Developing Genetic Engineering Techniques for Control of Seed Size and Yield
Abstract
1. Introduction
2. Genetic Factors Controlling Seed Size
3. The Role of Signaling Pathways in Seed Size Regulation
3.1. The Ubiquitin-26s Proteasome Pathway
3.2. The Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway in Plants
3.3. The IKU Pathway
3.4. The G-Protein Signaling Pathway
3.5. Transcriptional Regulation
4. Seed Size Regulation by Phytohormones
4.1. Auxins
4.2. Cytokinins
4.3. Brassinosteroids
4.4. Jasmonic Acid
5. Possible Strategies Used to Maintain Seed Size
6. Conclusions and Future Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Purugganan, M.D. Evolutionary insights into the nature of plant domestication. Curr. Biol. 2019, 29, R705–R714. [Google Scholar] [CrossRef] [PubMed]
- Arun, N.; Dalai, A.K. Environmental and socioeconomic impact assessment of biofuels from lignocellulosic biomass. In Lignocellulosic Biomass to Liquid Biofuels; Elsevier: Amsterdam, The Netherlands, 2020; pp. 283–299. [Google Scholar]
- Pachapur, P.K.; Pachapur, V.L.; Brar, S.K.; Galvez, R.; Le Bihan, Y.; Surampalli, R.Y. Food security and sustainability. Sustain. Fundam. Appl. 2020, 357–374. [Google Scholar]
- Ding, G.; Zhao, Z.; Liao, Y.; Hu, Y.; Shi, L.; Long, Y.; Xu, F. Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus. Ann. Bot. 2012, 109, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Yadava, S.K.; Ramchiary, N. Advanced Molecular Breeding for Yield. In The Brassica juncea Genome; Springer: Berlin/Heidelberg, Germany, 2022; pp. 399–412. [Google Scholar]
- Chaudhury, A.M.; Koltunow, A.; Payne, T.; Luo, M.; Tucker, M.R.; Dennis, E.; Peacock, W. Control of early seed development. Annu. Rev. Cell Dev. Biol. 2001, 17, 677–699. [Google Scholar] [CrossRef] [PubMed]
- Haughn, G.; Chaudhury, A. Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 2005, 10, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Armenta-Medina, A.; Gillmor, C.S.; Gao, P.; Mora-Macias, J.; Kochian, L.V.; Xiang, D.; Datla, R. Developmental and genomic architecture of plant embryogenesis: From model plant to crops. Plant Commun. 2021, 2, 100136. [Google Scholar] [CrossRef]
- Locascio, A.; Roig-Villanova, I.; Bernardi, J.; Varotto, S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: A focus on auxin. Front. Plant Sci. 2014, 5, 412. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Pedreschi, R.; Barkla, B.J.; Bindschedler, L.V.; Cramer, R.; Sarkar, A.; Renaut, J.; Job, D.; Rakwal, R. Translational plant proteomics: A perspective. J. Proteom. 2012, 75, 4588–4601. [Google Scholar] [CrossRef]
- Moles, A.T.; Ackerly, D.D.; Webb, C.O.; Tweddle, J.C.; Dickie, J.B.; Westoby, M. A brief history of seed size. Science 2005, 307, 576–580. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Blankestijn-de Vries, H.; Hanhart, C.J.; Koornneef, M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 4710–4717. [Google Scholar] [CrossRef]
- Song, X.-J.; Huang, W.; Shi, M.; Zhu, M.-Z.; Lin, H.-X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Tang, M.Q.; Chen, S.; Zheng, X.F.; Mo, H.X.; Li, S.J.; Wang, Z.; Zhu, K.M.; Ding, L.N.; Liu, S.Y. Down-regulation of BnDA1, whose gene locus is associated with the seeds weight, improves the seeds weight and organ size in Brassica napus. Plant Biotechnol. J. 2017, 15, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.-C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.-Q. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Wang, D.; Sun, W.; Yuan, Z.; Sun, Q.; Fan, K.; Zhang, C.; Yu, S. Identification of a novel QTL and candidate gene associated with grain size using chromosome segment substitution lines in rice. Sci. Rep. 2021, 11, 189. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef]
- Wang, A.; Garcia, D.; Zhang, H.; Feng, K.; Chaudhury, A.; Berger, F.; Peacock, W.J.; Dennis, E.S.; Luo, M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010, 63, 670–679. [Google Scholar] [CrossRef]
- Zhang, B.; Li, C.; Li, Y.; Yu, H. Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis. Nat. Plants 2020, 6, 1146–1157. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Kang, X.; Zhao, X.; Zhang, X.; Ni, M. SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 2009, 21, 106–117. [Google Scholar] [CrossRef]
- Kang, X.; Li, W.; Zhou, Y.; Ni, M. A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet. 2013, 9, e1003347. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Gerald, J.N.F.; Berger, F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 2005, 17, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gong, X.; Zhang, B.; Liang, Z.; Wong, C.E.; See, B.Y.H.; Yu, H. TOP1α, UPF1, and TTG2 regulate seed size in a parental dosage–dependent manner. PLoS Biol. 2020, 18, e3000930. [Google Scholar] [CrossRef]
- Li, Z.; Chi, H.; Liu, C.; Zhang, T.; Han, L.; Li, L.; Pei, X.; Long, Y. Genome-wide identification and functional characterization of LEA genes during seed development process in linseed flax (Linum usitatissimum L.). BMC Plant Biol. 2021, 21, 193. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.-S.; Xu, M.-K.; Li, D.; Zhou, M.-M.; Jiang, J.-H. Soluble sugar accumulation can influence seed size via AN3–YDA gene cascade. J. Agric. Food Chem. 2017, 65, 4121–4132. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.-S.; Xu, M.-K.; Wan, W.; Wang, J.-Y. Integration of environmental and developmental (or metabolic) control of seed mass by sugar and ethylene metabolisms in Arabidopsis. J. Agric. Food Chem. 2018, 66, 3477–3488. [Google Scholar] [CrossRef]
- Bauer, M.J.; Fischer, R.L. Genome demethylation and imprinting in the endosperm. Curr. Opin. Plant Biol. 2011, 14, 162–167. [Google Scholar] [CrossRef]
- Ren, D.; Wang, X.; Yang, M.; Yang, L.; He, G.; Deng, X.W. A new regulator of seed size control in Arabidopsis identified by a genome-wide association study. New Phytol. 2019, 222, 895–906. [Google Scholar] [CrossRef]
- Pien, S.; Grossniklaus, U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Et Biophys. Acta (BBA)-Gene Struct. Expr. 2007, 1769, 375–382. [Google Scholar] [CrossRef]
- Kang, I.-H.; Steffen, J.G.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 2008, 20, 635–647. [Google Scholar] [CrossRef]
- Köhler, C.; Hennig, L.; Spillane, C.; Pien, S.; Gruissem, W.; Grossniklaus, U. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003, 17, 1540–1553. [Google Scholar] [CrossRef] [PubMed]
- Kono, A.; Umeda-Hara, C.; Adachi, S.; Nagata, N.; Konomi, M.; Nakagawa, T.; Uchimiya, H.; Umeda, M. The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell 2007, 19, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Johnston, A.J.; Matveeva, E.; Kirioukhova, O.; Grossniklaus, U.; Gruissem, W. A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr. Biol. 2008, 18, 1680–1686. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zheng, L.; Pu, L.; Ma, X.; Wang, X.; Wu, Y.; Ming, H.; Wang, Q.; Zhang, G. ENO2 affects the seed size and weight by adjusting cytokinin content and forming ENO2-bZIP75 complex in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 1331. [Google Scholar] [CrossRef]
- Xiao, W.; Brown, R.C.; Lemmon, B.E.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 2006, 142, 1160–1168. [Google Scholar] [CrossRef]
- Jullien, P.E.; Katz, A.; Oliva, M.; Ohad, N.; Berger, F. Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr. Biol. 2006, 16, 486–492. [Google Scholar] [CrossRef]
- Kinoshita, T.; Miura, A.; Choi, Y.; Kinoshita, Y.; Cao, X.; Jacobsen, S.E.; Fischer, R.L.; Kakutani, T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 2004, 303, 521–523. [Google Scholar] [CrossRef]
- Tiwari, S.; Schulz, R.; Ikeda, Y.; Dytham, L.; Bravo, J.; Mathers, L.; Spielman, M.; Guzmán, P.; Oakey, R.J.; Kinoshita, T. MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 2008, 20, 2387–2398. [Google Scholar] [CrossRef]
- Pires, N.D.; Bemer, M.; Müller, L.M.; Baroux, C.; Spillane, C.; Grossniklaus, U. Quantitative genetics identifies cryptic genetic variation involved in the paternal regulation of seed development. PLoS Genet. 2016, 12, e1005806. [Google Scholar] [CrossRef]
