Ferritin Heavy Chain Binds Peroxiredoxin 6 and Inhibits Cell Proliferation and Migration
Abstract
1. Introduction
2. Results
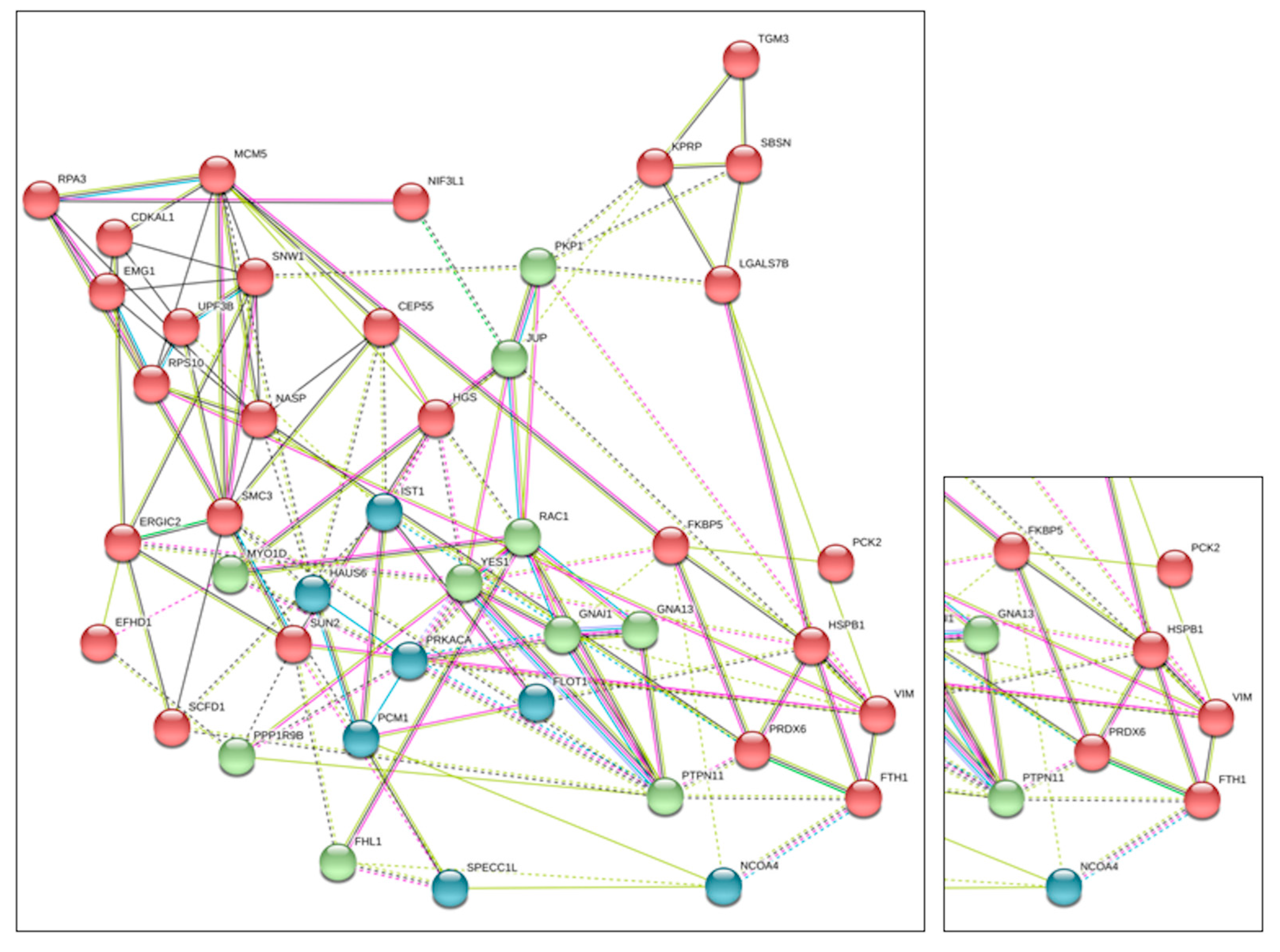
2.1. Identification of FTH1 Interacting Proteins
2.2. FTH1/PRDX6 Interaction in HEK-293T and NCI-H460 Cells
2.3. Molecular Recognition in FTH1/PRDX6 Complex
2.4. FTH1 Inhibits Proliferation and Migration in PRDX6 Overexpressed NCI-H460 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Transient Transfection and Cell Lysis
4.3. Isolation of Protein Complexes by Immunoprecipitation
4.4. Mass Spectrometry Analysis and Protein Identification
4.5. Co-Immunoprecipitation
4.6. Western Blotting Analysis
4.7. Molecular Modeling
4.8. Immunofluorescence Assay
4.9. Proliferation Assays
4.10. Wound Healing Assay
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Theil, E.C. Ferritin: Structure, Gene Regulation, and Cellular Function in Animals, Plants, and Microorganisms. Annu. Rev. Biochem. 1987, 56, 289–315. [Google Scholar] [CrossRef]
- Surguladze, N.; Patton, S.; Cozzi, A.; Fried, M.G.; Connor, J.R. Characterization of Nuclear Ferritin and Mechanism of Translocation. Biochem. J. 2005, 388, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Levi, S.; Corsi, B.; Bosisio, M.; Invernizzi, R.; Volz, A.; Sanford, D.; Arosio, P.; Drysdale, J. A Human Mitochondrial Ferritin Encoded by an Intronless Gene. J. Biol. Chem. 2001, 276, 24437–24440. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Levi, S. Ferritin, Iron Homeostasis, and Oxidative Damage. Free Radic. Biol. Med. 2002, 33, 457–463. [Google Scholar] [CrossRef]
- Theil, E.C. Ferritin: At the Crossroads of Iron and Oxygen Metabolism. J. Nutr. 2003, 133, 1549S–1553S. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, F.; Colombo, M.; Staempfli, S.; Santoro, C.; Marone, M.; Frank, R.; Delius, H.; Cortese, R. Structure of Gene and Pseudogenes of Human Apoferritin H. Nucleic Acids Res. 1986, 14, 721. [Google Scholar] [CrossRef]
- Santoro, C.; Marone, M.; Ferrone, M.; Costanzo, F.; Colombo, M.; Minganti, C.; Cortese, R.; Silengo, L. Cloning of the Gene Coding for Human L Apoferritin. Nucleic Acids Res. 1986, 14, 2863. [Google Scholar] [CrossRef][Green Version]
- Bevilacqua, M.A.; Faniello, M.C.; Russo, T.; Cimino, F.; Costanzo, F. Transcriptional Regulation of the Human H Ferritin-Encoding Gene (FERH) in G418-Treated Cells: Role of the B-Box-Binding Factor. Gene 1994, 141, 287–291. [Google Scholar] [CrossRef]
- Miller, L.L.; Miller, S.C.; Torti, S.v.; Tsuji, Y.; Torti, F.M. Iron-Independent Induction of Ferritin H Chain by Tumor Necrosis Factor. Proc. Natl. Acad. Sci. USA 1991, 88, 4946–4950. [Google Scholar] [CrossRef]
