Identification of Key Genes during Ethylene-Induced Adventitious Root Development in Cucumber (Cucumis sativus L.)
Abstract
:1. Introduction
2. Results
2.1. DEGs, GO, and KEGG Analysis
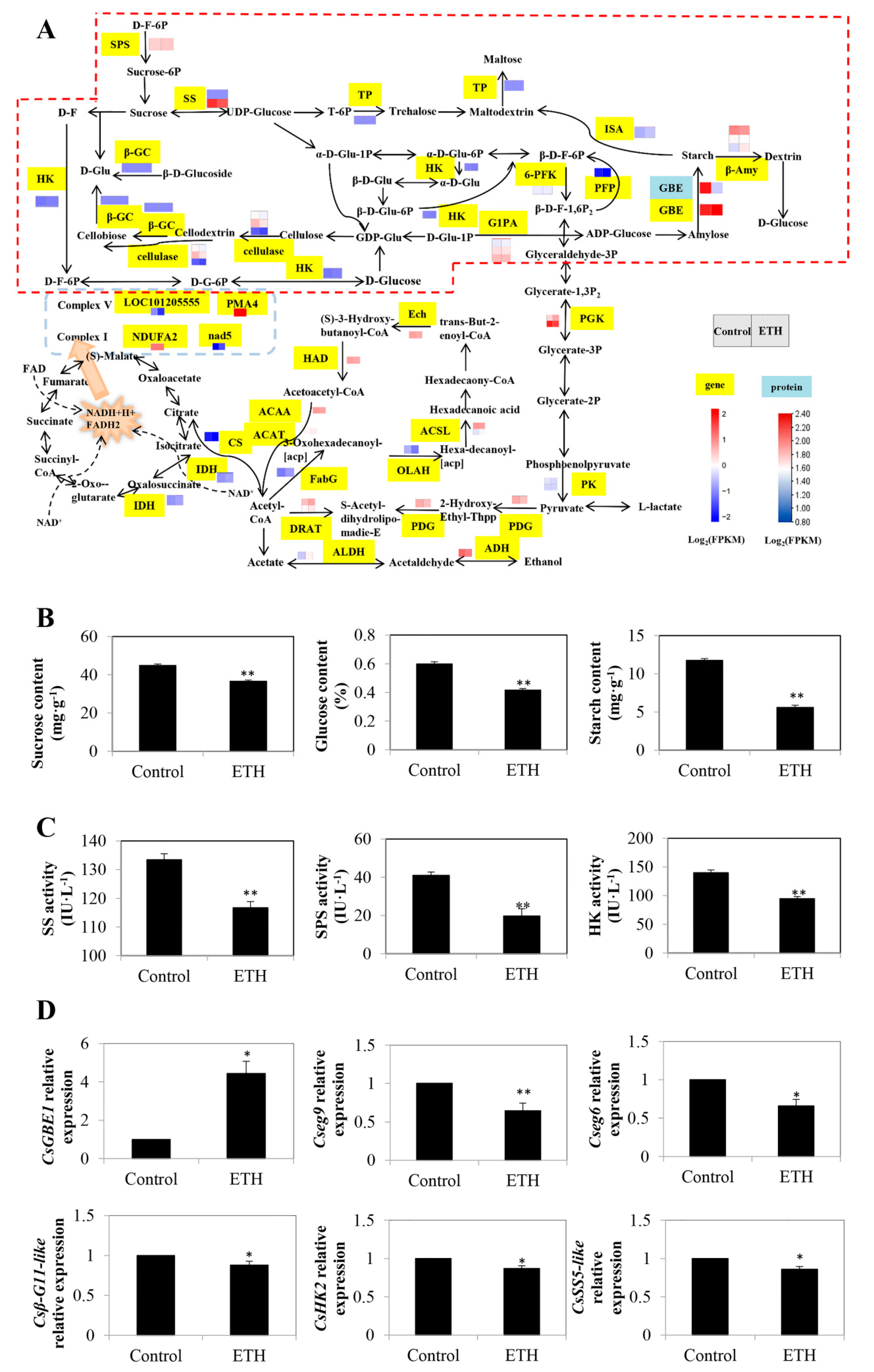
2.2. Carbon Metabolism
2.2.1. Starch and Sucrose Metabolism
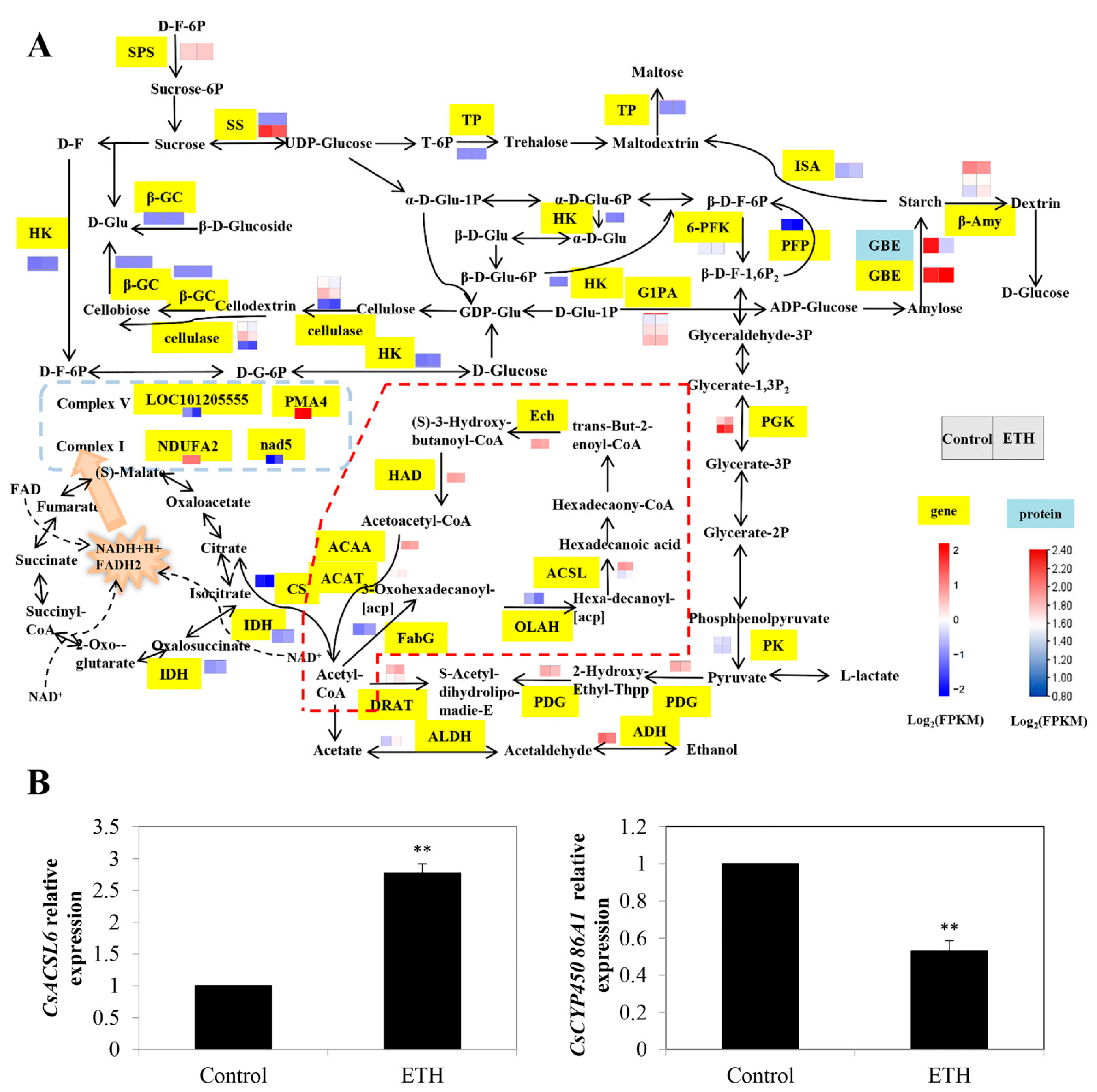
2.2.2. Glycolysis/Gluconeogenesis, Citrate Cycle (TCA Cycle) and Oxidative Phosphorylation
2.2.3. Fatty Acid Biosynthesis and Fatty Acid Degradation
2.3. Secondary Metabolism
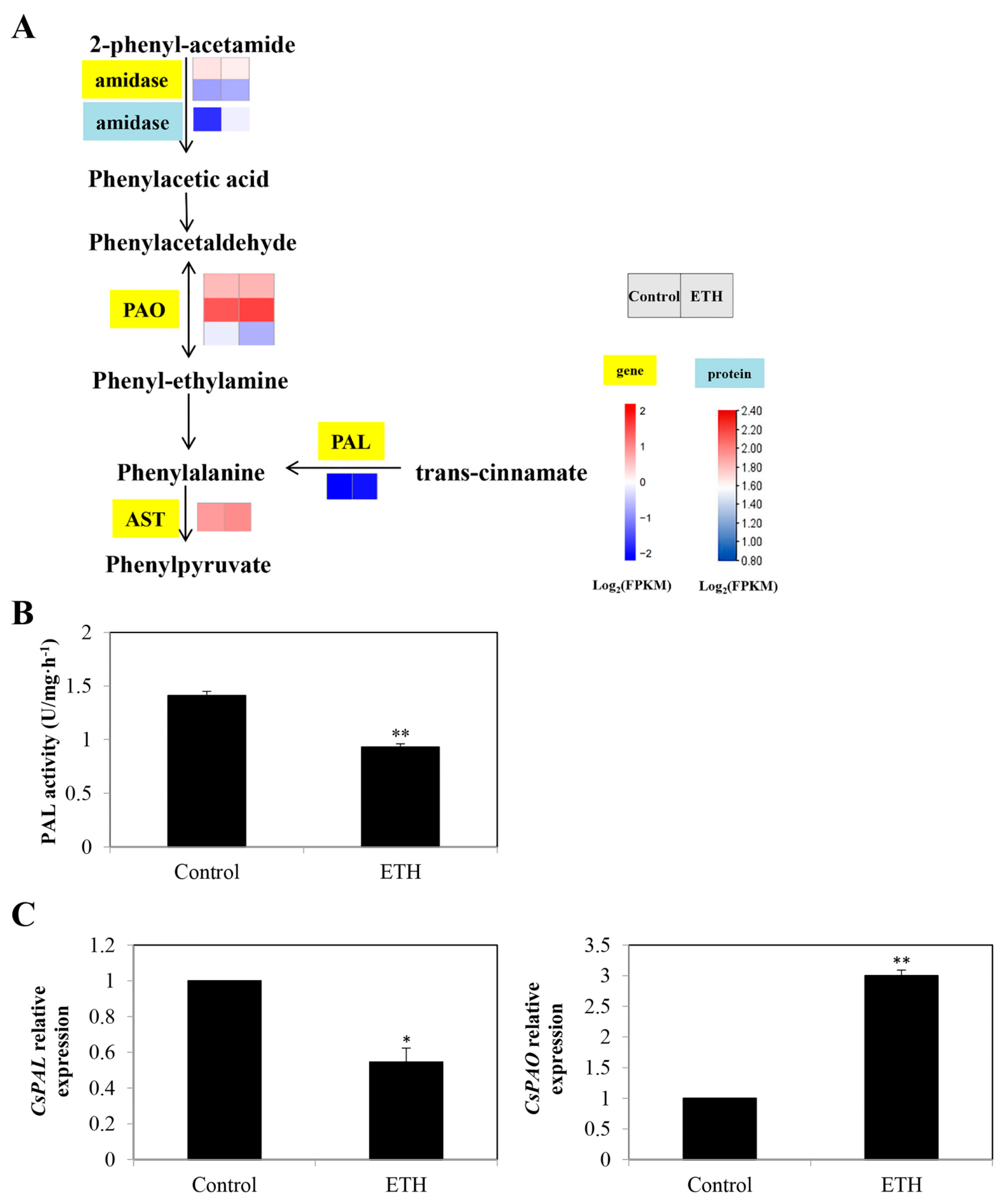
2.3.1. Phenylalanine Metabolism
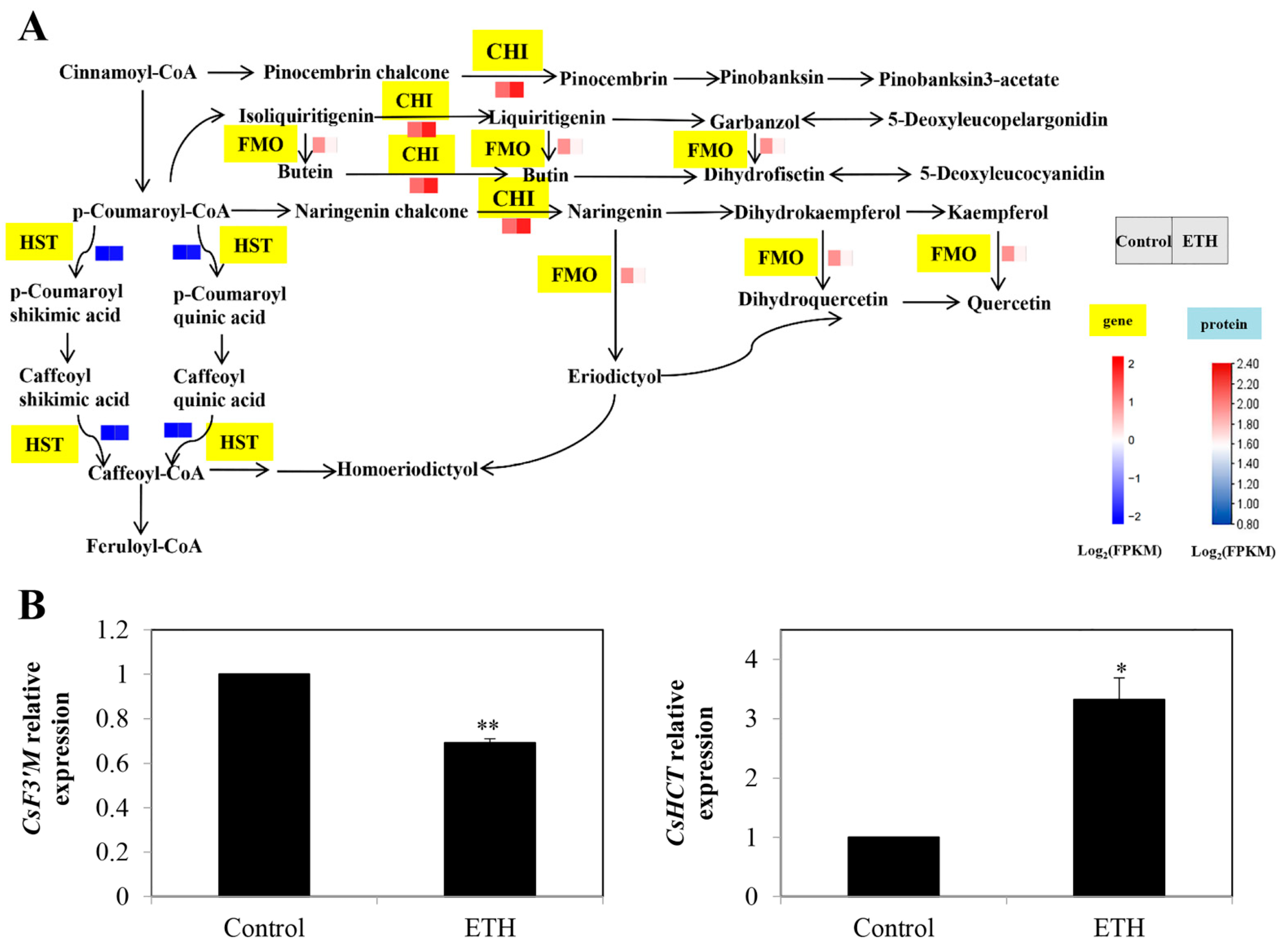
2.3.2. Flavonoid Biosynthesis
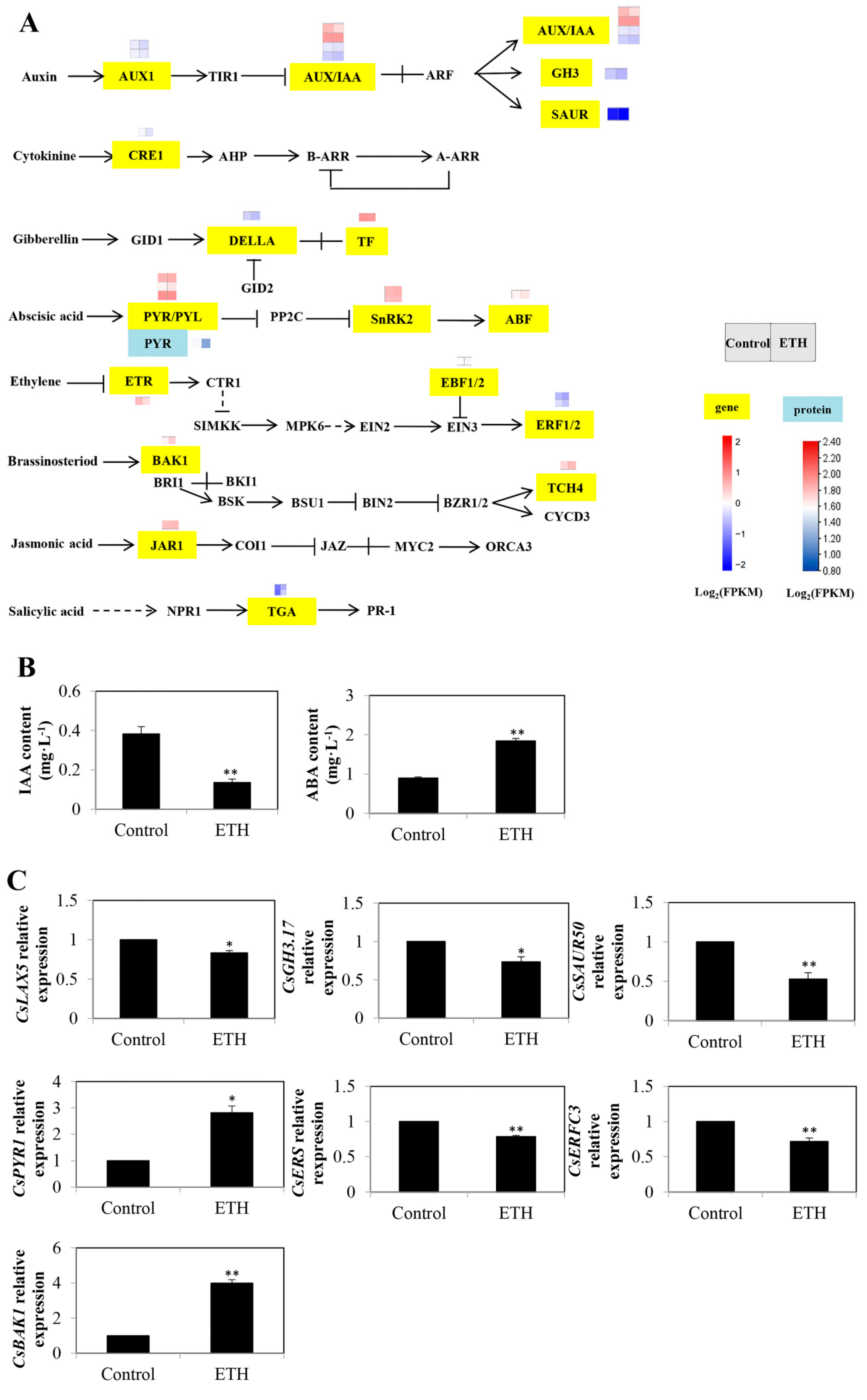
2.4. Plant Hormone Signal Transduction
3. Discussion
3.1. Carbon Metabolism in ETH-Induced Adventitious Root Development
3.2. Secondary Metabolite Biosynthesis Pathway in ETH-Induced Adventitious Root Development
3.3. Plant Hormone Signal Transduction in ETH-Induced Adventitious Root Development
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Chemicals and Treatments
4.3. Gene Expression Profiling Using RNA-Seq
4.3.1. RNA Extraction
4.3.2. cDNA Synthesis and Illumina Sequencing
4.3.3. Bioinformatics Analysis
4.4. Determination of Sucrose, Glucose, and Starch Content
4.5. Enzyme Assays
4.5.1. Determination of Sucrose Synthase (SS), Sucrose–Phosphate Synthase (SPS), and Hexokinase (HK) Activities
4.5.2. Determination of Phenylalanine Ammonia-Lyase (PAL) Activity
4.6. Determination of IAA and ABA Contents
4.7. RT-qPCR Assays
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, X.-T.; Jin, X.; Liao, W.-B.; Dawuda, M.M.; Li, X.-P.; Wang, M.; Niu, L.-J.; Ren, P.-J.; Zhu, Y.-C. Nitric oxide is involved in ethylene-induced adventitious root development in cucumber (Cucumis sativus L.) explants. Sci. Hortic. 2017, 215, 65–71. [Google Scholar] [CrossRef]
- Zhu, Y.; Liao, W. The metabolic constituent and rooting-related enzymes responses of marigold explants to hydrogen gas during adventitious root development. Theor. Exp. Plant Physiol. 2017, 29, 77–85. [Google Scholar] [CrossRef]
- Li, X.-P.; Xu, Q.-Q.; Liao, W.-B.; Ma, Z.-J.; Xu, X.-T.; Wang, M.; Ren, P.-J.; Niu, L.-J.; Jin, X.; Zhu, Y.-C. Hydrogen peroxide is involved in abscisic acid-induced adventitious rooting in cucumber (Cucumis sativus L.) under drought stress. J. Plant Biol. 2016, 59, 536–548. [Google Scholar] [CrossRef]
- Geiss, G.; Gutierrez, L.; Bellini, C. Adventitious root formation: New insights and perspectives. Annu. Plant Rev. 2009, 37, 127–156. [Google Scholar]