- Beauzamy, L.; Fourquin, C.; Dubrulle, N.; Boursiac, Y.; Boudaoud, A.; Ingram, G. Endosperm turgor pressure decreases during early Arabidopsis seed development. Development 2016, 143, 3295–3299. [Google Scholar] [CrossRef]
- Neff, M. Improving seed size, seed weight and seedling emergence in Camelina sativa by overexpressing the Atsob3-6 gene variant. Transgenic Res. 2020, 29, 409–418. [Google Scholar]
- Mosher, R.A.; Melnyk, C.W.; Kelly, K.A.; Dunn, R.M.; Studholme, D.J.; Baulcombe, D.C. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 2009, 460, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Taleski, M.; Chapman, K.; Imin, N.; Djordjevic, M.A.; Groszmann, M. The peptide hormone receptor CEPR1 functions in the reproductive tissue to control seed size and yield. Plant Physiol. 2020, 183, 620–636. [Google Scholar] [CrossRef]
- Sornay, E.; Forzani, C.; Forero-Vargas, M.; Dewitte, W.; Murray, J.A. Activation of CYCD 7; 1 in the central cell and early endosperm overcomes cell-cycle arrest in the Arabidopsis female gametophyte, and promotes early endosperm and embryo development. Plant J. 2015, 84, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Ingouff, M.; Jullien, P.E.; Berger, F. The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell 2006, 18, 3491–3501. [Google Scholar] [CrossRef]
- Tonosaki, K.; Ono, A.; Kunisada, M.; Nishino, M.; Nagata, H.; Sakamoto, S.; Kijima, S.T.; Furuumi, H.; Nonomura, K.-I.; Sato, Y.; et al. Mutation of the imprinted gene OsEMF2a induces autonomous endosperm development and delayed cellularization in rice. Plant Cell 2021, 33, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef]
- Verma, A.; Prakash, G.; Ranjan, R.; Tyagi, A.K.; Agarwal, P. Silencing of an ubiquitin ligase increases grain width and weight in Indica rice. Front. Genet. 2021, 11, 1650. [Google Scholar] [CrossRef]
- Takano-Kai, N.; Jiang, H.; Kubo, T.; Sweeney, M.; Matsumoto, T.; Kanamori, H.; Padhukasahasram, B.; Bustamante, C.; Yoshimura, A.; Doi, K. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 2009, 182, 1323–1334. [Google Scholar] [CrossRef]
- Lyu, J.; Wang, D.; Duan, P.; Liu, Y.; Huang, K.; Zeng, D.; Zhang, L.; Dong, G.; Li, Y.; Xu, R. Control of grain size and weight by the GSK2-LARGE1/OML4 pathway in rice. Plant Cell 2020, 32, 1905–1918. [Google Scholar] [CrossRef]
- Bai, X.; Luo, L.; Yan, W.; Kovi, M.R.; Zhan, W.; Xing, Y. Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 2010, 11, 16. [Google Scholar] [CrossRef] [PubMed]
- Chun, Y.; Fang, J.; Zafar, S.A.; Shang, J.; Zhao, J.; Yuan, S.; Li, X. MINI SEED 2 (MIS2) Encodes a Receptor-like Kinase that Controls Grain Size and Shape in Rice. Rice 2020, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Dhatt, B.K.; Miller, M.; Folsom, J.J.; Wang, Z.; Krassovskaya, I.; Liu, K.; Sandhu, J.; Yu, H.; Zhang, C. MADS78 and MADS79 are essential regulators of early seed development in rice. Plant Physiol. 2020, 182, 933–948. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, Z.; Gao, L.; Wang, L.; Gao, M.; Jiao, Z.; Qiao, H.; Yang, J.; Chen, M.; Yao, L. Genome-wide identification and characterization of microRNAs in developing grains of Zea mays L. PLoS ONE 2016, 11, e0153168. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Lee, J.; Kichey, T.; Gerentes, D.; Zivy, M.; Tatout, C.; Dubois, F.; Balliau, T.; Valot, B.; Davanture, M. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 2006, 18, 3252–3274. [Google Scholar] [CrossRef]
- Li, Q.; Yang, X.; Bai, G.; Warburton, M.L.; Mahuku, G.; Gore, M.; Dai, J.; Li, J.; Yan, J. Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. Appl. Genet. 2010, 120, 753–763. [Google Scholar] [CrossRef]
- Li, Q.; Li, L.; Yang, X.; Warburton, M.L.; Bai, G.; Dai, J.; Li, J.; Yan, J. Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 2010, 10, 143. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, W.; Du, D.; Pu, L.; Zhang, C. Overexpression of a maize BR transcription factor ZmBZR1 in Arabidopsis enlarges organ and seed size of the transgenic plants. Plant Sci. 2020, 292, 110378. [Google Scholar] [CrossRef]
- Su, Z.; Hao, C.; Wang, L.; Dong, Y.; Zhang, X. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2011, 122, 211–223. [Google Scholar] [CrossRef]
- Liu, N.; Wu, S.; Li, Z.; Khan, A.Q.; Hu, H.; Zhang, X.; Tu, L. Repression of microRNA 160 results in retarded seed integument growth and smaller final seed size in cotton. Crop J. 2020, 8, 602–612. [Google Scholar] [CrossRef]
- Geravandi, M.; Cheghamirza, K.; Farshadfar, E.; Gepts, P. QTL analysis of seed size and yield-related traits in an inter-genepool population of common bean (Phaseolus vulgaris L.). Sci. Hortic. 2020, 274, 109678. [Google Scholar] [CrossRef]
- Khan, M.H.; Hu, L.; Zhu, M.; Zhai, Y.; Khan, S.U.; Ahmar, S.; Amoo, O.; Zhang, K.; Fan, C.; Zhou, Y. Targeted mutagenesis of EOD3 gene in Brassica napus L. regulates seed production. J. Cell. Physiol. 2021, 236, 1996–2007. [Google Scholar] [CrossRef] [PubMed]
- Hina, A.; Cao, Y.; Song, S.; Li, S.; Sharmin, R.A.; Elattar, M.A.; Bhat, J.A.; Zhao, T. High-resolution mapping in two RIL populations refines major “QTL Hotspot” regions for seed size and shape in soybean (Glycine max L.). Int. J. Mol. Sci. 2020, 21, 1040. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.X.; Paddock, K.J.; Zhang, Z.; Stacey, M.G. GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol. 2021, 229, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Alyr, M.H.; Pallu, J.; Sambou, A.; Nguepjop, J.R.; Seye, M.; Tossim, H.-A.; Djiboune, Y.R.; Sane, D.; Rami, J.-F.; Fonceka, D. Fine-Mapping of a Wild Genomic Region Involved in Pod and Seed Size Reduction on Chromosome A07 in Peanut (Arachis hypogaea L.). Genes 2020, 11, 1402. [Google Scholar] [CrossRef]
- Scott, R.J.; Spielman, M.; Bailey, J.; Dickinson, H.G. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 1998, 125, 3329–3341. [Google Scholar] [CrossRef]
- Adams, S.; Vinkenoog, R.; Spielman, M.; Dickinson, H.G.; Scott, R.J. Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 2000, 127, 2493–2502. [Google Scholar] [CrossRef]
- Garcia, D.; Saingery, V.; Chambrier, P.; Mayer, U.; Jurgens, G.; Berger, F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003, 131, 1661–1670. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, C.; Hearn, D.J.; Kang, I.-H.; Punwani, J.A.; Skaggs, M.I.; Drews, G.N.; Schumaker, K.S.; Yadegari, R. Identification of transcription-factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biol. 2010, 10, 110. [Google Scholar] [CrossRef]
- Zuo, J.; Li, J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 2014, 48, 99–118. [Google Scholar] [CrossRef]
- Gehring, M.; Satyaki, P. Endosperm and imprinting, inextricably linked. Plant Physiol. 2017, 173, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Jahnke, S.; Scholten, S. Epigenetic resetting of a gene imprinted in plant embryos. Curr. Biol. 2009, 19, 1677–1681. [Google Scholar] [CrossRef] [PubMed]
- Nodine, M.D.; Bartel, D.P. Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 2012, 482, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Taylor, J.M.; Spriggs, A.; Zhang, H.; Wu, X.; Russell, S.; Singh, M.; Koltunow, A. A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 2011, 7, e1002125. [Google Scholar] [CrossRef]
- Waters, A.J.; Bilinski, P.; Eichten, S.R.; Vaughn, M.W.; Ross-Ibarra, J.; Gehring, M.; Springer, N.M. Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc. Natl. Acad. Sci. USA 2013, 110, 19639–19644. [Google Scholar] [CrossRef]
- Jullien, P.E.; Kinoshita, T.; Ohad, N.; Berger, F. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 2006, 18, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Köhler, C. Evolution, function, and regulation of genomic imprinting in plant seed development. J. Exp. Bot. 2012, 63, 4713–4722. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Stewart Jr, C.N. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Vielle-Calzada, J.-P.; Hoeppner, M.A.; Gagliano, W.B. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 1998, 280, 446–450. [Google Scholar] [CrossRef]
