| Knockout in primary hepatocytes |
| [106] | ||
| Adipose tissue | Knockout mice |
| [107,108] | |
| [107,108] | |||
| Skeletal muscle | Rev-Erbα/β double-knockout mice |
| [109] | |
| Knockout mice |
| [59,110,111] | ||
| Upregulation in Mice |
| [110] | ||
| Upregulation in C2C12 myocytes |
| [59] | ||
| Upregulation in myoblasts |
| [110] | ||
| Kidney | Rev-Erbα/β knockout mice |
| [112] | |
| Rev-Erbβ knockout mice |
| [112] | ||
| B | ||||
| Rev-Erb ligands | Effector organ | Animal Model | Effects | Reference |
| SR9009 (Agonist) | Heart | TAC mice |
| [113] |
| Mice |
| [97] | ||
| LDL-receptor deficient mice fed with a western diet |
| [98] | ||
| GSK4112 (Agonist) | Lungs | Human small airway epithelial cells and mouse lung fibroblasts |
| [100,114] |
| HFL-1 cells |
| [115] | ||
| SR9009 (Agonist) | Liver | Mice |
| [63] |
| [116] | |||
| HSC |
| [117] | ||
| Mice |
| [106] | ||
| NASH mice |
| [118,119] | ||
| Ccl4-induced fibrosis in mice and Rat HSCs |
| [120,121] | ||
| SR6452 (Agonist) | Adipose tissue | 3T3L1 cells |
| [122] |
| SR9011 (Agonist) | Mice |
| [58] | |
| SR9009 (Agonist) | Mice |
| [58] | |
| SR6452 (Agonist) | Skeletal muscle | Mice |
| [110] |
| SR8278 (Antagonist) | Dystrophic mice |
| [123] | |
| GSK1362 (Inverse agonist | Kidney | HEK293 cells |
| [114] |
| SR8278 (Antagonist) | Mice |
| [112] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raza, G.S.; Sodum, N.; Kaya, Y.; Herzig, K.-H. Role of Circadian Transcription Factor Rev-Erb in Metabolism and Tissue Fibrosis. Int. J. Mol. Sci. 2022, 23, 12954. https://doi.org/10.3390/ijms232112954
Raza GS, Sodum N, Kaya Y, Herzig K-H. Role of Circadian Transcription Factor Rev-Erb in Metabolism and Tissue Fibrosis. International Journal of Molecular Sciences. 2022; 23(21):12954. https://doi.org/10.3390/ijms232112954
Chicago/Turabian StyleRaza, Ghulam Shere, Nalini Sodum, Yagmur Kaya, and Karl-Heinz Herzig. 2022. "Role of Circadian Transcription Factor Rev-Erb in Metabolism and Tissue Fibrosis" International Journal of Molecular Sciences 23, no. 21: 12954. https://doi.org/10.3390/ijms232112954
APA StyleRaza, G. S., Sodum, N., Kaya, Y., & Herzig, K.-H. (2022). Role of Circadian Transcription Factor Rev-Erb in Metabolism and Tissue Fibrosis. International Journal of Molecular Sciences, 23(21), 12954. https://doi.org/10.3390/ijms232112954






