Hypusinated eIF5A Promotes Ribosomal Frameshifting during Decoding of ODC Antizyme mRNA in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Results
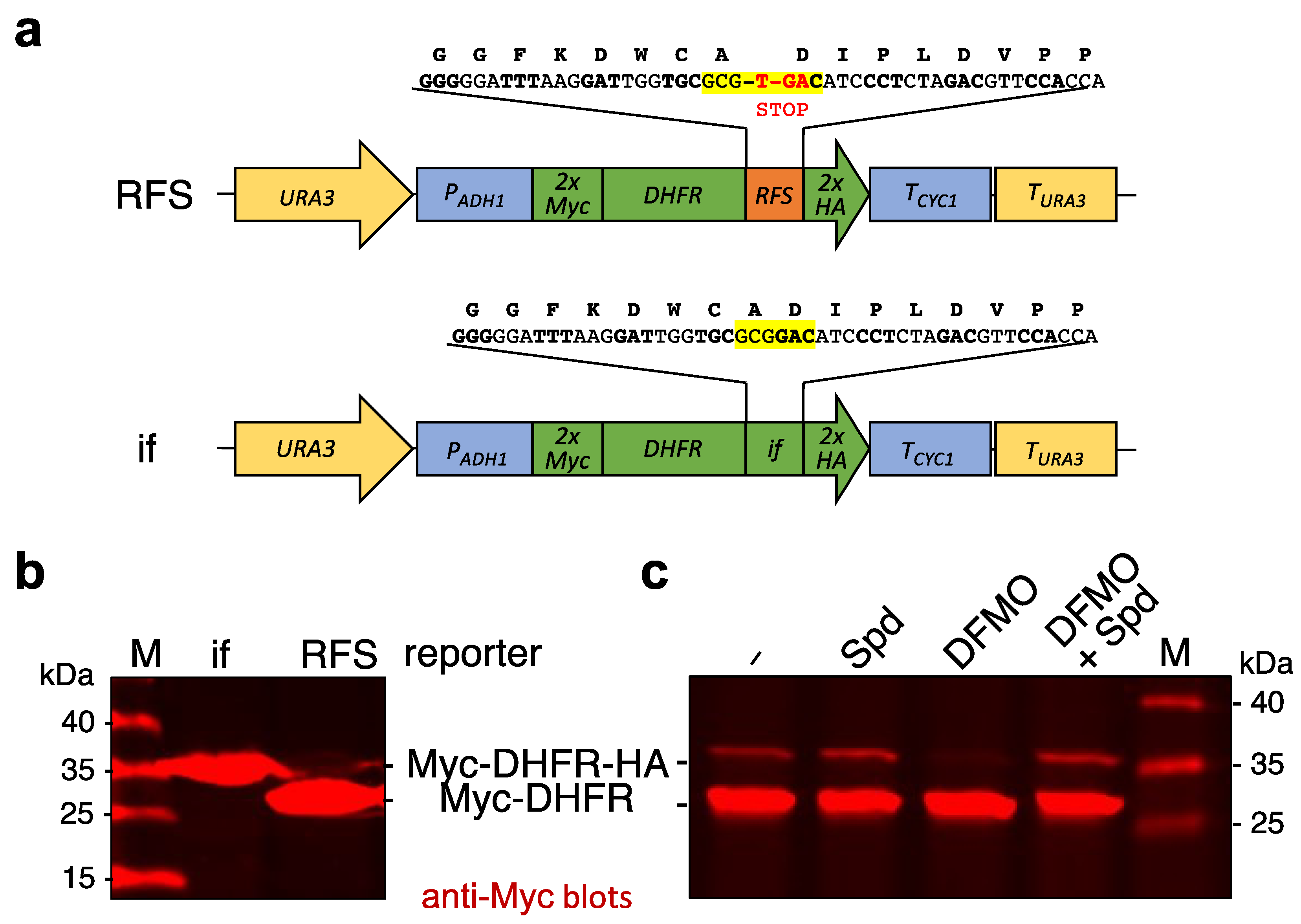
2.1. Reporter Constructs with the OAZ1 RFS Site
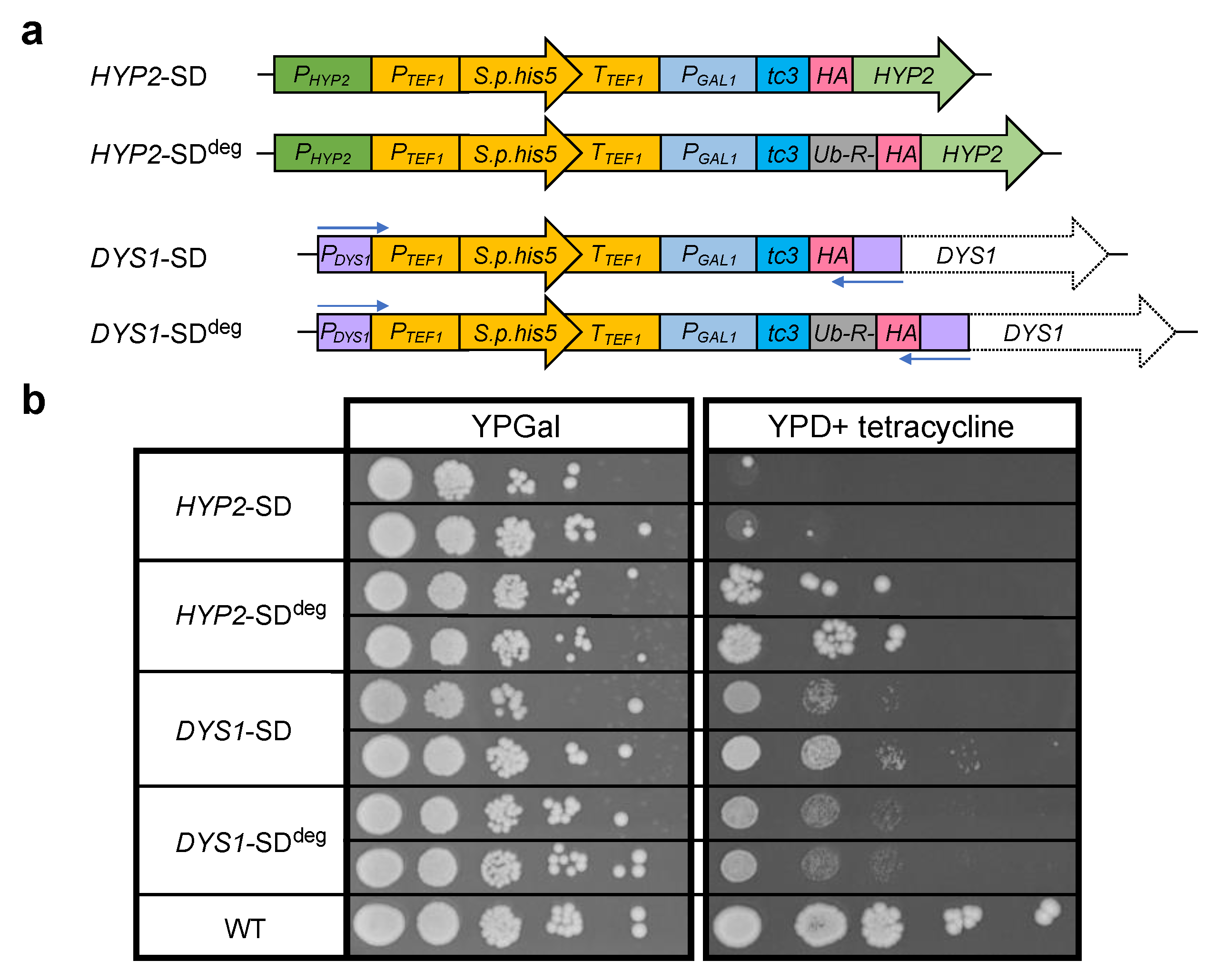
2.2. Generation of Strains with Conditional HYP2 or DYS1 Alleles
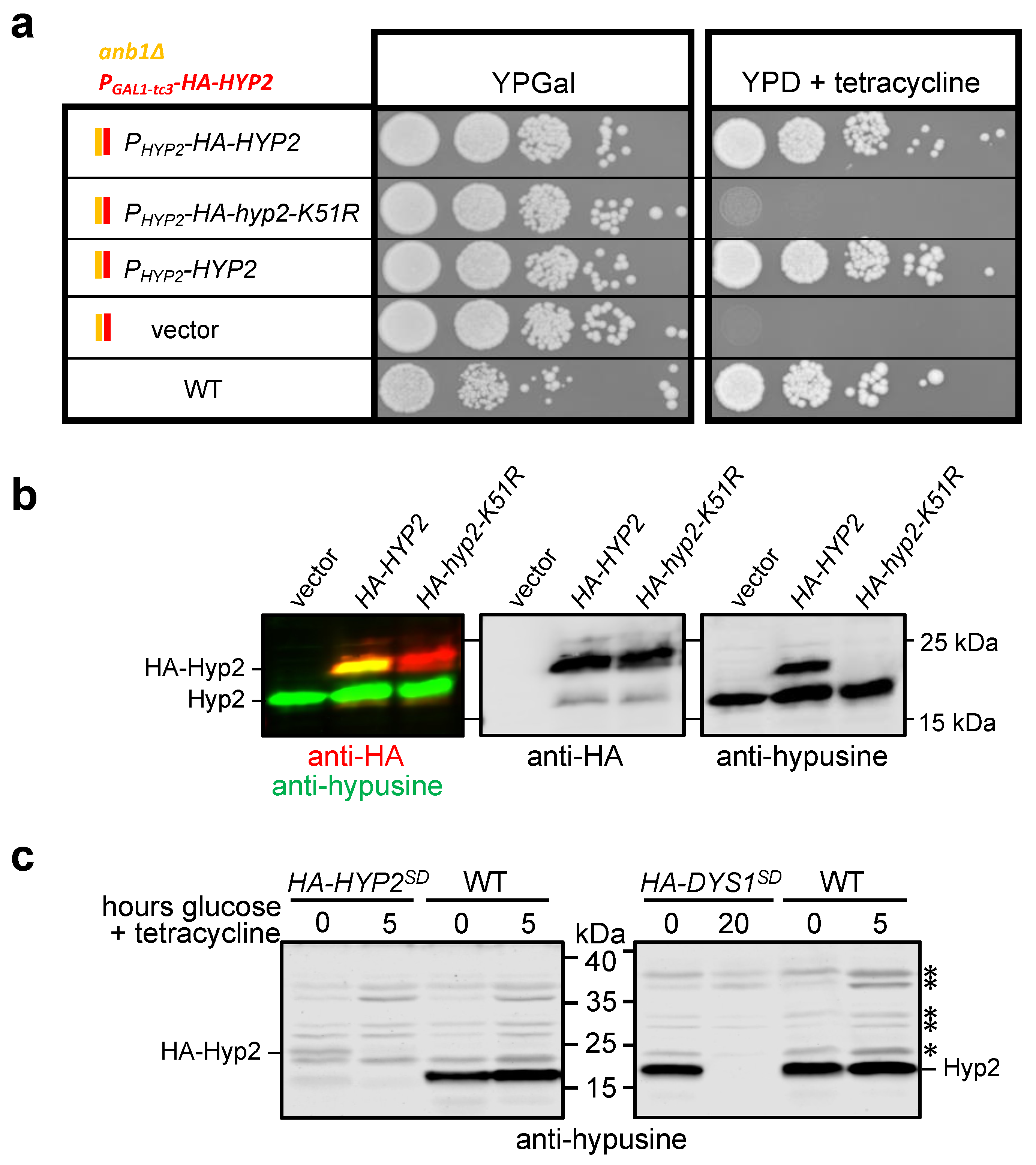
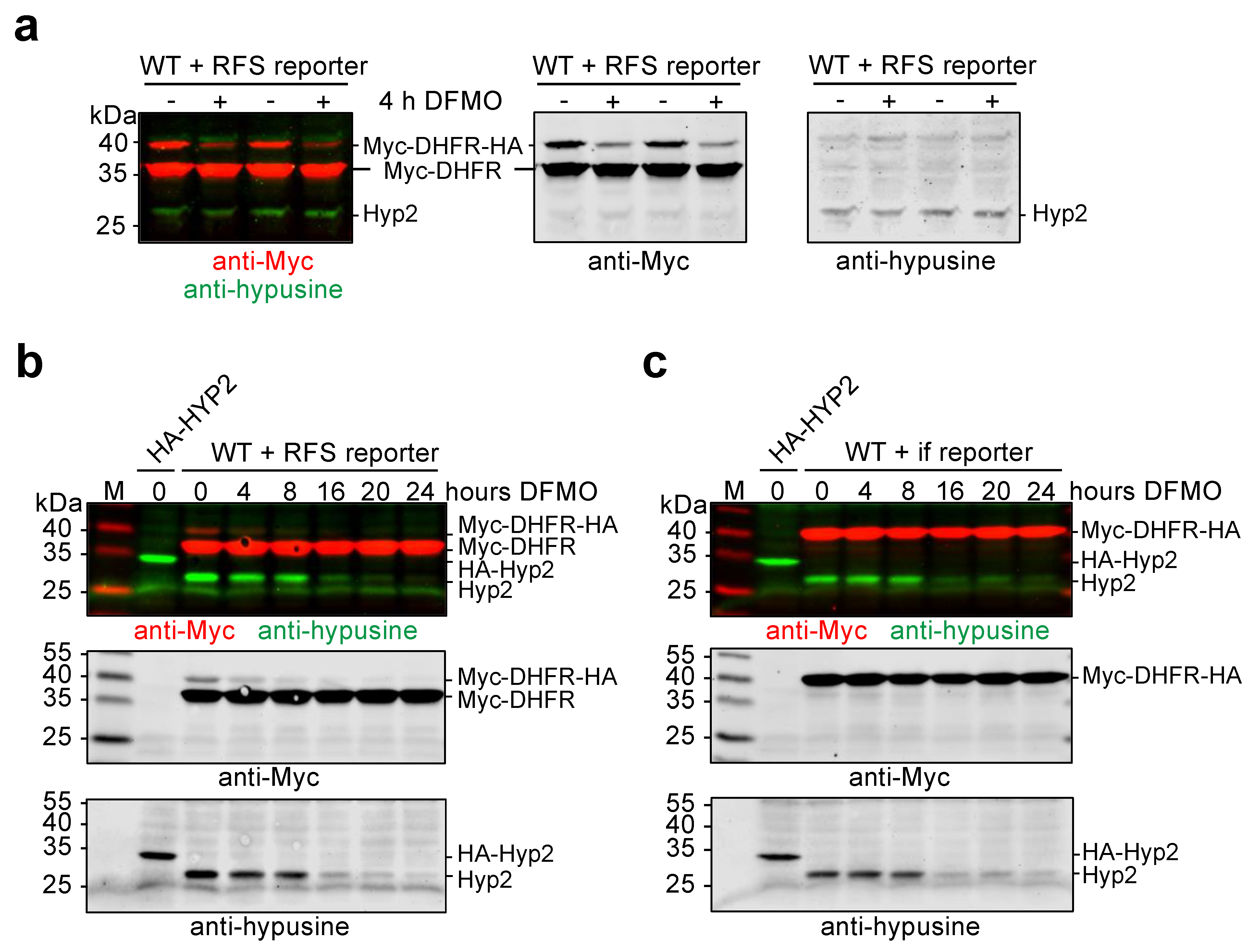
2.3. Control and Detection of Hypusinated Hyp2
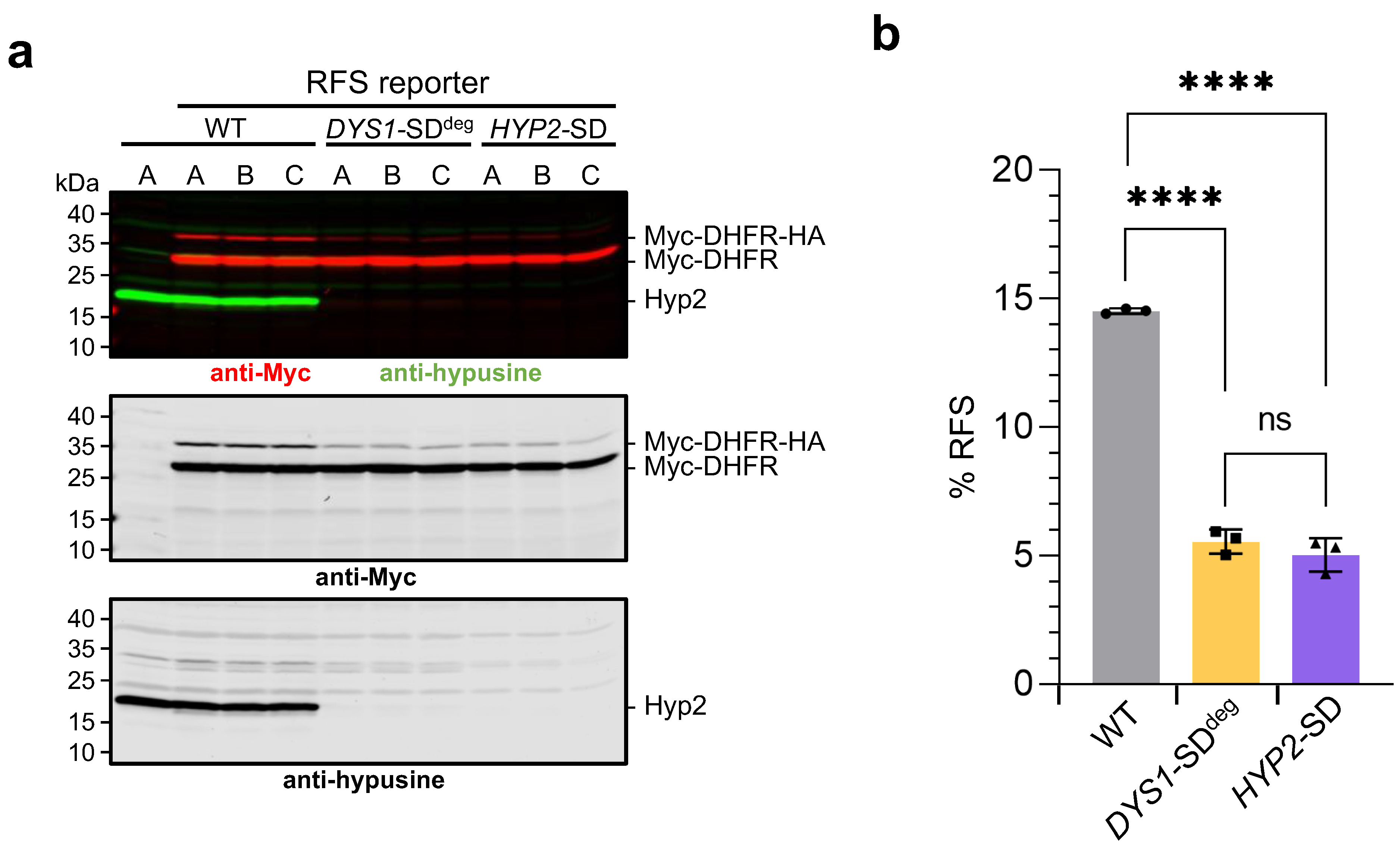
2.4. Hypusinated Hyp2 Promotes Ribosomal Frameshifting during Decoding of the OAZ1 RFS Site
2.5. Polyamines Affect RFS Not only by Promoting Hyp2 Hypusination
3. Discussion
3.1. Reporter Constructs to Study Effects of Polyamine Depletion on Decoding of OAZ1 RFS Site
3.2. Design of HYP2 and DYS1 Shutdown Alleles
3.3. Hypusinated Hyp2 Is Required for Efficient Translation across the OAZ1 RFS Site
3.4. Polyamines Are Required to Promote Translation across the OAZ1 RFS Site
3.5. Polyamines Act at Multiple Levels in a Feedback Control of Their Own Synthesis
4. Materials and Methods
4.1. Yeast Methods
4.2. Generation of Ribosomal Frameshift (RFS) Reporters
4.3. Generation of HYP2 Shutdown (SD) Strains
4.4. Generation of DYS1 Shutdown (SD) Strains
4.5. Analysis of Ribosomal Frameshift Efficiency in HYP2 and DYS1 Shutdown (SD) Strains
4.6. Effect of DFMO-Induced Polyamine Depletion on Ribosomal Frameshift Efficiency
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tabor, C.W.; Tabor, H. Polyamines. Annu. Rev. Biochem. 1984, 53, 749–790. [Google Scholar] [CrossRef]
- Michael, A.J. Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 2016, 473, 2315–2329. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, B.; Hittelman, A.; Daneshvar, L.; Basu, H.S.; Marton, L.J.; Feuerstein, B.G. A new model for disruption of the ornithine decarboxylase gene, SPE1, in Saccharomyces cerevisiae exhibits growth arrest and genetic instability at the MAT locus. Biochem. J. 1995, 312, 83–90. [Google Scholar] [CrossRef][Green Version]