- Rajkumar, M.S.; Gupta, K.; Khemka, N.K.; Garg, R.; Jain, M. DNA methylation reprogramming during seed development and its functional relevance in seed size/weight determination in chickpea. Commun. Biol. 2020, 3, 340. [Google Scholar] [CrossRef]
- Kumar, S.; Mohapatra, T. Dynamics of DNA Methylation and Its Functions in Plant Growth and Development. Front. Plant Sci. 2021, 12, 596236. [Google Scholar] [CrossRef] [PubMed]
- Lu, C. Breeding and utilization of two-line intersubspecific hybrid rice Liangyou Peijiu. Hybrid Rice 2000, 15, 4–5. [Google Scholar]
- Zhou, S.; Xing, M.; Zhao, Z.; Gu, Y.; Xiao, Y.; Liu, Q.; Xue, H. DNA methylation modification in heterosis initiation through analyzing rice hybrid contemporary seeds. Crop J. 2021, 9, 1179–1190. [Google Scholar] [CrossRef]
- Hsieh, T.-F.; Shin, J.; Uzawa, R.; Silva, P.; Cohen, S.; Bauer, M.J.; Hashimoto, M.; Kirkbride, R.C.; Harada, J.J.; Zilberman, D. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 2011, 108, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.-Q.; Zhang, Y.-J.; Zhou, S.-R.; Hu, W.-Y.; Wu, X.-T.; Ye, Y.-J.; Wu, X.-X.; Xiao, Y.-P.; Li, X.; Xue, H.-W. Global analysis reveals the crucial roles of DNA methylation during rice seed development. Plant Physiol. 2015, 168, 1417–1432. [Google Scholar] [CrossRef] [PubMed]
- Creff, A.; Ali, O.; Bayle, V.; Ingram, G.; Landrein, B. Endosperm turgor pressure both promotes and restricts seed growth and size. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dhaka, N.; Sharma, R. MicroRNA-mediated regulation of agronomically important seed traits: A treasure trove with shades of grey! Crit. Rev. Biotechnol. 2021, 41, 594–608. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Z.; Song, X.; Han, L.; Wang, L.; Zhang, J.; Long, Y.; Pei, X. Identification and Characterization of microRNAs in the Developing Seed of Linseed Flax (Linum usitatissimum L.). Int. J. Mol. Sci. 2020, 21, 2708. [Google Scholar] [CrossRef]
- Parreira, J.R.; Cappuccio, M.; Balestrazzi, A.; Fevereiro, P.; Araújo, S.d.S. MicroRNAs expression dynamics reveal post-transcriptional mechanisms regulating seed development in Phaseolus vulgaris L. Hortic. Res. 2021, 8, 18. [Google Scholar] [CrossRef]
- He, X.-J.; Hsu, Y.-F.; Zhu, S.; Liu, H.-L.; Pontes, O.; Zhu, J.; Cui, X.; Wang, C.-S.; Zhu, J.-K. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009, 23, 2717–2722. [Google Scholar] [CrossRef]
- Mosher, R.A.; Melnyk, C.W. siRNAs and DNA methylation: Seedy epigenetics. Trends Plant Sci. 2010, 15, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Xiao, Y.; Yang, W.; Ye, W.; Wang, B.; Younas, M.; Wu, J.; Liu, K. Association mapping of six yield-related traits in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2014, 127, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, L.; Liu, S.; Li, Z.; Huang, R.; Li, Y.; Cheng, H.; Li, X.; Zhou, B.; Wu, S. The genetic basis of natural variation in kernel size and related traits using a four-way cross population in maize. PLoS ONE 2016, 11, e0153428. [Google Scholar] [CrossRef] [PubMed]
- Liller, C.B.; Walla, A.; Boer, M.P.; Hedley, P.; Macaulay, M.; Effgen, S.; von Korff, M.; van Esse, G.W.; Koornneef, M. Fine mapping of a major QTL for awn length in barley using a multiparent mapping population. Theor. Appl. Genet. 2017, 130, 269–281. [Google Scholar] [CrossRef]
- Pu, C.X.; Ma, Y.; Wang, J.; Zhang, Y.C.; Jiao, X.W.; Hu, Y.H.; Wang, L.L.; Zhu, Z.G.; Sun, D.; Sun, Y. Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J. 2012, 70, 940–953. [Google Scholar] [CrossRef] [PubMed]
- De Veylder, L.; Beeckman, T.; Inzé, D. The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 2007, 8, 655–665. [Google Scholar] [CrossRef]
- Johnston, A.J.; Kirioukhova, O.; Barrell, P.J.; Rutten, T.; Moore, J.M.; Baskar, R.; Grossniklaus, U.; Gruissem, W. Dosage-sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genet. 2010, 6, e1000988. [Google Scholar] [CrossRef]
- Collins, E.S.; Balchand, S.K.; Faraci, J.L.; Wadsworth, P.; Lee, W.-L. Cell cycle–regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell 2012, 23, 3380–3390. [Google Scholar] [CrossRef]
- Liu, H.; Xu, H.; Qiu, W.; Li, X.; Zhang, H.; Zhu, Y.; Li, C.; Wang, L. Roles of bZIP transcription factors in plant growth and development and abiotic stress response. Acta Agric. Zhejiangensis 2019, 31, 1205–1214. [Google Scholar]
- Lee, H.; Guo, Y.; Ohta, M.; Xiong, L.; Stevenson, B.; Zhu, J.K. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 2002, 21, 2692–2702. [Google Scholar] [CrossRef]
- Andriotis, V.M.; Kruger, N.J.; Pike, M.J.; Smith, A.M. Plastidial glycolysis in developing Arabidopsis embryos. New Phytol. 2010, 185, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Parry, G.; Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell 2004, 16, 3181–3195. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Schwager, K. Regulated proteolysis and plant development. Plant Cell Rep. 2004, 23, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Park, S.-H.; Soh, M.Y.; Chua, N.-H. Ubiquitin-specific proteases UBP12 and UBP13 promote shade avoidance response by enhancing PIF7 stability. Proc. Natl. Acad. Sci. USA 2021, 118, e2103633118. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.J.; Casalongué, C.A.; Terrile, M.C. 28—Ubiquitin-proteasome system as part of nitric oxide sensing in plants. In Nitric Oxide in Plant Biology; Pratap Singh, V., Singh, S., Tripathi, D.K., Romero-Puertas, M.C., Sandalio, L.M., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 653–687. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Morreale, F.E.; Walden, H. Types of ubiquitin ligases. Cell 2016, 165, 248–248.e1. [Google Scholar] [CrossRef]
- Xia, T.; Li, N.; Dumenil, J.; Li, J.; Kamenski, A.; Bevan, M.W.; Gao, F.; Li, Y. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 2013, 25, 3347–3359. [Google Scholar] [CrossRef]
- Li, N.; Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016, 33, 23–32. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef]
- Disch, S.; Anastasiou, E.; Sharma, V.K.; Laux, T.; Fletcher, J.C.; Lenhard, M. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 2006, 16, 272–279. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, F.; Zhang, H.; He, H.; Ma, L.; Deng, X.W. Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 2008, 55, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, N.; Chen, L.; Xu, Y.; Li, Y.; Zhang, Y.; Li, C.; Li, Y. The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 2014, 26, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Wang, S.; Li, Y.; Zaitlin, D.; Pierce, A.J.; Smalle, J.A. Loss of 26S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiol. 2009, 150, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Eloy, N.B.; Gonzalez, N.; Van Leene, J.; Maleux, K.; Vanhaeren, H.; De Milde, L.; Dhondt, S.; Vercruysse, L.; Witters, E.; Mercier, R. SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proc. Natl. Acad. Sci. USA 2012, 109, 13853–13858. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.; De Veylder, L. The Plant Anaphase-Promoting Complex/Cyclosome. Annu. Rev. Cell Dev. Biol. 2022, 38, 25–48. [Google Scholar] [CrossRef] [PubMed]
- Vanhaeren, H.; Inzé, D.; Gonzalez, N. Plant growth beyond limits. Trends Plant Sci. 2016, 21, 102–109. [Google Scholar] [CrossRef]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Ying, J.-Z.; Gao, J.-P.; Shan, J.-X.; Zhu, M.-Z.; Shi, M.; Lin, H.-X. Dissecting the genetic basis of extremely large grain shape in rice cultivar ‘JZ1560’. J. Genet. Genom. 2012, 39, 325–333. [Google Scholar] [CrossRef]
- Keren, I.; Lacroix, B.; Kohrman, A.; Citovsky, V. Histone deubiquitinase OTU1 epigenetically regulates DA1 and DA2, which control Arabidopsis seed and organ size. Iscience 2020, 23, 100948. [Google Scholar] [CrossRef]