- KeÇpczyński, J.; KeÇpczyńska, E. Ethylene in seed dormancy and germination. Physiol. Plant. 1997, 101, 720–726. [Google Scholar] [CrossRef]
- Hu, Y.; Vandenbussche, F.; Van Der Straeten, D. Regulation of seedling growth by ethylene and the ethylene–auxin crosstalk. Planta 2017, 245, 467–489. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; He, L.; Huang, R. The Coordination of Ethylene and Other Hormones in Primary Root Development. Front. Plant Sci. 2019, 10, 874. [Google Scholar] [CrossRef]
- Qi, X.; Li, Q.; Ma, X.; Qian, C.; Wang, H.; Ren, N.; Shen, C.; Huang, S.; Xu, X.; Xu, Q.; et al. Waterlogging-induced adventitious root formation in cucumber is regulated by ethylene and auxin through reactive oxygen species signalling. Plant Cell Environ. 2019, 42, 1458–1470. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, C.; Zou, B.; Wang, C.; Xu, W.; Cui, C.; Qu, S. Morphological, Transcriptomic and Hormonal Characterization of Trimonoecious and Subandroecious Pumpkin (Cucurbita maxima) Suggests Important Roles of Ethylene in Sex Expression. Int. J. Mol. Sci. 2019, 20, 3185. [Google Scholar] [CrossRef] [Green Version]
- An, J.; Almasaud, R.A.; Bouzayen, M.; Zouine, M.; Chervin, C. Auxin and ethylene regulation of fruit set. Plant Sci. 2020, 292, 110381. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Z.; Sun, Y.; Wang, S.; Wang, T.; Wang, Y.; Xu, K.; Zhang, X.; Xu, X.; Han, Z.; et al. ERF4 affects fruit firmness through TPL4 by reducing ethylene production. Plant J. 2020, 103, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Seidi, F.; Zhang, T.; Jin, Y.; Xiao, H. Ethylene scavengers for the preservation of fruits and vegetables: A review. Food Chem. 2021, 337, 127750. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Keunen, E.; Schellingen, K.; Vangronsveld, J.; Cuypers, A. Ethylene and Metal Stress: Small Molecule, Big Impact. Front. Plant Sci. 2016, 7, 23. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Liao, W.B.; Yu, J.H.; Ren, P.J.; Dawuda, M.; Wang, M.; Niu, L.J.; Li, X.P.; Xu, X.T. Nitric oxide is involved in ethylene-induced adventitious rooting in marigold (Tagetes erecta L.). Can. J. Plant Sci. 2017, 97, 620–631. [Google Scholar]
- Lyu, J.; Wu, Y.; Jin, X.; Tang, Z.; Liao, W.; Dawuda, M.M.; Hu, L.; Xie, J.; Yu, J.; Calderón-Urrea, A. Proteomic analysis reveals key proteins involved in ethylene-induced adventitious root development in cucumber (Cucumis sativus L.). PeerJ 2021, 9, e10887. [Google Scholar] [CrossRef]