- Wallace, H.M.; Fraser, A.V.; Hughes, A. A perspective of polyamine metabolism. Biochem. J. 2003, 376, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Minois, N.; Carmona-Gutierrez, D.; Madeo, F. Polyamines in aging and disease. Aging (Albany NY) 2011, 3, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Palanimurugan, R.; Kurian, L.; Hegde, V.; Hofmann, K.; Dohmen, R.J. Co-translational Polyamine Sensing by Nascent ODC Antizyme. In Regulatory Nascent Polypeptides; Ito, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 203–222. [Google Scholar]
- Igarashi, K.; Kashiwagi, K. The functional role of polyamines in eukaryotic cells. Int. J. Biochem. Cell Biol. 2019, 107, 104–115. [Google Scholar] [CrossRef]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Buttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef]
- Zhang, H.; Alsaleh, G.; Feltham, J.; Sun, Y.; Napolitano, G.; Riffelmacher, T.; Charles, P.; Frau, L.; Hublitz, P.; Yu, Z.; et al. Polyamines Control eIF5A Hypusination, TFEB Translation, and Autophagy to Reverse B Cell Senescence. Mol. Cell 2019, 76, 110–125.e119. [Google Scholar] [CrossRef] [PubMed]
- Nowotarski, S.L.; Woster, P.M.; Casero, R.A., Jr. Polyamines and cancer: Implications for chemotherapy and chemoprevention. Expert Rev. Mol. Med. 2013, 15, e3. [Google Scholar] [CrossRef]
- Pegg, A.E. S-Adenosylmethionine decarboxylase. Essays Biochem. 2009, 46, 25–45. [Google Scholar] [CrossRef] [PubMed]
- Soda, K. Polyamine Metabolism and Gene Methylation in Conjunction with One-Carbon Metabolism. Int. J. Mol. Sci. 2018, 19, 3106. [Google Scholar] [CrossRef] [PubMed]