- Yin, P.; Ma, Q.; Wang, H.; Feng, D.; Wang, X.; Pei, Y.; Wen, J.; Tadege, M.; Niu, L.; Lin, H. SMALL LEAF AND BUSHY1 controls organ size and lateral branching by modulating the stability of BIG SEEDS1 in Medicago truncatula. New Phytol. 2020, 226, 1399–1412. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Yin, Q.; Zhou, L.; Meng, L.; Hu, W.; Li, F.; Li, Y.; Han, K.; Zhang, S.; Fu, S. Metagenomic sequencing reveals viral abundance and diversity in mosquitoes from the Shaanxi-Gansu-Ningxia region, China. PLoS Negl. Trop. Dis. 2021, 15, e0009381. [Google Scholar] [CrossRef] [PubMed]
- Bent, A.F. Plant mitogen-activated protein kinase cascades: Negative regulatory roles turn out positive. Proc. Natl. Acad. Sci. USA 2001, 98, 784–786. [Google Scholar] [CrossRef] [PubMed]
- Tena, G.; Asai, T.; Chiu, W.-L.; Sheen, J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001, 4, 392–400. [Google Scholar] [CrossRef]
- Pedley, K.F.; Martin, G.B. Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 2005, 8, 541–547. [Google Scholar] [CrossRef]
- Chen, J.; Wang, L.; Yuan, M. Update on the Roles of Rice MAPK Cascades. Int. J. Mol. Sci. 2021, 22, 1679. [Google Scholar] [CrossRef]
- Jagodzik, P.; Tajdel-Zielinska, M.; Ciesla, A.; Marczak, M.; Ludwikow, A. Mitogen-Activated Protein Kinase Cascades in Plant Hormone Signaling. Front. Plant Sci. 2018, 9, 1387. [Google Scholar] [CrossRef]
- Duan, P.; Rao, Y.; Zeng, D.; Yang, Y.; Xu, R.; Zhang, B.; Dong, G.; Qian, Q.; Li, Y. SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 2014, 77, 547–557. [Google Scholar] [CrossRef]
- Liu, S.; Hua, L.; Dong, S.; Chen, H.; Zhu, X.; Jiang, J.e.; Zhang, F.; Li, Y.; Fang, X.; Chen, F. Os MAPK 6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 2015, 84, 672–681. [Google Scholar] [CrossRef]
- Xu, R.; Yu, H.; Wang, J.; Duan, P.; Zhang, B.; Li, J.; Li, Y.; Xu, J.; Lyu, J.; Li, N.; et al. A mitogen-activated protein kinase phosphatase influences grain size and weight in rice. Plant J. Cell Mol. Biol. 2018, 95, 937–946. [Google Scholar] [CrossRef]
- Guo, T.; Chen, K.; Dong, N.-Q.; Shi, C.-L.; Ye, W.-W.; Gao, J.-P.; Shan, J.-X.; Lin, H.-X. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 2018, 30, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, N.; Del Carpio, D.P.; Hofmann, A.; Mizuta, Y.; Kurihara, D.; Higashiyama, T.; Uchida, N.; Torii, K.U.; Colombo, L.; Groth, G. A peptide pair coordinates regular ovule initiation patterns with seed number and fruit size. Curr. Biol. 2020, 30, 4352–4361.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yao, W.; Wang, F.; Su, Y.; Zhang, D.; Hu, S.; Zhang, X. AGC protein kinase AGC1-4 mediates seed size in Arabidopsis. Plant Cell Rep. 2020, 39, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Han, X.; Yu, S.; Liu, Z.; Guo, D.; He, Y.; Li, W.; Tao, Y.; Sun, C.; Xu, P. OsINV3 and its homolog, OsINV2, control grain size in rice. Int. J. Mol. Sci. 2020, 21, 2199. [Google Scholar] [CrossRef]
- He, C.; Wang, J.; Dong, R.; Guan, H.; Liu, T.; Liu, C.; Liu, Q.; Wang, L. Overexpression of an Antisense RNA of Maize Receptor-Like Kinase Gene ZmRLK7 Enlarges the Organ and Seed Size of Transgenic Arabidopsis Plants. Front. Plant Sci. 2020, 11, 579120. [Google Scholar] [CrossRef]
- Xiao, W.; Hu, S.; Zou, X.; Cai, R.; Liao, R.; Lin, X.; Yao, R.; Guo, X. Lectin receptor-like kinase LecRK-VIII. 2 is a missing link in MAPK signaling-mediated yield control. Plant Physiol. 2021, 187, 303–320. [Google Scholar] [CrossRef]
- Sun, X.; Shantharaj, D.; Kang, X.; Ni, M. Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 2010, 13, 611–620. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Zhao, X.Y.; Shao, X.X.; Wang, F.; Zhou, C.; Liu, Y.G.; Zhang, Y.; Zhang, X.S. Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 2014, 26, 1053–1068. [Google Scholar] [CrossRef]
- Xi, X.; Hu, Z.; Nie, X.; Meng, M.; Xu, H.; Li, J. Cross Inhibition of MPK10 and WRKY10 Participating in the Growth of Endosperm in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 521. [Google Scholar] [CrossRef]
- Xu, R.; Li, N.; Li, Y. Control of grain size by G protein signaling in rice. J. Integr. Plant Biol. 2019, 61, 533–540. [Google Scholar] [CrossRef]
- Ullah, H.; Chen, J.-G.; Young, J.C.; Im, K.-H.; Sussman, M.R.; Jones, A.M. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 2001, 292, 2066–2069. [Google Scholar] [CrossRef] [PubMed]
- Lease, K.A.; Wen, J.; Li, J.; Doke, J.T.; Liscum, E.; Walker, J.C. A mutant Arabidopsis heterotrimeric G-protein β subunit affects leaf, flower, and fruit development. Plant Cell 2001, 13, 2631–2641. [Google Scholar] [PubMed]
- Li, S.; Liu, Y.; Zheng, L.; Chen, L.; Li, N.; Corke, F.; Lu, Y.; Fu, X.; Zhu, Z.; Bevan, M.W. The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol. 2012, 194, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, D.; Trusov, Y.; Zhang, W.; Acharya, B.R.; Sheahan, M.B.; McCurdy, D.W.; Assmann, S.M.; Botella, J.R. An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 2011, 67, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Roy Choudhury, S.; Riesselman, A.J.; Pandey, S. Constitutive or seed-specific overexpression of A rabidopsis G-protein γ subunit 3 (AGG 3) results in increased seed and oil production and improved stress tolerance in C amelina sativa. Plant Biotechnol. J. 2014, 12, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Utsunomiya, Y.; Samejima, C.; Takayanagi, Y.; Izawa, Y.; Yoshida, T.; Sawada, Y.; Fujisawa, Y.; Kato, H.; Iwasaki, Y. Suppression of the rice heterotrimeric G protein β-subunit gene, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J. 2011, 67, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, Y.; Kato, T.; Ohki, S.; Ishikawa, A.; Kitano, H.; Sasaki, T.; Asahi, T.; Iwasaki, Y. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 1999, 96, 7575–7580. [Google Scholar] [CrossRef]
- Ashikari, M.; Wu, J.; Yano, M.; Sasaki, T.; Yoshimura, A. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 1999, 96, 10284–10289. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, M.; Liang, J. RGB1 Regulates Grain Development and Starch Accumulation through Its Effect on OsYUC11-Mediated Auxin Biosynthesis in Rice Endosperm Cells. Front. Plant Sci. 2021, 12, 585174. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lu, L.; Liu, H.; Bai, X.; Zhou, X.; Wu, B.; Yuan, M.; Yang, L.; Xing, Y. A minor QTL, SG3, encoding an R2R3-MYB protein, negatively controls grain length in rice. Theor. Appl. Genet. 2020, 133, 2387–2399. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wang, C.; Fu, Y.; Wang, J.; Liu, Q.; Zhang, X.; Yan, C.; Qian, Q.; Wang, K. QTL editing confers opposing yield performance in different rice varieties. J. Integr. Plant Biol. 2018, 60, 89–93. [Google Scholar] [CrossRef]
- Liu, Q.; Han, R.; Wu, K.; Zhang, J.; Ye, Y.; Wang, S.; Chen, J.; Pan, Y.; Li, Q.; Xu, X.; et al. G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 2018, 9, 852. [Google Scholar] [CrossRef]
- Gao, Y.; Gu, H.; Leburu, M.; Li, X.; Wang, Y.; Sheng, J.; Fang, H.; Gu, M.; Liang, G. The heterotrimeric G protein β subunit RGB1 is required for seedling formation in rice. Rice 2019, 12, 53. [Google Scholar] [CrossRef]
- Yang, W.; Wu, K.; Wang, B.; Liu, H.; Guo, S.; Guo, X.; Luo, W.; Sun, S.; Ouyang, Y.; Fu, X. The RING E3 ligase CLG1 targets GS3 for degradation via the endosome pathway to determine grain size in rice. Mol. Plant 2021, 14, 1699–1713. [Google Scholar] [CrossRef]
- Ohto, M.-a.; Fischer, R.L.; Goldberg, R.B.; Nakamura, K.; Harada, J.J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3123–3128. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Omidyar, P.K.; Gee, Z.; Okamuro, J.K. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3117–3122. [Google Scholar] [CrossRef]
- Ohto, M.-a.; Floyd, S.K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex. Plant Reprod. 2009, 22, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, L.; Xu, R.; Cui, R.; Hao, J.; Sun, C.; Li, Y. Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. Plant Cell 2015, 27, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.; Gerentes, D.; Perez, P.; Smyth, D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar]