- Bai, T.; Dong, Z.; Zheng, X.; Song, S.; Jiao, J.; Wang, M.; Song, C. Auxin and Its Interaction with Ethylene Control Adventitious Root Formation and Development in Apple Rootstock. Front. Plant Sci. 2020, 11, 574881. [Google Scholar] [CrossRef]
- Yu, J.; Niu, L.; Yu, J.; Liao, W.; Xie, J.; Lv, J.; Feng, Z.; Hu, L.; Dawuda, M.M. The Involvement of Ethylene in Calcium-Induced Adventitious Root Formation in Cucumber under Salt Stress. Int. J. Mol. Sci. 2019, 20, 1047. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Xie, J.; Yu, J. Physiological and transcriptomic responses of Lanzhou Lily (Lilium davidii, var. unicolor) to cold stress. PLoS ONE 2020, 15, e0227921. [Google Scholar] [CrossRef]
- Lin, Z.; Wang, Z.; Zhang, X.; Liu, Z.; Li, G.; Wang, S.; Ding, Y. Complementary Proteome and Transcriptome Profiling in Developing Grains of a Notched-Belly Rice Mutant Reveals Key Pathways Involved in Chalkiness Formation. Plant Cell Physiol. 2017, 58, 560–573. [Google Scholar] [CrossRef]
- Domergue, J.; Abadie, C.; Limami, A.; Way, D.; Tcherkez, G. Seed quality and carbon primary metabolism. Plant Cell Environ. 2019, 42, 2776–2788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.H.; Liu, Y.X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, I.; Ricard, B.; Rolin, D.; Maucourt, M.; Andrieu, M.-H.; Bizid, E.; Smiti, S.; Brouquisse, R. Effect of hexokinase activity on tomato root metabolism during prolonged hypoxia. Plant Cell Environ. 2007, 30, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Andriunas, F.A.; Zhang, H.-M.; Weber, H.; McCurdy, D.W.; Offler, C.E.; Patrick, J.W. Glucose and ethylene signalling pathways converge to regulate trans-differentiation of epidermal transfer cells in Vicia narbonensis cotyledons. Plant J. 2011, 68, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Jang, J.-C.; Jones, T.L.; Sheen, J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. USA 1998, 95, 10294–10299. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.-H.; Sheen, J.; Yoo, S.-D. Low Glucose Uncouples Hexokinase1-Dependent Sugar Signaling from Stress and Defense Hormone Abscisic Acid and C2H4 Responses in Arabidopsis. Plant Physiol. 2010, 152, 1180–1182. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.; Zemach, H.; Shabtai, S.; Aloni, R.; Yang, J.; Zhang, P.; Sergeeva, L.; Ligterink, W.; Firon, N. Proximal and Distal Parts of Sweetpotato Adventitious Roots Display Differences in Root Architecture, Lignin, and Starch Metabolism and Their Developmental Fates. Front. Plant Sci. 2021, 11, 609923. [Google Scholar] [CrossRef]