- Kern, A.D.; Oliveira, M.A.; Coffino, P.; Hackert, M.L. Structure of mammalian ornithine decarboxylase at 1.6 A resolution: Stereochemical implications of PLP-dependent amino acid decarboxylases. Struct. Fold Des. 1999, 7, 567–581. [Google Scholar] [CrossRef]
- Palanimurugan, R.; Scheel, H.; Hofmann, K.; Dohmen, R.J. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. Embo. J. 2004, 23, 4857–4867. [Google Scholar] [CrossRef] [PubMed]
- Kahana, C. The antizyme family for regulating polyamines. J. Biol. Chem. 2018, 293, 18730–18735. [Google Scholar] [CrossRef]
- Coffino, P. Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2001, 2, 188–194. [Google Scholar] [CrossRef]
- Gödderz, D.; Schäfer, E.; Palanimurugan, R.; Dohmen, R.J. The N-terminal unstructured domain of yeast ODC functions as a transplantable and replaceable ubiquitin-independent degron. J. Mol. Biol. 2011, 407, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Matsufuji, S.; Kameji, T.; Hayashi, S.; Igarashi, K.; Tamura, T.; Tanaka, K.; Ichihara, A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 1992, 360, 597–599. [Google Scholar] [CrossRef]
- Kurian, L.; Palanimurugan, R.; Gödderz, D.; Dohmen, R.J. Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA. Nature 2011, 477, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Matsufuji, S.; Matsufuji, T.; Miyazaki, Y.; Murakami, Y.; Atkins, J.F.; Gesteland, R.F.; Hayashi, S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 1995, 80, 51–60. [Google Scholar] [CrossRef]
- Ivanov, I.P.; Atkins, J.F. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: Close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007, 35, 1842–1858. [Google Scholar] [CrossRef]