- Costanzo, F.; Bevilacqua, M.A.; Giordano, M.; Cimino, F. Expression of Genes of Ferritin Subunits in Human Hepatoma Cell Lines. Biochem. Biophys. Res. Commun. 1989, 161, 902–909. [Google Scholar] [CrossRef]
- Munro, H.N. Iron Regulation of Ferritin Gene Expression. J. Cell. Biochem. 1990, 44, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Klausner, R.D.; Rouault, T.A.; Harford, J.B. Regulating the Fate of MRNA: The Control of Cellular Iron Metabolism. Cell 1993, 72, 19–28. [Google Scholar] [CrossRef]
- Kakhlon, O.; Cabantchik, Z.I. The Labile Iron Pool: Characterization, Measurement, and Participation in Cellular Processes(1). Free Radic. Biol. Med. 2002, 33, 1037–1046. [Google Scholar] [CrossRef]
- Tian, Y.; Lu, J.; Hao, X.; Li, H.; Zhang, G.; Liu, X.; Li, X.; Zhao, C.; Kuang, W.; Chen, D.; et al. FTH1 Inhibits Ferroptosis Through Ferritinophagy in the 6-OHDA Model of Parkinson’s Disease. Neurotherapeutics 2020, 17, 1796–1812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.H.; Tian, H.Y.; Gao, X.; Lei, W.W.; Hu, Y.; Wang, D.M.; Pan, X.C.; Yu, M.L.; Xu, G.J.; Zhao, F.K.; et al. Ferritin Heavy Chain-Mediated Iron Homeostasis and Subsequent Increased Reactive Oxygen Species Production Are Essential for Epithelial-Mesenchymal Transition. Cancer Res. 2009, 69, 5340–5348. [Google Scholar] [CrossRef] [PubMed]
- Coffman, L.G.; Parsonage, D.; D’Agostino, R.; Torti, F.M.; Torti, S.v. Regulatory Effects of Ferritin on Angiogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Storr, H.L.; Kind, B.; Parfitt, D.A.; Chapple, J.P.; Lorenz, M.; Koehler, K.; Huebner, A.; Clark, A.J.L. Deficiency of Ferritin Heavy-Chain Nuclear Import in Triple a Syndrome Implies Nuclear Oxidative Damage as the Primary Disease Mechanism. Mol. Endocrinol. 2009, 23, 2086–2094. [Google Scholar] [CrossRef]
- Li, R.; Luo, C.; Mines, M.; Zhang, J.; Fan, G.H. Chemokine CXCL12 Induces Binding of Ferritin Heavy Chain to the Chemokine Receptor CXCR4, Alters CXCR4 Signaling, and Induces Phosphorylation and Nuclear Translocation of Ferritin Heavy Chain. J. Biol. Chem. 2006, 281, 37616–37627. [Google Scholar] [CrossRef]
- Lee, J.H.; Jang, H.; Cho, E.J.; Youn, H.D. Ferritin Binds and Activates P53 under Oxidative Stress. Biochem. Biophys. Res. Commun. 2009, 389, 399–404. [Google Scholar] [CrossRef]
- Bellelli, R.; Federico, G.; Matte’, A.; Colecchia, D.; Iolascon, A.; Chiariello, M.; Santoro, M.; de Franceschi, L.; Carlomagno, F. NCOA4 Deficiency Impairs Systemic Iron Homeostasis. Cell Rep. 2016, 14, 411–421. [Google Scholar] [CrossRef]