- Seung, D. Amylose in starch: Towards an understanding of biosynthesis, structure and function. New Phytol. 2020, 228, 1490–1504. [Google Scholar] [CrossRef]
- Megguer, C.A.; Fugate, K.K.; Lafta, A.M.; Ferrareze, J.P.; Deckard, E.L.; Campbell, L.G.; Lulai, E.C.; Finger, F.L. Glycolysis Is Dynamic and Relates Closely to Respiration Rate in Stored Sugarbeet Roots. Front. Plant Sci. 2017, 8, 861. [Google Scholar] [CrossRef] [Green Version]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef]
- Lim, G.-H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty acid- and lipid-mediated signaling in plant defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- Höfer, R.; Briesen, I.; Beck, M.; Pinot, F.; Schreiber, L.; Franke, R. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 2008, 59, 2347–2360. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xu, J.; Li, L.; Guo, Z.; Si, Q.; Zhu, G.; Wang, X.; Guo, W. GbCYP86A1-1 from Gossypium barbadense positively regulates defence against Verticillium dahliae by cell wall modification and activation of immune pathways. Plant Biotechnol. J. 2020, 18, 222–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, I.; Hosmani, P.S.; Rus, A.; Lahner, B.; Borevitz, J.O.; Muthukumar, B.; Mickelbart, M.V.; Schreiber, L.; Franke, R.B.; Salt, D.E. Root Suberin Forms an Extracellular Barrier That Affects Water Relations and Mineral Nutrition in Arabidopsis. PLoS Genet. 2009, 5, e1000492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Sun, H.; Wang, H.; Yang, X.; Xu, Y.; Yang, Z.; Xu, C.; Li, P. Integrating transcriptome, co-expression and QTL-seq analysis reveals that primary root growth in maize is regulated via flavonoid biosynthesis and auxin signal transduction. J. Exp. Bot. 2021, 72, 4773–4795. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Thwe, A.A.; Li, X.; Kim, Y.J.; Kim, J.K.; Arasu, M.V.; Al-Dhabi, N.A.; Park, S.U. Triterpene and Flavonoid Biosynthesis and Metabolic Profiling of Hairy Roots, Adventitious Roots, and Seedling Roots of Astragalus membranaceus. J. Agric. Food Chem. 2015, 63, 8862–8869. [Google Scholar] [CrossRef]