- Petros, L.M.; Howard, M.T.; Gesteland, R.F.; Atkins, J.F. Polyamine sensing during antizyme mRNA programmed frameshifting. Biochem. Biophys. Res. Commun. 2005, 338, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.T.; Shirts, B.H.; Zhou, J.; Carlson, C.L.; Matsufuji, S.; Gesteland, R.F.; Weeks, R.S.; Atkins, J.F. Cell culture analysis of the regulatory frameshift event required for the expression of mammalian antizymes. Genes Cells 2001, 6, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Gutierrez, E.; Shin, B.S. The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 413–425. [Google Scholar] [CrossRef]
- Saini, P.; Eyler, D.E.; Green, R.; Dever, T.E. Hypusine-containing protein eIF5A promotes translation elongation. Nature 2009, 459, 118–121. [Google Scholar] [CrossRef]
- Schuller, A.P.; Wu, C.C.; Dever, T.E.; Buskirk, A.R.; Green, R. eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 2017, 66, 194–205.e195. [Google Scholar] [CrossRef]
- Pelechano, V.; Alepuz, P. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 2017, 45, 7326–7338. [Google Scholar] [CrossRef]
- Manjunath, H.; Zhang, H.; Rehfeld, F.; Han, J.; Chang, T.C.; Mendell, J.T. Suppression of Ribosomal Pausing by eIF5A Is Necessary to Maintain the Fidelity of Start Codon Selection. Cell Rep. 2019, 29, 3134–3146.e3136. [Google Scholar] [CrossRef]
- Schwelberger, H.G.; Kang, H.A.; Hershey, J.W. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J. Biol. Chem. 1993, 268, 14018–14025. [Google Scholar] [CrossRef]
- Schnier, J.; Schwelberger, H.G.; Smit-McBride, Z.; Kang, H.A.; Hershey, J.W. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 3105–3114. [Google Scholar] [CrossRef]
- Gutierrez, E.; Shin, B.S.; Woolstenhulme, C.J.; Kim, J.R.; Saini, P.; Buskirk, A.R.; Dever, T.E. eIF5A Promotes Translation of Polyproline Motifs. Mol. Cell 2013, 51, 35–45. [Google Scholar] [CrossRef]
- Schrader, R.; Young, C.; Kozian, D.; Hoffmann, R.; Lottspeich, F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J. Biol. Chem. 2006, 281, 35336–35346. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Wolff, E.C.; Folk, J.E. Hypusine: Its post-translational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Biofactors 1993, 4, 95–104. [Google Scholar]