- Mizukami, Y.; Fischer, R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Liu, H.; Zhou, T.; Gu, B.; Huang, X.; Shangguan, Y.; Zhu, J.; Li, Y.; Zhao, Y.; Wang, Y. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 2013, 25, 3360–3376. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Den Boer, B.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [PubMed]
- Berger, F.; Chaudhury, A. Parental memories shape seeds. Trends Plant Sci. 2009, 14, 550–556. [Google Scholar] [CrossRef]
- Aya, K.; Hobo, T.; Sato-Izawa, K.; Ueguchi-Tanaka, M.; Kitano, H.; Matsuoka, M. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol. 2014, 55, 897–912. [Google Scholar] [CrossRef]
- Hirano, K.; Yoshida, H.; Aya, K.; Kawamura, M.; Hayashi, M.; Hobo, T.; Sato-Izawa, K.; Kitano, H.; Ueguchi-Tanaka, M.; Matsuoka, M. SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Mol. Plant 2017, 10, 590–604. [Google Scholar] [CrossRef]
- Taketa, S.; Hattori, M.; Takami, T.; Himi, E.; Sakamoto, W. Barley albino lemma 1 resulted from mutations in a Golden2-like gene reduces seed weight. Plant Cell Physiol. 2021, 62, 447–457. [Google Scholar] [CrossRef]
- Alam, I.; Wu, X.; Yu, Q.; Ge, L. Comprehensive Genomic Analysis of G2-like Transcription Factor Genes and Their Role in Development and Abiotic Stresses in Arabidopsis. Diversity 2022, 14, 228. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, Y.; Fang, Y.; Zeng, L.; Xu, J.; Yu, H.; Shi, Z.; Pan, J.; Zhang, D.; Kang, S. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Wang, H. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2015, 2, 15203. [Google Scholar] [CrossRef]
- Sun, P.; Zhang, W.; Wang, Y.; He, Q.; Shu, F.; Liu, H.; Wang, J.; Wang, J.; Yuan, L.; Deng, H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016, 58, 836–847. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, L.; Zheng, J.; Chen, F.; Wang, T.; Wang, M.; Tao, Y.; Wang, H.; Hong, Z.; Huang, Y. A missense mutation in Large Grain Size 1 increases grain size and enhances cold tolerance in rice. J. Exp. Bot. 2019, 70, 3851–3866. [Google Scholar] [CrossRef]
- Shimano, S.; Hibara, K.-I.; Furuya, T.; Arimura, S.-I.; Tsukaya, H.; Itoh, J.-I. Conserved functional control, but distinct regulation, of cell proliferation in rice and Arabidopsis leaves revealed by comparative analysis of GRF-INTERACTING FACTOR 1 orthologs. Development 2018, 145, dev159624. [Google Scholar] [CrossRef]
- He, Z.; Zeng, J.; Ren, Y.; Chen, D.; Li, W.; Gao, F.; Cao, Y.; Luo, T.; Yuan, G.; Wu, X. OsGIF1 positively regulates the sizes of stems, leaves, and grains in rice. Front. Plant Sci. 2017, 8, 1730. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Wu, K.; Ye, Y.; Yu, J.; Zhang, J.; Liu, Q.; Hu, M.; Li, H.; Tong, Y. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021, 134, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Jing, D.; Wang, S.; Xu, F.; Bao, C.; Luo, M.; Guo, Q. The Putative Role of the NAC Transcription Factor EjNACL47 in Cell Enlargement of Loquat (Eriobotrya japonica Lindl.). Horticulturae 2021, 7, 323. [Google Scholar] [CrossRef]
- Mathew, I.E.; Das, S.; Mahto, A.; Agarwal, P. Three rice NAC transcription factors heteromerize and are associated with seed size. Front. Plant Sci. 2016, 7, 1638. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, N.; Zhang, Y.; Li, Y. Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat. Commun. 2020, 11, 1846. [Google Scholar] [CrossRef]
- Yang, T.; Guo, L.; Ji, C.; Wang, H.; Wang, J.; Zheng, X.; Xiao, Q.; Wu, Y. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell 2021, 33, 104–128. [Google Scholar] [CrossRef]
- Swinnen, G.; Mauxion, J.-P.; Baekelandt, A.; De Clercq, R.; Van Doorsselaere, J.; Inzé, D.; Gonzalez, N.; Goossens, A.; Pauwels, L. SlKIX8 and SlKIX9 are negative regulators of leaf and fruit growth in tomato. bioRxiv 2021, 188, 382–396. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Pan, Y.H.; Gao, L.J.; Liang, Y.T.; Zhao, Y.; Liang, H.F.; Chen, W.W.; Yang, X.H.; Qing, D.J.; Gao, J.; Wu, H. OrMKK3 Influences Morphology and Grain Size in Rice. J. Plant Biol. 2021, 1–14. [Google Scholar] [CrossRef]
- Huang, L.; Hua, K.; Xu, R.; Zeng, D.; Wang, R.; Dong, G.; Zhang, G.; Lu, X.; Fang, N.; Wang, D. The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell 2021, 33, 1212–1228. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, M.A.; Nelson, D.C.; Weijers, D. Evolution of plant hormone response pathways. Annu. Rev. Plant Biol. 2020, 71, 327–353. [Google Scholar] [CrossRef]
- Figueiredo, D.D.; Köhler, C. Auxin: A molecular trigger of seed development. Genes Dev. 2018, 32, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Stone, S.L.; Braybrook, S.A.; Paula, S.L.; Kwong, L.W.; Meuser, J.; Pelletier, J.; Hsieh, T.-F.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3151–3156. [Google Scholar] [CrossRef]
- Wójcikowska, B.; Jaskóła, K.; Gąsiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440. [Google Scholar] [CrossRef]
- Junker, A.; Mönke, G.; Rutten, T.; Keilwagen, J.; Seifert, M.; Thi, T.M.N.; Renou, J.P.; Balzergue, S.; Viehöver, P.; Hähnel, U. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 2012, 71, 427–442. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.; Vivian-Smith, A.; Johnson, S.D.; Koltunow, A.M. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 2006, 18, 1873–1886. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.R.; Arreola, A.; Gallagher, T.L.; Gasser, C.S. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 2012, 139, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.W.; Wu, M.-F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three auxin response factors promote hypocotyl elongation. Plant Physiol. 2018, 178, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Yu, Y.; Liu, X.; Zhang, X.S.; Su, Y.H. The Arabidopsis MATERNAL EFFECT EMBRYO ARREST45 protein modulates maternal auxin biosynthesis and controls seed size by inducing AINTEGUMENTA. Plant Cell 2021, 33, 1907–1926. [Google Scholar] [CrossRef]
- Shi, L.; Song, J.; Guo, C.; Wang, B.; Guan, Z.; Yang, P.; Chen, X.; Zhang, Q.; King, G.J.; Wang, J. A CACTA-like transposable element in the upstream region of BnaA9. CYP 78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 2019, 98, 524–539. [Google Scholar] [CrossRef]
- Guo, L.; Ma, M.; Wu, L.; Zhou, M.; Li, M.; Wu, B.; Li, L.; Liu, X.; Jing, R.; Chen, W. Modified expression of TaCYP78A5 enhances grain weight with yield potential by accumulating auxin in wheat (Triticum Aestivum L.). Plant Biotechnol. J. 2022, 20, 168–182. [Google Scholar] [CrossRef]
- Rijavec, T.; Jain, M.; Dermastia, M.; Chourey, P.S. Spatial and temporal profiles of cytokinin biosynthesis and accumulation in developing caryopses of maize. Ann. Bot. 2011, 107, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ding, J.; Lin, S.; Liu, D.; Gu, T.; Wu, H.; Trigiano, R.N.; McAvoy, R.; Huang, J.; Li, Y. Evolution and roles of cytokinin genes in angiosperms 2: Do ancient CKXs play housekeeping roles while non-ancient CKXs play regulatory roles? Hortic. Res. 2020, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Holubová, K.; Hensel, G.; Vojta, P.; Tarkowski, P.; Bergougnoux, V.; Galuszka, P. Modification of barley plant productivity through regulation of cytokinin content by reverse-genetics approaches. Front. Plant Sci. 2018, 9, 1676. [Google Scholar] [CrossRef]
- Gasparis, S.; Przyborowski, M.; Kała, M.; Nadolska-Orczyk, A. Knockout of the HvCKX1 or HvCKX3 gene in barley (Hordeum vulgare L.) by RNA-guided Cas9 nuclease affects the regulation of cytokinin metabolism and root morphology. Cells 2019, 8, 782. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; He, J.; Liu, L.; Deng, Q.; Yao, X.; Liu, C.; Qiao, Y.; Li, P.; Ming, F. OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol. J. 2020, 18, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chang, C.; Zhang, H.-P.; Wang, S.-X.; Sun, G.; Xiao, S.-H.; Ma, C.-X. Identification of a novel allele of TaCKX6a02 associated with grain size, filling rate and weight of common wheat. PLoS ONE 2015, 10, e0144765. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhao, J.; Song, J.; Jameson, P.E. Cytokinin dehydrogenase: A genetic target for yield improvement in wheat. Plant Biotechnol. J. 2020, 18, 614–630. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, S.; Park, G.; Cho, H.; Choi, D.; Umeda, M.; Choi, Y.; Hwang, D.; Hwang, I. C YTO K ININ-RESPONSIVE G ROWTH REGULATOR regulates cell expansion and cytokinin-mediated cell cycle progression. Plant Physiol. 2021, 186, 1734–1746. [Google Scholar] [CrossRef]