- Cheniany, M.; Ganjeali, A. Developmental role of phenylalanine-ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) genes during adventitious rooting of Juglans regia L. microshoots. Acta Biol. Hung. 2016, 67, 379–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.-Z.; Fan, W.-J.; Lin, Y.-J.; Zhang, F.-F.; Gupta, D.K. Differential expression of the PAL gene family in rice seedlings exposed to chromium by microarray analysis. Ecotoxicology 2018, 27, 325–335. [Google Scholar] [CrossRef]
- Mehrabanjoubani, P.; Abdolzadeh, A.; Sadeghipour, H.R.; Aghdasi, M.; Bagherieh-Najjar, M.B.; Barzegargolchini, B. Silicon increases cell wall thickening and lignification in rice (Oryza sativa) root tip under excess Fe nutrition. Plant Physiol. Biochem. 2019, 144, 264–273. [Google Scholar] [CrossRef]
- Ye, Z.; Yu, J.; Yan, W.; Zhang, J.; Yang, D.; Yao, G.; Liu, Z.; Wu, Y.; Hou, X. Integrative iTRAQ-based proteomic and transcriptomic analysis reveals the accumulation patterns of key metabolites associated with oil quality during seed ripening of Camellia oleifera. Hortic. Res. 2021, 8, 157. [Google Scholar] [CrossRef]
- Sharma, A.; Badola, P.K.; Bhatia, C.; Sharma, D.; Trivedi, P.K. Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Nat. Plants 2020, 6, 1262–1274. [Google Scholar] [CrossRef] [PubMed]
- Della Rovere, F.; Fattorini, L.; D’Angeli, S.; Veloccia, A.; Del Duca, S.; Cai, G.; Falasca, G.; Altamura, M.M. Arabidopsis SHR and SCR transcription factors and AUX1 auxin influx carrier control the switch between adventitious rooting and xylogenesis in planta and in in vitro cultured thin cell layers. Ann. Bot. 2015, 115, 617–628. [Google Scholar] [CrossRef]
- Wei, K.; Ruan, L.; Wang, L.; Cheng, H. Auxin-Induced Adventitious Root Formation in Nodal Cuttings of Camellia sinensis. Int. J. Mol. Sci. 2019, 20, 4817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Costa, C.T.; Offringa, R.; Fett-Neto, A.G. The role of auxin transporters and receptors in adventitious rooting of Arabidopsis thaliana pre-etiolated flooded seedlings. Plant Sci. 2020, 290, 110294. [Google Scholar] [CrossRef]
- Harkey, A.F.; Watkins, J.M.; Olex, A.L.; DiNapoli, K.T.; Lewis, D.R.; Fetrow, J.S.; Binder, B.M.; Muday, G.K. Identification of Transcriptional and Receptor Networks That Control Root Responses to Ethylene. Plant Physiol. 2018, 176, 2095–2118. [Google Scholar] [CrossRef] [Green Version]
- Ahkami, A.H.; Melzer, M.; Ghaffari, M.R.; Pollmann, S.; Ghorbani Javid, M.; Shahinnia, F.; Hajirezaei, M.R.; Druege, U. Distribution of indole-3-acetic acid in Petunia hybrida shoot tip cuttings and relationship between auxin transport, carbohydrate metabolism and adventitious root formation. Planta 2013, 238, 499–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffens, B.; Wang, J.; Sauter, M. Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 2005, 223, 604–612. [Google Scholar] [CrossRef] [PubMed]