- Chattopadhyay, M.K.; Park, M.H.; Tabor, H. Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc. Natl. Acad. Sci. USA 2008, 105, 6554–6559. [Google Scholar] [PubMed]
- Park, M.H.; Wolff, E.C. Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem. 2018, 293, 18710–18718. [Google Scholar] [CrossRef] [PubMed]
- Galvao, F.C.; Rossi, D.; Silveira Wda, S.; Valentini, S.R.; Zanelli, C.F. The deoxyhypusine synthase mutant dys1-1 reveals the association of eIF5A and Asc1 with cell wall integrity. PLoS ONE 2013, 8, e60140. [Google Scholar] [CrossRef]
- Park, M.H. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J. Biochem. 2006, 139, 161–169. [Google Scholar] [CrossRef]
- Thompson, G.M.; Cano, V.S.; Valentini, S.R. Mapping eIF5A binding sites for Dys1 and Lia1: In vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 2003, 555, 464–468. [Google Scholar] [CrossRef]
- Jao, D.L.; Chen, K.Y. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J. Cell. Biochem. 2006, 97, 583–598. [Google Scholar] [CrossRef]
- Sasaki, K.; Abid, M.R.; Miyazaki, M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996, 384, 151–154. [Google Scholar] [CrossRef]
- Almeida, O.P., Jr.; Toledo, T.R.; Rossi, D.; Rossetto, D.D.; Watanabe, T.F.; Galvao, F.C.; Medeiros, A.I.; Zanelli, C.F.; Valentini, S.R. Hypusine Modification Of The Ribosome-Binding Protein Eif5a, A Target For New Anti-Inflammatory Drugs: Understanding The Action Of The Inhibitor GC7 on a Murine Macrophage Cell Line. Curr. Pharm. Des. 2013, 20, 284–292. [Google Scholar]