- Werner, S.; Bartrina, I.; Novák, O.; Strnad, M.; Werner, T.; Schmülling, T. The cytokinin status of the epidermis regulates aspects of vegetative and reproductive development in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 613488. [Google Scholar] [CrossRef]
- Nedvěd, D.; Hošek, P.; Klíma, P.; Hoyerová, K. Differential subcellular distribution of cytokinins: How does membrane transport fit into the big picture? Int. J. Mol. Sci. 2021, 22, 3428. [Google Scholar] [CrossRef]
- Day, R.C.; Herridge, R.P.; Ambrose, B.A.; Macknight, R.C. Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 2008, 148, 1964–1984. [Google Scholar] [CrossRef]
- Belmonte, M.F.; Kirkbride, R.C.; Stone, S.L.; Pelletier, J.M.; Bui, A.Q.; Yeung, E.C.; Hashimoto, M.; Fei, J.; Harada, C.M.; Munoz, M.D. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 2013, 110, E435–E444. [Google Scholar] [CrossRef]
- Song, J.; Jiang, L.; Jameson, P.E. Expression patterns of Brassica napus genes implicate IPT, CKX, sucrose transporter, cell wall invertase, and amino acid permease gene family members in leaf, flower, silique, and seed development. J. Exp. Bot. 2015, 66, 5067–5082. [Google Scholar] [CrossRef]
- Ma, Q.-H.; Wang, X.-M.; Wang, Z.-M. Expression of isopentenyl transferase gene controlled by seed-specific lectin promoter in transgenic tobacco influences seed development. J. Plant Growth Regul. 2008, 27, 68–76. [Google Scholar] [CrossRef]
- Motyka, V.; Vaňková, R.; Čapková, V.; Petrášek, J.; Kamínek, M.; Schmülling, T. Cytokinin-induced upregulation of cytokinin oxidase activity in tobacco includes changes in enzyme glycosylation and secretion. Physiol. Plant. 2003, 117, 11–21. [Google Scholar] [CrossRef]
- Yu, H.Q.; Sun, F.A.; Feng, W.Q.; Lu, F.Z.; Li, W.C.; Fu, F.L. The BES1/BZR1 transcription factors regulate growth, development and stress resistance in plants. Yi Chuan 2019, 41, 206–214. [Google Scholar]
- Song, J.; Jiang, L.; Jameson, P.E. Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biol. 2012, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Dong, H.; Mu, J.; Ren, B.; Zheng, B.; Ji, Z.; Yang, W.-C.; Liang, Y.; Zuo, J. Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 2010, 22, 1232–1248. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, B.; Yuan, D.; Duan, M.; Qian, Q.; Tang, L.; Wang, B.; Liu, X.; Zhang, J.; Wang, J. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA 2013, 110, 3167–3172. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Zhao, B.; Lv, M.; Feng, Z.; Campbell, T.; Liscum, E.; Li, J. TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol. Plant 2016, 9, 582–592. [Google Scholar] [CrossRef]
- Tong, H.; Chu, C. Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 2018, 23, 1016–1028. [Google Scholar] [CrossRef]
- Xiong, M.; Yu, J.; Wang, J.; Gao, Q.; Huang, L.; Chen, C.; Zhang, C.; Fan, X.; Zhao, D.; Liu, Q.-Q. Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module. Plant Physiol. 2022, 189, 402–418. [Google Scholar] [CrossRef]
- Wu, Y.; Fu, Y.; Zhao, S.; Gu, P.; Zhu, Z.; Sun, C.; Tan, L. Clustered primary branch 1, a new allele of DWARF 11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 2016, 14, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-y.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.; Wan, J.; Zhai, H.; Takatsuto, S. Brassinosteroids regulate grain filling in rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Fujioka, S.; Shimada, Y.; Takatsuto, S.; Agetsuma, M.; Yoshida, S.; Watanabe, Y. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002, 32, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Nomura, T.; Ooka, H.; Ishizaka, M.; Yokota, T.; Sugimoto, K.; Okabe, K.; Kajiwara, H.; Satoh, K.; Yamamoto, K. Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 2002, 130, 1152–1161. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.-B.; Huang, H.-Y.; Hu, Y.-W.; Zhu, S.-W.; Wang, Z.-Y.; Lin, W.-H. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef]
- Zhu, X.; Liang, W.; Cui, X.; Chen, M.; Yin, C.; Luo, Z.; Zhu, J.; Lucas, W.J.; Wang, Z.; Zhang, D. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J. 2015, 82, 570–581. [Google Scholar] [CrossRef]
- Tanabe, S.; Ashikari, M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Yano, M.; Yoshimura, A.; Kitano, H.; Matsuoka, M.; Fujisawa, Y. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 2005, 17, 776–790. [Google Scholar] [CrossRef]
- Feng, Z.; Wu, C.; Wang, C.; Roh, J.; Zhang, L.; Chen, J.; Zhang, S.; Zhang, H.; Yang, C.; Hu, J. SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J. Exp. Bot. 2016, 67, 4241–4253. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 2005, 17, 2243–2254. [Google Scholar] [CrossRef]
- Tian, P.; Liu, J.; Mou, C.; Shi, C.; Zhang, H.; Zhao, Z.; Lin, Q.; Wang, J.; Wang, J.; Zhang, X. GW5-Like, a homolog of GW5, negatively regulates grain width, weight and salt resistance in rice. J. Integr. Plant Biol. 2019, 61, 1171–1185. [Google Scholar] [CrossRef]
- Yang, C.; Shen, W.; He, Y.; Tian, Z.; Li, J. OVATE family protein 8 positively mediates brassinosteroid signaling through interacting with the GSK3-like kinase in rice. PLoS Genet. 2016, 12, e1006118. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, G.; Liu, D.; Niu, M.; Tong, H.; Chu, C. GSK2 stabilizes OFP3 to suppress brassinosteroid responses in rice. Plant J. 2020, 102, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, R.; Li, J.; Huang, X.; Zeng, X.; Zhao, D. DWARF and SMALL SEED1, a Novel Allele of OsDWARF, Controls Rice Plant Architecture, Seed Size, and Chlorophyll Biosynthesis. Phyton 2021, 90, 111. [Google Scholar] [CrossRef]
- Dong, N.; Yin, W.; Liu, D.; Zhang, X.; Yu, Z.; Huang, W.; Liu, J.; Yang, Y.; Meng, W.; Niu, M. Regulation of brassinosteroid signaling and salt resistance by SERK2 and potential utilization for crop improvement in rice. Front. Plant Sci. 2020, 11, 621859. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef]
- Li, C.; Liu, G.; Xu, C.; Lee, G.I.; Bauer, P.; Ling, H.-Q.; Ganal, M.W.; Howe, G.A. The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 2003, 15, 1646–1661. [Google Scholar] [CrossRef]
- Lyons, R.; Manners, J.M.; Kazan, K. Jasmonate biosynthesis and signaling in monocots: A comparative overview. Plant Cell Rep. 2013, 32, 815–827. [Google Scholar] [CrossRef]
- Browse, J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 2009, 70, 1539–1546. [Google Scholar] [CrossRef]
- Goetz, S.; Hellwege, A.; Stenzel, I.; Kutter, C.; Hauptmann, V.; Forner, S.; McCaig, B.; Hause, G.; Miersch, O.; Wasternack, C. Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiol. 2012, 158, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, Y.; McCaig, B.C.; Wingerd, B.A.; Wang, J.; Whalon, M.E.; Pichersky, E.; Howe, G.A. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 2004, 16, 126–143. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef]
- Acosta, I.F.; Laparra, H.; Romero, S.P.; Schmelz, E.; Hamberg, M.; Mottinger, J.P.; Moreno, M.A.; Dellaporta, S.L. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 2009, 323, 262–265. [Google Scholar] [CrossRef]
- DeLong, A.; Calderon-Urrea, A.; Dellaporta, S.L. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 1993, 74, 757–768. [Google Scholar] [CrossRef]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.; Ponce, M. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef]
- Ali, M.S.; Baek, K.-H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef]
- Mehra, P.; Pandey, B.K.; Verma, L.; Prusty, A.; Singh, A.P.; Sharma, S.; Malik, N.; Bennett, M.J.; Parida, S.K.; Giri, J.; et al. OsJAZ11 regulates spikelet and seed development in rice. Plant Direct 2022, 6, e401. [Google Scholar] [CrossRef]