- García-Andrade, J.; González, B.; Gonzalez-Guzman, M.; Rodriguez, P.L.; Vera, P. The Role of ABA in Plant Immunity is Mediated through the PYR1 Receptor. Int. J. Mol. Sci. 2020, 21, 5852. [Google Scholar] [CrossRef]
- Wang, Z.; Ren, Z.; Cheng, C.; Wang, T.; Ji, H.; Zhao, Y.; Deng, Z.; Zhi, L.; Lu, J.; Wu, X.; et al. Counteraction of ABA-Mediated Inhibition of Seed Germination and Seedling Establishment by ABA Signaling Terminator in Arabidopsis. Mol. Plant 2020, 13, 1284–1297. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.-J.; Gao, J.; Wang, P.; Duan, C.-G.; Zhu, X.; Zhu, J.-K. The ABA Receptor PYL8 Promotes Lateral Root Growth by Enhancing MYB77-Dependent Transcription of Auxin-Responsive Genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, S.; Fujii, N.; Takahashi, H. The Ethylene-Regulated Expression of CS-ETR2 and CS-ERS Genes in Cucumber Plants and Their Possible Involvement with Sex Expression in Flowers. Plant Cell Physiol. 2000, 41, 608–616. [Google Scholar] [CrossRef] [Green Version]
- Hua, J.; Sakai, H.; Nourizadeh, S.; Chen, Q.G.; Bleecker, A.B.; Ecker, J.R.; Meyerowitz, E.M. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 1998, 10, 1321–1332. [Google Scholar] [CrossRef] [PubMed]
- Müssig, C.; Shin, G.-H.; Altmann, T. Brassinosteroids Promote Root Growth in Arabidopsis. Plant Physiol. 2003, 133, 1261–1271. [Google Scholar] [CrossRef] [Green Version]
- Maharjan, P.M.; Dilkes, B.P.; Fujioka, S.; Pěnčík, A.; Ljung, K.; Burow, M.; Halkier, B.A.; Choe, S. Arabidopsis gulliver1/superroot2-7 identifies a metabolic basis for auxin and brassinosteroid synergy. Plant J. 2014, 80, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Chen, F.; Du, H.; Zhang, X.; Wang, X.; Yao, G.; Xu, B. Graphene oxide and indole-3-acetic acid cotreatment regulates the root growth of Brassica napus L. via multiple phytohormone pathways. BMC Plant Biol. 2020, 20, 101. [Google Scholar] [CrossRef] [Green Version]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.-P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Li, C.; Liu, H.; Yang, J.; Huang, P.; Liao, W. The Involvement of Glucose in Hydrogen Gas-Medicated Adventitious Rooting in Cucumber. Plants 2021, 10, 1937. [Google Scholar] [CrossRef]