- Liu, F.; Du, Z.Y.; He, J.L.; Liu, X.Q.; Yu, Q.B.; Wang, Y.X. FTH1 Binds to Daxx and Inhibits Daxx-Mediated Cell Apoptosis. Mol. Biol. Rep. 2012, 39, 873–879. [Google Scholar] [CrossRef] [PubMed]
- di Sanzo, M.; Gaspari, M.; Misaggi, R.; Romeo, F.; Falbo, L.; de Marco, C.; Agosti, V.; Quaresima, B.; Barni, T.; Viglietto, G.; et al. H Ferritin Gene Silencing in a Human Metastatic Melanoma Cell Line: A Proteomic Analysis. J. Proteome Res. 2011, 10, 5444–5453. [Google Scholar] [CrossRef] [PubMed]
- Sottile, R.; Federico, G.; Garofalo, C.; Tallerico, R.; Faniello, M.C.; Quaresima, B.; Cristiani, C.M.; di Sanzo, M.; Cuda, G.; Ventura, V.; et al. Iron and Ferritin Modulate MHC Class I Expression and NK Cell Recognition. Front. Immunol. 2019, 10, 224. [Google Scholar] [CrossRef] [PubMed]
- Biamonte, F.; Zolea, F.; Bisognin, A.; di Sanzo, M.; Saccoman, C.; Scumaci, D.; Aversa, I.; Panebianco, M.; Faniello, M.C.; Bortoluzzi, S.; et al. H-Ferritin-Regulated MicroRNAs Modulate Gene Expression in K562 Cells. PLoS ONE 2015, 10, e0122105. [Google Scholar] [CrossRef]
- di Sanzo, M.; Chirillo, R.; Aversa, I.; Biamonte, F.; Santamaria, G.; Giovannone, E.D.; Faniello, M.C.; Cuda, G.; Costanzo, F. ShRNA Targeting of Ferritin Heavy Chain Activates H19/MiR-675 Axis in K562 Cells. Gene 2018, 657, 92–99. [Google Scholar] [CrossRef]
- Zolea, F.; Biamonte, F.; Candeloro, P.; di Sanzo, M.; Cozzi, A.; di Vito, A.; Quaresima, B.; Lobello, N.; Trecroci, F.; di Fabrizio, E.; et al. H Ferritin Silencing Induces Protein Misfolding in K562 Cells: A Raman Analysis. Free Radic. Biol. Med. 2015, 89, 614–623. [Google Scholar] [CrossRef]
- Aversa, I.; Zolea, F.; Ieranò, C.; Bulotta, S.; Trotta, A.M.; Faniello, M.C.; de Marco, C.; Malanga, D.; Biamonte, F.; Viglietto, G.; et al. Epithelial-to-Mesenchymal Transition in FHC-Silenced Cells: The Role of CXCR4/CXCL12 Axis. J. Exp. Clin. Cancer Res. 2017, 36, 104. [Google Scholar] [CrossRef]
- Lobello, N.; Biamonte, F.; Pisanu, M.E.; Faniello, M.C.; Jakopin, Ž.; Chiarella, E.; Giovannone, E.D.; Mancini, R.; Ciliberto, G.; Cuda, G.; et al. Ferritin Heavy Chain Is a Negative Regulator of Ovarian Cancer Stem Cell Expansion and Epithelial to Mesenchymal Transition. Oncotarget 2016, 7, 62019–62033. [Google Scholar] [CrossRef]
- Fisher, A.B. Peroxiredoxin 6: A Bifunctional Enzyme with Glutathione Peroxidase and Phospholipase A2 Activities. Antioxid. Redox Signal. 2011, 15, 831–844. [Google Scholar] [CrossRef]
- Li, H.; Zhang, D.; Li, B.; Zhen, H.; Chen, W.; Men, Q. PRDX6 Overexpression Promotes Proliferation, Invasion, and Migration of A549 Cells in Vitro and in Vivo. Cancer Manag. Res. 2021, 13, 1245–1255. [Google Scholar] [CrossRef]