- Gnan, S.; Priest, A.; Kover, P.X. The genetic basis of natural variation in seed size and seed number and their trade-off using Arabidopsis thaliana MAGIC lines. Genetics 2014, 198, 1751–1758. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Crouch, J.H. Marker-assisted selection in plant breeding: From publications to practice. Crop Sci. 2008, 48, 391–407. [Google Scholar] [CrossRef]
- William, H.M.; Morris, M.; Warburton, M.; Hoisington, D.A. Technical, economic and policy considerations on marker-assisted selection in crops: Lessons from the experience at an international agricultural research centre. Marker-Assist. Sel. 2007, 19, 381. [Google Scholar]
- Crosbie, T.M.; Eathington, S.R.; Johnson Sr, G.R.; Edwards, M.; Reiter, R.; Stark, S.; Mohanty, R.G.; Oyervides, M.; Buehler, R.E.; Walker, A.K. Plant breeding: Past, present, and future. In Plant Breeding: The Arnel R. Hallauer International Symposium; Wiley: Hoboken, NJ, USA, 2006; pp. 3–50. [Google Scholar]
- Eathington, S.R.; Crosbie, T.M.; Edwards, M.D.; Reiter, R.S.; Bull, J.K. Molecular markers in a commercial breeding program. Crop Sci. 2007, 47, S-154–S-163. [Google Scholar] [CrossRef]
- Gregorio, G.; Islam, M.; Vergara, G.; Thirumeni, S. Recent advances in rice science to design salinity and other abiotic stress tolerant rice varieties. SABRAO J. Breed. Genet. 2013, 45, 31–40. [Google Scholar]
- Heffner, E.L.; Sorrells, M.E.; Jannink, J.-L. Genomic selection for crop improvement. Crop Sci. 2009, 49, 1–12. [Google Scholar] [CrossRef]
- Desta, Z.A.; Ortiz, R. Genomic selection: Genome-wide prediction in plant improvement. Trends Plant Sci. 2014, 19, 592–601. [Google Scholar] [CrossRef]
- Heslot, N.; Jannink, J.L.; Sorrells, M.E. Perspectives for genomic selection applications and research in plants. Crop Sci. 2015, 55, 1–12. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Mihalyov, P.D.; Lewien, M.J.; Pumphrey, M.O.; Carter, A.H. Combining genomic and phenomic information for predicting grain protein content and grain yield in spring wheat. Front. Plant Sci. 2021, 12, 170. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Tao, H.-J.; Zhao, X.-Q.; Jie, X.; Li, G.-M.; Hu, S.-K.; Dong, G.-J.; Shi, Z.-Y.; Wu, L.-W.; Jiang, H. Molecular improvement of grain weight and yield in rice by using GW6 gene. Rice Sci. 2014, 21, 127–132. [Google Scholar] [CrossRef]
- Sun, L.; Ma, D.; Yu, H.; Zhou, F.; Li, Y.; Luo, L.; Gao, G.; Zhang, Q.; Xu, C.; He, Y. Identification of quantitative trait loci for grain size and the contributions of major grain-size QTLs to grain weight in rice. Mol. Breed. 2013, 31, 451–461. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, X.; Lan, H.; Huang, J.; Wang, J.; Zhang, H. The additive effects of GS3 and qGL3 on rice grain length regulation revealed by genetic and transcriptome comparisons. BMC Plant Biol. 2015, 15, 156. [Google Scholar] [CrossRef]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.-S.; Li, Q.-F.; Zhang, C.-Q.; Zhang, C.; Yang, Q.-Q.; Pan, L.-X.; Ren, X.-Y.; Lu, J.; Gu, M.-H.; Liu, Q.-Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef]
- Dong, N.-Q.; Sun, Y.; Guo, T.; Shi, C.-L.; Zhang, Y.-M.; Kan, Y.; Xiang, Y.-H.; Zhang, H.; Yang, Y.-B.; Li, Y.-C. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef]
- Song, X.J.; Kuroha, T.; Ayano, M.; Furuta, T.; Nagai, K.; Komeda, N.; Segami, S.; Miura, K.; Ogawa, D.; Kamura, T. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 76–81. [Google Scholar] [CrossRef]
- Ruan, B.; Shang, L.; Zhang, B.; Hu, J.; Wang, Y.; Lin, H.; Zhang, A.; Liu, C.; Peng, Y.; Zhu, L. Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 2020, 227, 629–640. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef]
- Shi, C.L.; Dong, N.Q.; Guo, T.; Ye, W.W.; Shan, J.X.; Lin, H.X. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020, 103, 1174–1188. [Google Scholar] [CrossRef]
- Yu, J.; Xiong, H.; Zhu, X.; Zhang, H.; Li, H.; Miao, J.; Wang, W.; Tang, Z.; Zhang, Z.; Yao, G. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 2017, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Miao, J.; Zhang, Z.; Xiong, H.; Zhu, X.; Sun, X.; Pan, Y.; Liang, Y.; Zhang, Q.; Abdul Rehman, R.M.; et al. Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 2018, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Lin, Y.-S.; Song, X.-J.; Shen, J.-B.; Huang, W.; Shan, J.-X.; Zhu, M.-Z.; Jiang, L.; Gao, J.-P.; Lin, H.-X. The novel quantitative trait locus GL3. 1 controls rice grain size and yield by regulating Cyclin-T1; 3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef]
- Ying, J.-Z.; Ma, M.; Bai, C.; Huang, X.-H.; Liu, J.-L.; Fan, Y.-Y.; Song, X.-J. TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant 2018, 11, 750–753. [Google Scholar] [CrossRef]
- Xia, D.; Zhou, H.; Liu, R.; Dan, W.; Li, P.; Wu, B.; Chen, J.; Wang, L.; Gao, G.; Zhang, Q. GL3. 3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol. Plant 2018, 11, 754–756. [Google Scholar] [CrossRef]
- Wu, W.; Liu, X.; Wang, M.; Meyer, R.; Luo, X.; Ndjiondjop, M.; Tan, L.; Zhang, J.; Wu, J.; Cai, H. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 2017, 3, 17064. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.-I.; Onishi, A. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Wang, A.; Hou, Q.; Si, L.; Huang, X.; Luo, J.; Lu, D.; Zhu, J.; Shangguan, Y.; Miao, J.; Xie, Y. The PLATZ transcription factor GL6 affects grain length and number in rice. Plant Physiol. 2019, 180, 2077–2090. [Google Scholar] [CrossRef] [PubMed]
- Savadi, S.; Naresh, V.; Kumar, V.; Bhat, S.R. Seed-specific overexpression of Arabidopsi s DGAT1 in Indian mustard (Brassica Juncea) increases seed oil content and seed weight. Botany 2016, 94, 177–184. [Google Scholar] [CrossRef]
- Kim, H.; Park, J.H.; Kim, A.Y.; Suh, M.C. Functional analysis of diacylglycerol acyltransferase1 genes from Camelina sativa and effects of CsDGAT1B overexpression on seed mass and storage oil content in C. sativa. Plant Biotechnol. Rep. 2016, 10, 141–153. [Google Scholar] [CrossRef]
- Liu, B.; Hua, C.; Song, G.; Wu, M.; Cui, R.; Zhang, A.; Liu, Y.; Huang, L.; Yan, A.; Ali, I. The SPATULA transcription factor regulates seed oil content by controlling seed specific genes in Arabidopsis thaliana. Plant Growth Regul. 2017, 82, 111–121. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Vankova, R.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS ONE 2012, 7, e42411. [Google Scholar] [CrossRef] [PubMed]
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2016, 67, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Dai, A.; Wei, H.; Yang, S.; Wang, B.; Jiang, N.; Feng, X. ArabidopsisKLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 2016, 90, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Kim, M.-H.; Kanno, Y.; Seo, M.; Kamiya, Y.; Imai, R. Arabidopsis COLD SHOCK DOMAIN PROTEIN 2 influences ABA accumulation in seed and negatively regulates germination. Biochem. Biophys. Res. Commun. 2015, 456, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.G.; Sun, Q.B.; Kang, X.J.; Chen, C.B.; Ni, M. SHORT HYPOCOTYL UNDER BLUE 1 or HAIKU 2 mixepression alters canola and Arabidopsis seed development. New Phytol. 2016, 209, 636–649. [Google Scholar] [CrossRef]
- Liu, L.; Tong, H.; Xiao, Y.; Che, R.; Xu, F.; Hu, B.; Liang, C.; Chu, J.; Li, J.; Chu, C. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 11102–11107. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Yu, J.; Wang, H.; Luth, D.; Bai, G.; Wang, K.; Chen, R. Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. USA 2016, 113, 12414–12419. [Google Scholar] [CrossRef]