- Pattemore, J.A. RNA Extraction from Cereal Vegetative Tissue. In Cereal Genomics; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1099, pp. 17–21. [Google Scholar] [CrossRef]
- Wei, Q.; Jiao, C.; Guo, L.; Ding, Y.; Cao, J.; Feng, J.; Dong, X.; Mao, L.; Sun, H.; Yu, F.; et al. Exploring key cellular processes and candidate genes regulating the primary thickening growth of Moso underground shoots. New Phytol. 2017, 214, 81–96. [Google Scholar] [CrossRef]
- Davies, J.; Denyer, T.; Hadfield, J. Bioanalyzer chips can be used interchangeably for many analyses of DNA or RNA. BioTechniques 2016, 60, 197–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, A.J.; Culver, H.R.; Prieto, T.R.; Martinez, P.J.; Sinha, J.; Bryant, S.J.; Bowman, C.N. Messenger RNA enrichment using synthetic oligo (T) click nucleic acids. Chem. Commun. 2020, 56, 13987–13990. [Google Scholar] [CrossRef]
- Degenkolbe, T.; Hannah, M.A.; Freund, S.; Hincha, D.K.; Heyer, A.G.; Köhl, K.I. A quality-controlled microarray method for gene expression profiling. Anal. Biochem. 2005, 346, 217–224. [Google Scholar] [CrossRef]
- Annaluru, N.; Muller, H.; Ramalingam, S.; Kandavelou, K.; London, V.; Richardson, S.M.; Dymond, J.S.; Cooper, E.M.; Bader, J.S.; Boeke, J.D.; et al. Assembling DNA Fragments by USER Fusion. In Gene Synthesis; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 852, pp. 77–95. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Tauzin, A.S.; Giardina, T. Sucrose and invertases, a part of the plant defense response to the biotic stresses. Front. Plant Sci. 2014, 5, 293. [Google Scholar] [CrossRef]
- Smith, A.M.; Zeeman, S. Quantification of starch in plant tissues. Nat. Protoc. 2006, 1, 1342–1345. [Google Scholar] [CrossRef]
- Sun, L.; Li, C.; Zhu, J.; Jiang, C.; Li, Y.; Ge, Y. Influences of postharvest ATP treatment on storage quality and enzyme activity in sucrose metabolism of Malus domestica. Plant Physiol. Biochem. 2020, 156, 87–94. [Google Scholar] [CrossRef] [PubMed]
- González-Mendoza, D.; Troncoso-Rojas, R.; Gonzalez-Soto, T.; Grimaldo-Juarez, O.; Ceceña-Duran, C.; Duran-Hernandez, D.; Gutiérrez-Miceli, F. Changes in the phenylalanine ammonia lyase activity, total phenolic compounds, and flavonoids in Prosopis glandulosa treated with cadmium and copper. An. Acad. Bras. Cienc. 2018, 90, 1465–1472. [Google Scholar] [CrossRef] [Green Version]