- Lee, Y.B.; Park, M.H.; Folk, J.E. Diamine and triamine analogs and derivatives as inhibitors of deoxyhypusine synthase: Synthesis and biological activity. J. Med. Chem. 1995, 38, 3053–3061. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Becker, T.; Heuer, A.; Braunger, K.; Shanmuganathan, V.; Pech, M.; Berninghausen, O.; Wilson, D.N.; Beckmann, R. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 2016, 44, 1944–1951. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, S.; Cleveland, J.L. Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 2016, 48, 2353–2362. [Google Scholar] [CrossRef] [PubMed]
- Mathews, M.B.; Hershey, J.W. The translation factor eIF5A and human cancer. Biochim. Biophys. Acta 2015, 1849, 836–844. [Google Scholar] [CrossRef]
- Scuoppo, C.; Miething, C.; Lindqvist, L.; Reyes, J.; Ruse, C.; Appelmann, I.; Yoon, S.; Krasnitz, A.; Teruya-Feldstein, J.; Pappin, D.; et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature 2012, 487, 244–248. [Google Scholar] [CrossRef]
- Liang, Y.; Piao, C.; Beuschel, C.B.; Toppe, D.; Kollipara, L.; Bogdanow, B.; Maglione, M.; Lutzkendorf, J.; See, J.C.K.; Huang, S.; et al. eIF5A hypusination, boosted by dietary spermidine, protects from premature brain aging and mitochondrial dysfunction. Cell Rep. 2021, 35, 108941. [Google Scholar] [CrossRef]
- Madeo, F.; Eisenberg, T.; Buttner, S.; Ruckenstuhl, C.; Kroemer, G. Spermidine: A novel autophagy inducer and longevity elixir. Autophagy 2010, 6, 160–162. [Google Scholar] [CrossRef]
- Kim, H.I.; Schultz, C.R.; Chandramouli, G.V.R.; Geerts, D.; Risinger, J.I.; Bachmann, A.S. Pharmacological targeting of polyamine and hypusine biosynthesis reduces tumour activity of endometrial cancer. J. Drug Target. 2022, 30, 623–633. [Google Scholar] [CrossRef]
- Holbert, C.E.; Cullen, M.T.; Casero, R.A., Jr.; Stewart, T.M. Polyamines in cancer: Integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 2022, 22, 467–480. [Google Scholar] [CrossRef]
- Dohmen, R.J.; Wu, P.; Varshavsky, A. Heat-inducible degron: A method for constructing temperature-sensitive mutants. Science 1994, 263, 1273–1276. [Google Scholar] [CrossRef]