- Huang, W.S.; Huang, C.Y.; Hsieh, M.C.; Kuo, Y.H.; Tung, S.Y.; Shen, C.H.; Hsieh, Y.Y.; Teng, C.C.; Lee, K.C.; Lee, K.F.; et al. Expression of PRDX6 Correlates with Migration and Invasiveness of Colorectal Cancer Cells. Cell. Physiol. Biochem. 2018, 51, 2616–2630. [Google Scholar] [CrossRef] [PubMed]
- Misaggi, R.; di Sanzo, M.; Cosentino, C.; Bond, H.M.; Scumaci, D.; Romeo, F.; Stellato, C.; Giurato, G.; Weisz, A.; Quaresima, B.; et al. Identification of H Ferritin-Dependent and Independent Genes in K562 Differentiating Cells by Targeted Gene Silencing and Expression Profiling. Gene 2014, 535, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Aung, W.; Hasegawa, S.; Furukawa, T.; Saga, T. Potential Role of Ferritin Heavy Chain in Oxidative Stress and Apoptosis in Human Mesothelial and Mesothelioma Cells: Implications for Asbestos-Induced Oncogenesis. Carcinogenesis 2007, 28, 2047–2052. [Google Scholar] [CrossRef]
- Chang, X.Z.; Li, D.Q.; Hou, Y.F.; Wu, J.; Lu, J.S.; Di, G.H.; Jin, W.; Ou, Z.L.; Shen, Z.Z.; Shao, Z.M. Identification of the Functional Role of Peroxiredoxin 6 in the Progression of Breast Cancer. Breast. Cancer Res. 2007, 9, R76. [Google Scholar] [CrossRef]
- Lee, S.B.; Ho, J.N.; Yoon, S.H.; Kang, G.Y.; Hwang, S.G.; Um, H.D. Peroxiredoxin 6 Promotes Lung Cancer Cell Invasion by Inducing Urokinase-Type Plasminogen Activator via P38 Kinase, Phosphoinositide 3-Kinase, and Akt. Mol. Cells 2009, 28, 583–588. [Google Scholar] [CrossRef]
- Hu, X.; Lu, E.; Pan, C.; Xu, Y.; Zhu, X. Overexpression and Biological Function of PRDX6 in Human Cervical Cancer. J. Cancer 2020, 11, 2390–2400. [Google Scholar] [CrossRef]
- López-Grueso, M.J.; Lagal, D.J.; García-Jiménez, Á.F.; Tarradas, R.M.; Carmona-Hidalgo, B.; Peinado, J.; Requejo-Aguilar, R.; Bárcena, J.A.; Padilla, C.A. Knockout of PRDX6 Induces Mitochondrial Dysfunction and Cell Cycle Arrest at G2/M in HepG2 Hepatocarcinoma Cells. Redox Biol. 2020, 37, 101737. [Google Scholar] [CrossRef]
- Zolea, F.; Biamonte, F.; Battaglia, A.M.; Faniello, M.C.; Cuda, G.; Costanzo, F. Caffeine Positively Modulates Ferritin Heavy Chain Expression in H460 Cells: Effects on Cell Proliferation. PLoS ONE 2016, 11, e0163078. [Google Scholar] [CrossRef] [PubMed]
- Venuto, S.; Monteonofrio, L.; Cozzolino, F.; Monti, M.; Appolloni, I.; Mazza, T.; Canetti, D.; Giambra, V.; Panelli, P.; Fusco, C.; et al. TRIM8 Interacts with KIF11 and KIFC1 and Controls Bipolar Spindle Formation and Chromosomal Stability. Cancer Lett. 2020, 473, 98–106. [Google Scholar] [CrossRef]