- Qi, N.; Hou, X.; Wang, C.; Li, C.; Huang, D.; Li, Y.; Wang, N.; Liao, W. Methane-rich water induces bulblet formation of scale cuttings in Lilium davidii var. unicolor by regulating the signal transduction of phytohormones and their levels. Physiol. Plant. 2021, 172, 1919–1930. [Google Scholar] [PubMed]
- Tsubone, M.; Kubota, F.; Saitou, K.; Kadowaki, M. Enhancement of tuberous root production and adenosine 5’-diphosphate pyrophosphorylase (AGPase) activity in sweet potato (Ipomoea batatas Lam.) by exogenous injection of sucrose solution. J. Agron. Crop Sci. 2000, 184, 181–186. [Google Scholar] [CrossRef]
- Stals, A.; Werbrouck, H.; Baert, L.; Botteldoorn, N.; Herman, L.; Uyttendaele, M.; Van Coillie, E. Laboratory efforts to eliminate contamination problems in the real-time RT-PCR detection of noroviruses. J. Microbiol. Methods 2009, 77, 72–76. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| KEGGID | Pathway | p Value | Total DEGs | Up-Regulated DEGs | Down-Regulated DEGs |
|---|---|---|---|---|---|
| csv00591 | Linoleic acid metabolism | 0.006637889 | 6 | 4 | 2 |
| csv00500 | Starch and sucrose metabolism | 0.007112864 | 17 | 2 | 15 |
| csv00909 | Sesquiterpenoid and triterpenoid biosynthesis | 0.007603753 | 5 | 2 | 3 |
| csv00071 | Fatty acid degradation | 0.017687328 | 8 | 6 | 2 |
| csv00950 | Isoquinoline alkaloid biosynthesis | 0.018821384 | 4 | 4 | 0 |
| csv00360 | Phenylalanine metabolism | 0.021066898 | 7 | 5 | 2 |
| csv04075 | Plant hormone signal transduction | 0.026635092 | 26 | 13 | 13 |
| csv01212 | Fatty acid metabolism | 0.032091296 | 9 | 7 | 2 |
| csv00040 | Pentose and glucuronate interconversions | 0.037498837 | 10 | 2 | 8 |
| csv00073 | Cutin, suberine and wax biosynthesis | 0.039280085 | 5 | 0 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Y.; Wang, C.; Zhang, M.; Wei, L.; Liao, W. Identification of Key Genes during Ethylene-Induced Adventitious Root Development in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2022, 23, 12981. https://doi.org/10.3390/ijms232112981
Deng Y, Wang C, Zhang M, Wei L, Liao W. Identification of Key Genes during Ethylene-Induced Adventitious Root Development in Cucumber (Cucumis sativus L.). International Journal of Molecular Sciences. 2022; 23(21):12981. https://doi.org/10.3390/ijms232112981
Chicago/Turabian StyleDeng, Yuzheng, Chunlei Wang, Meiling Zhang, Lijuan Wei, and Weibiao Liao. 2022. "Identification of Key Genes during Ethylene-Induced Adventitious Root Development in Cucumber (Cucumis sativus L.)" International Journal of Molecular Sciences 23, no. 21: 12981. https://doi.org/10.3390/ijms232112981