- Faden, F.; Ramezani, T.; Mielke, S.; Almudi, I.; Nairz, K.; Froehlich, M.S.; Höckendorff, J.; Brandt, W.; Hoehenwarter, W.; Dohmen, R.J.; et al. Phenotypes on demand via switchable target protein degradation in multicellular organisms. Nat. Commun. 2016, 7, 12202. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.J.R. Studying Essential Genes: Generating and Using Promoter Fusions and Conditional Alleles, 2nd ed.; Stansfield, I., Stark, M.J.R., Eds.; Academic Press: Cambridge, MA, USA, 2007; Volume 36, p. 24. [Google Scholar]
- Kötter, P.; Weigand, J.E.; Meyer, B.; Entian, K.D.; Suess, B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009, 37, e120. [Google Scholar] [CrossRef] [PubMed]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Ramos, P.C.; Höckendorff, J.; Johnson, E.S.; Varshavsky, A.; Dohmen, R.J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 1998, 92, 489–499. [Google Scholar] [CrossRef]
- Puleston, D.J.; Buck, M.D.; Klein Geltink, R.I.; Kyle, R.L.; Caputa, G.; O’Sullivan, D.; Cameron, A.M.; Castoldi, A.; Musa, Y.; Kabat, A.M.; et al. Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 2019, 30, 352–363.e358. [Google Scholar] [CrossRef]
- Dever, T.E.; Ivanov, I.P. Roles of polyamines in translation. J. Biol. Chem. 2018, 293, 18719–18729. [Google Scholar] [CrossRef]
- Beenukumar, R.R.; Gödderz, D.; Palanimurugan, R.; Dohmen, R.J. Polyamines directly promote antizyme-mediated degradation of ornithine-decarboxylase by the proteasome. Microb. Cell 2015, 2, 197–207. [Google Scholar] [CrossRef]
- Vindu, A.; Shin, B.S.; Choi, K.; Christenson, E.T.; Ivanov, I.P.; Cao, C.; Banerjee, A.; Dever, T.E. Translational autoregulation of the S. cerevisiae high-affinity polyamine transporter Hol1. Mol. Cell 2021, 81, 3904–3918.e6. [Google Scholar] [CrossRef]