- Scieuzo, C.; Salvia, R.; Franco, A.; Pezzi, M.; Cozzolino, F.; Chicca, M.; Scapoli, C.; Vogel, H.; Monti, M.; Ferracini, C.; et al. An Integrated Transcriptomic and Proteomic Approach to Identify the Main Torymus Sinensis Venom Components. Sci. Rep. 2021, 11, 5032. [Google Scholar] [CrossRef]
- Battaglia, A.M.; Sacco, A.; Perrotta, I.D.; Faniello, M.C.; Scalise, M.; Torella, D.; Levi, S.; Costanzo, F.; Biamonte, F. Iron Administration Overcomes Resistance to Erastin-Mediated Ferroptosis in Ovarian Cancer Cells. Front. Oncol. 2022, 12, 868351. [Google Scholar] [CrossRef] [PubMed]
- Vecchio, E.; Nisticò, N.; Golino, G.; Iaccino, E.; Maisano, D.; Mimmi, S.; Aloisio, A.; Renna, M.; Avagliano, A.; Arcucci, A.; et al. IBtkα Activates the β-Catenin-Dependent Transcription of MYC through Ubiquitylation and Proteasomal Degradation of GSK3β in Cancerous B Cells. Int. J. Mol. Sci. 2022, 23, 2044. [Google Scholar] [CrossRef] [PubMed]
- Biamonte, F.; Zolea, F.; Santamaria, G.; Battaglia, A.M.; Cuda, G.; Costanzo, F. Human Haematological and Epithelial Tumor-Derived Cell Lines Express Distinct Patterns of Onco-MicroRNAs. Cell. Mol. Biol. 2017, 63, 75–85. [Google Scholar] [CrossRef] [PubMed]
- di Caprio, R.; Ciano, M.; Montano, G.; Costanzo, P.; Cesaro, E. KAP1 Is a Novel Substrate for the Arginine Methyltransferase PRMT5. Biology 2015, 4, 41. [Google Scholar] [CrossRef]
- Sodaro, G.; Cesaro, E.; Montano, G.; Blasio, G.; Fiorentino, F.; Romano, S.; Jacquel, A.; Aurberger, P.; Costanzo, P. Role of ZNF224 in C-Myc Repression and Imatinib Responsiveness in Chronic Myeloid Leukemia. Oncotarget 2017, 9, 3417–3431. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Masuda, T.; Goto, F.; Yoshihara, T.; Mikami, B. The Universal Mechanism for Iron Translocation to the Ferroxidase Site in Ferritin, Which Is Mediated by the Well Conserved Transit Site. Biochem. Biophys. Res. Commun. 2010, 400, 94–99. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, W.; Kim, E.E.K. Crystal Structures of Human Peroxiredoxin 6 in Different Oxidation States. Biochem. Biophys. Res. Commun. 2016, 477, 717–722. [Google Scholar] [CrossRef]
- Goodford, P.J. A Computational Procedure for Determining Energetically Favorable Binding Sites on Biologically Important Macromolecules. J. Med. Chem. 1985, 28, 849–857. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455. [Google Scholar] [CrossRef]