- Savadi, S.; Naresh, V.; Kumar, V.; Dargan, S.; Gupta, N.C.; Chamola, R.; Bhat, S.R. Effect of overexpression of Arabidopsis thaliana SHB1 and KLUH genes on seed weight and yield contributing traits in Indian mustard (Brassica juncea L.(Czern.)). Indian J. Genet. Plant Breed 2015, 75, 349–356. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Joshi, R.K.; Zhao, K. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. J. 2020, 18, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, J.; He, Y.; Xu, M.; Zhang, J.; Du, W.; Zhao, Y.; Xia, L. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat. Biotechnol. 2019, 37, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kaya, H.B.; Zhang, N.; Rai, R.; Willmann, M.R.; Carpenter, S.C.; Read, A.C.; Martin, F.; Fei, Z.; Leach, J.E. Spelling changes and fluorescent tagging with prime editing vectors for plants. Front. Genome Ed. 2021, 3, 617553. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Hua, K.; Jiang, Y.; Tao, X.; Zhu, J.K. Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 2020, 18, 2167. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Chen, J.; Yan, L.; Xia, L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 2020, 13, 671–674. [Google Scholar] [CrossRef]
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of plant prime-editing systems for precise genome editing. Plant Commun. 2020, 1, 100043. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Jiang, Y.-Y.; Chai, Y.-P.; Lu, M.-H.; Han, X.-L.; Lin, Q.; Zhang, Y.; Zhang, Q.; Zhou, Y.; Wang, X.-C.; Gao, C. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020, 21, 257. [Google Scholar] [CrossRef]
- Lu, Y.; Tian, Y.; Shen, R.; Yao, Q.; Zhong, D.; Zhang, X.; Zhu, J.K. Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol. J. 2021, 19, 415. [Google Scholar] [CrossRef] [PubMed]
- Perroud, P.-F.; Guyon-Debast, A.; Veillet, F.; Kermarrec, M.-P.; Chauvin, L.; Chauvin, J.-E.; Gallois, J.-L.; Nogué, F. Prime Editing in the model plant Physcomitrium patens and its potential in the tetraploid potato. Plant Sci. 2022, 316, 111162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, L.; Zheng, G.; Jiang, W.; Deng, Z.; Wang, Z.; Lu, Y. CRISPR/dCas9-Mediated Multiplex Gene Repression in Streptomyces. Biotechnol. J. 2018, 13, e1800121. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Jurkowski, T.P.; Ravichandran, M.; Stepper, P. Synthetic epigenetics—Towards intelligent control of epigenetic states and cell identity. Clin. Epigenetics 2015, 7, 18. [Google Scholar] [CrossRef]
- Zafar, S.A.; Zaidi, S.S.-E.-A.; Gaba, Y.; Singla-Pareek, S.L.; Dhankher, O.P.; Li, X.; Mansoor, S.; Pareek, A. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Bot. 2020, 71, 470–479. [Google Scholar] [CrossRef]
- Ibrahim, S.; Saleem, B.; Rehman, N.; Zafar, S.A.; Naeem, M.K.; Khan, M.R. CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J. Adv. Res. 2021, 37, 33–41. [Google Scholar] [CrossRef]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR–Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering quantitative trait variation for crop improvement by genome editing. Cell 2017, 171, 470–480.e8. [Google Scholar] [CrossRef]
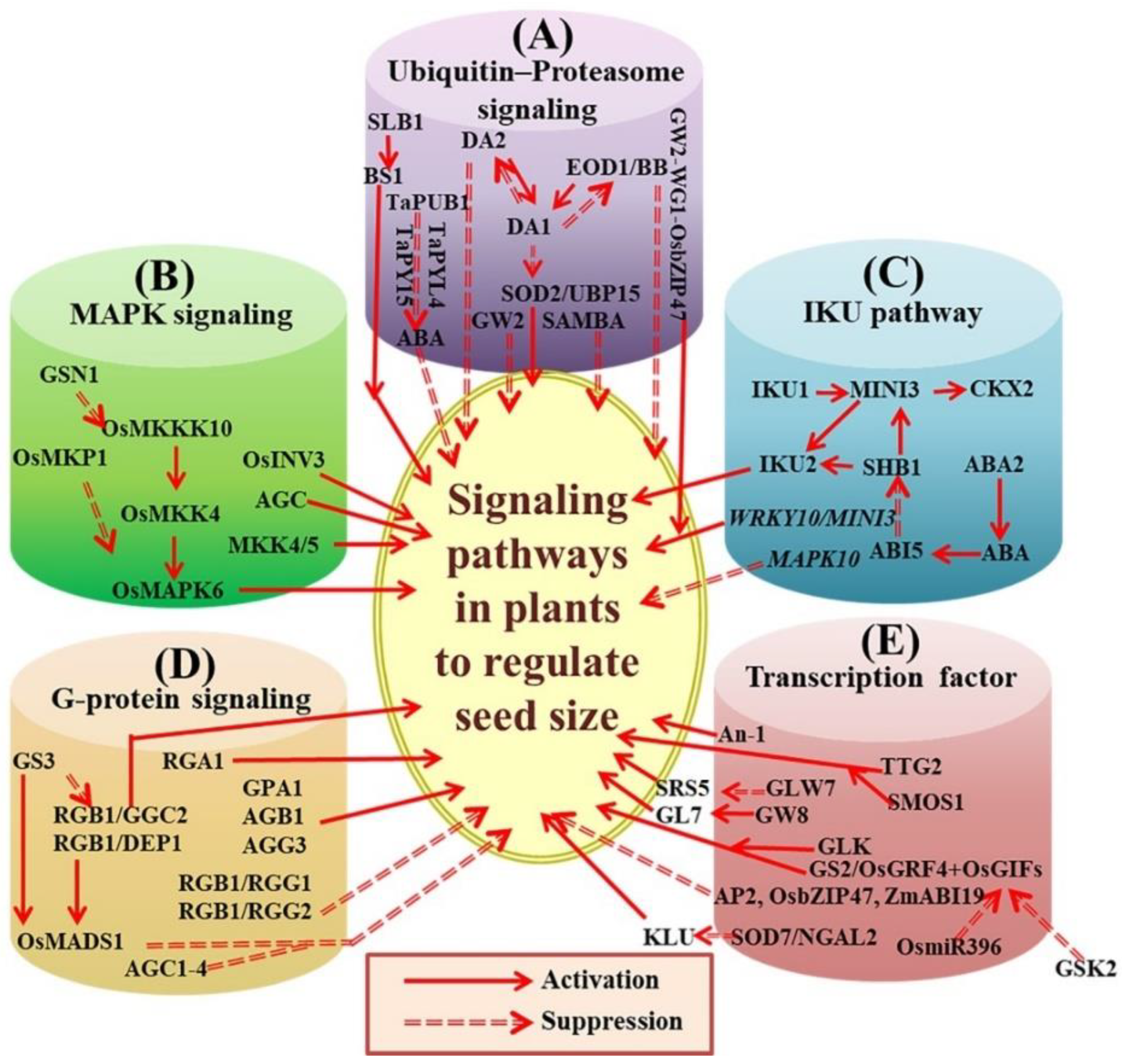


| Species | Genes or Mutants | Function | References |
|---|---|---|---|
| Arabidopsis | IKU1, IKU2, and MINI3 | Early cellularization of endosperm produces smaller seeds | [19,20] |
| Arabidopsis | TFL1 | Timely endosperm cellularization enhances seed size | [21] |
| Arabidopsis | SHB1 | Up-regulates the functions of IKU2 and MINI 3 and postpones endosperm cellularization, producing larger seeds | [22,23] |
| Arabidopsis | TTG2 | Identifies integument cell elongation and endosperm growth as the primary regulators of seed size | [24] |
| Arabidopsis | TOP1α and UPF1 | The molecular interplay among these three genes explains their biparental gametophytic effect on seed size regulation | [25] |
| Arabidopsis | LuLEA1 | Negatively regulates seed size and yield | [26] |
| Arabidopsis | AN3-YDA | Disrupts normal sucrose and glucose contents and alters seed size regulation in an3 or yda mutants | [27] |
| Arabidopsis | EIN3-YDA | Both are integral to a sugar-mediated metabolism cascade that regulates seed mass by maternally controlling embryo size | [28] |
| Arabidopsis | DME | Enhances DNA de-methylation activity and endosperm chromatin composition | [29] |
| Arabidopsis | CYCB1;4 | Overexpressed in transgenic plants to show increased seed size in response to faster cell cycle progression to increase grain size and yield | [30] |
| Arabidopsis | FIS2, FIS3, FIE, MEA, and MSI1 | Control endosperm development | [31] |
| Arabidopsis | AGL62 | Regulates endosperm development by the FIX | [32] |
| Arabidopsis | PHE1 | Controls seed abortion by mea mutant | [33] |
| Arabidopsis | CDKs and CYC | Control cell cycle development | [34] |
| Arabidopsis | RBR | Decreases the cell proliferation in the integuments and stops further development of seed coat | [35] |
| Arabidopsis | AtENO2 | Decreases the size of seed and mass with the declined concentration of cytokinin | [36] |
| Arabidopsis | MET1 and DDM1 | DNA methylation of maternal and paternal genome regulates seed size | [37] |
| Arabidopsis | FIS2, FWA, and MPC | Regulate parental imprinting by DNA methylation | [38,39,40] |
| Arabidopsis | MEA | It plays the role as a maternal buffer of paternal effects on seed development | [41] |
| Arabidopsis | PRC2 | Controls turgor pressure during seed development | [42] |
| Arabidopsis | AtSOB3 | Produces heavy and larger seeds with long hypocotyl | [43] |
| Arabidopsis | p4-siRNAs | Expressions are observed in maternal gametophytes and continue until endosperm development | [44] |
| Arabidopsis | CEPR1 | Promotes reproductive parts including seed size and yield | [45] |
| Arabidopsis | CYCD3:1 and CYCD7:1 | Promote early endosperm and embryo development | [46,47] |
| Arabidopsis | AtbZIP75 | Regulates seed development | [36] |
| Rice | OsEMF2a | Delayed cellularization and autonomic endosperm development | [48] |
| Rice | GLW7 | Controls grain size and yield | [49] |
| Rice | OsGW2 | Regulates grain size and reduces rice yield | [50] |
| Rice | GS3 | Regulates grain length | [51] |
| Rice | OML4 | Negatively regulates seed size by terminating cell expansion | [52] |
| Rice | qGL7 | Could display a single mendelian characteristic, suggested as producing diverse seed shapes | [53] |
| Rice | qGL11 | Regulates rice grain weight and length | [18] |
| Rice | MIS2 | Controls grain size by coordinately regulating epidermal cell size and cell number | [54] |
| Rice | MADS78 and MADS79 | Regulate endosperm cellularization and early seed formation in rice | [55] |
| Maize | miRNAs | Control embryogenesis via transcriptional regulation | [56] |
| Maize | GSI, qZmGW2-CHR4, zMgs3, and qZmGW2-CHR5 | Control kernel size and grain production | [57,58,59] |
| Maize | ZmBZR1 | It attaches to the promoter region of GRACE and KRP6 to control their expression and regulate seed size | [60] |
| Wheat | TaGW2 | Controls grain weight | [61] |
| Cotton | MicroRNA160 | Regulates smaller seed size | [62] |
| Bean | SL9.1 GA | Controls seed length | [63] |
| Rapeseed | BnaEOD3 | Negatively regulates the seed growth and development | [64] |
| Soybean | GmSWEET10a and GmSWEET10b | Change seed size, oil quantity, and protein contents | [15] |
| Soybean | qSL-13-3zy, -13-4zy, and qSW-13-4zy | Control seed size and yield | [65] |
| Soybean | GmKIX8-1 | Increases cell proliferation and size of aerial plant organs such as leaves and seeds with high expression of CYCLIN D3;1-10 | [66] |
| Peanut | SNE and SAP | Regulate seed and fruit size | [67] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alam, I.; Batool, K.; Huang, Y.; Liu, J.; Ge, L. Developing Genetic Engineering Techniques for Control of Seed Size and Yield. Int. J. Mol. Sci. 2022, 23, 13256. https://doi.org/10.3390/ijms232113256
Alam I, Batool K, Huang Y, Liu J, Ge L. Developing Genetic Engineering Techniques for Control of Seed Size and Yield. International Journal of Molecular Sciences. 2022; 23(21):13256. https://doi.org/10.3390/ijms232113256
Chicago/Turabian StyleAlam, Intikhab, Khadija Batool, Yuanyuan Huang, Junjie Liu, and Liangfa Ge. 2022. "Developing Genetic Engineering Techniques for Control of Seed Size and Yield" International Journal of Molecular Sciences 23, no. 21: 13256. https://doi.org/10.3390/ijms232113256
APA StyleAlam, I., Batool, K., Huang, Y., Liu, J., & Ge, L. (2022). Developing Genetic Engineering Techniques for Control of Seed Size and Yield. International Journal of Molecular Sciences, 23(21), 13256. https://doi.org/10.3390/ijms232113256