- Schultz, C.R.; Geerts, D.; Mooney, M.; El-Khawaja, R.; Koster, J.; Bachmann, A.S. Synergistic drug combination GC7/DFMO suppresses hypusine/spermidine-dependent eIF5A activation and induces apoptotic cell death in neuroblastoma. Biochem. J. 2018, 475, 531–545. [Google Scholar] [CrossRef]
- Heinemeyer, W.; Gruhler, A.; Mohrle, V.; Mahe, Y.; Wolf, D.H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 1993, 268, 5115–5120. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.; Messing, J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 1991, 100, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef]
- Pabst, S.; Döring, L.M.; Petreska, N.; Dohmen, R.J. Methods to study SUMO dynamics in yeast. Methods Enzymol. 2019, 618, 187–210. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halwas, K.; Döring, L.-M.; Oehlert, F.V.; Dohmen, R.J. Hypusinated eIF5A Promotes Ribosomal Frameshifting during Decoding of ODC Antizyme mRNA in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2022, 23, 12972. https://doi.org/10.3390/ijms232112972
Halwas K, Döring L-M, Oehlert FV, Dohmen RJ. Hypusinated eIF5A Promotes Ribosomal Frameshifting during Decoding of ODC Antizyme mRNA in Saccharomyces cerevisiae. International Journal of Molecular Sciences. 2022; 23(21):12972. https://doi.org/10.3390/ijms232112972
Chicago/Turabian StyleHalwas, Kai, Lennard-Maximilian Döring, Franziska Valentina Oehlert, and R. Jürgen Dohmen. 2022. "Hypusinated eIF5A Promotes Ribosomal Frameshifting during Decoding of ODC Antizyme mRNA in Saccharomyces cerevisiae" International Journal of Molecular Sciences 23, no. 21: 12972. https://doi.org/10.3390/ijms232112972
APA StyleHalwas, K., Döring, L.-M., Oehlert, F. V., & Dohmen, R. J. (2022). Hypusinated eIF5A Promotes Ribosomal Frameshifting during Decoding of ODC Antizyme mRNA in Saccharomyces cerevisiae. International Journal of Molecular Sciences, 23(21), 12972. https://doi.org/10.3390/ijms232112972