- Ortuso, F.; Langer, T.; Alcaro, S. GBPM: GRID-Based Pharmacophore Model: Concept and Application Studies to Protein-Protein Recognition. Bioinformatics 2006, 22, 1449–1455. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ortuso, F.; Mercatelli, D.; Guzzi, P.H.; Giorgi, F.M. Structural Genetics of Circulating Variants Affecting the SARS-CoV-2 Spike/Human ACE2 Complex. J. Biomol. Struct. Dyn. 2022, 40, 6545–6555. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, E.; Pastore, A.; Polverino, A.; Manna, L.; Divisato, G.; Quintavalle, C.; di Sanzo, M.; Faniello, M.C.; Grosso, M.; Costanzo, P. ZNF224 Is a Mediator of TGF-β pro-Oncogenic Function in Melanoma. Hum. Mol. Genet. 2021, 30, 2100–2109. [Google Scholar] [CrossRef] [PubMed]





| FTH1 Residues Interacting to | PRDX6_r | PRDX6_s | |
|---|---|---|---|
| PRDX6_r | PRDX6_s | Residues Interacting with FTH1 | |
| Thr5 | Gly4 | ||
| Asp9 | |||
| Gln14 | Asn13 | ||
| Arg22 | Glu15 | ||
| Tyr32 a | Arg24 | ||
| Tyr39 | Asp31 | ||
| Asp42 | Arg53 | ||
| Lys53 | Lys56 | ||
| Arg63 | Tyr89 | ||
| Glu64 | Asn90 | ||
| Glu67 | Glu109 | ||
| Lys71 a | Lys122 | ||
| Asn74a | Lys125 | ||
| Arg79 | Lys141 | ||
| Gln83 | Lys142 | ||
| Asp84 | Asp158 | ||
| Lys86 a | Glu172 | ||
| Lys87 a,b | Arg174 | ||
| Asp89 | Asp180 | ||
| Asp91 a | Lys182 | ||
| Lys108 | Asp183 | ||
| Glu116 | Gly184 | ||
| Asn125 | Asp185 | ||
| Asn139 | Ser186 | ||
| Lys143 | Lys200 | ||
| Lys157 | Lys204 | ||
| Lys209 | |||
| Tyr220 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Sanzo, M.; Cozzolino, F.; Battaglia, A.M.; Aversa, I.; Monaco, V.; Sacco, A.; Biamonte, F.; Palmieri, C.; Procopio, F.; Santamaria, G.; et al. Ferritin Heavy Chain Binds Peroxiredoxin 6 and Inhibits Cell Proliferation and Migration. Int. J. Mol. Sci. 2022, 23, 12987. https://doi.org/10.3390/ijms232112987
Di Sanzo M, Cozzolino F, Battaglia AM, Aversa I, Monaco V, Sacco A, Biamonte F, Palmieri C, Procopio F, Santamaria G, et al. Ferritin Heavy Chain Binds Peroxiredoxin 6 and Inhibits Cell Proliferation and Migration. International Journal of Molecular Sciences. 2022; 23(21):12987. https://doi.org/10.3390/ijms232112987
Chicago/Turabian StyleDi Sanzo, Maddalena, Flora Cozzolino, Anna Martina Battaglia, Ilenia Aversa, Vittoria Monaco, Alessandro Sacco, Flavia Biamonte, Camillo Palmieri, Francesca Procopio, Gianluca Santamaria, and et al. 2022. "Ferritin Heavy Chain Binds Peroxiredoxin 6 and Inhibits Cell Proliferation and Migration" International Journal of Molecular Sciences 23, no. 21: 12987. https://doi.org/10.3390/ijms232112987
APA StyleDi Sanzo, M., Cozzolino, F., Battaglia, A. M., Aversa, I., Monaco, V., Sacco, A., Biamonte, F., Palmieri, C., Procopio, F., Santamaria, G., Ortuso, F., Pucci, P., Monti, M., & Faniello, M. C. (2022). Ferritin Heavy Chain Binds Peroxiredoxin 6 and Inhibits Cell Proliferation and Migration. International Journal of Molecular Sciences, 23(21), 12987. https://doi.org/10.3390/ijms232112987










