Genome-Wide Identification, Evolution and Expressional Analysis of OSCA Gene Family in Barley (Hordeum vulgare L.)
Abstract
:1. Introduction
2. Results
2.1. Identification of OSCA Gene in Barley
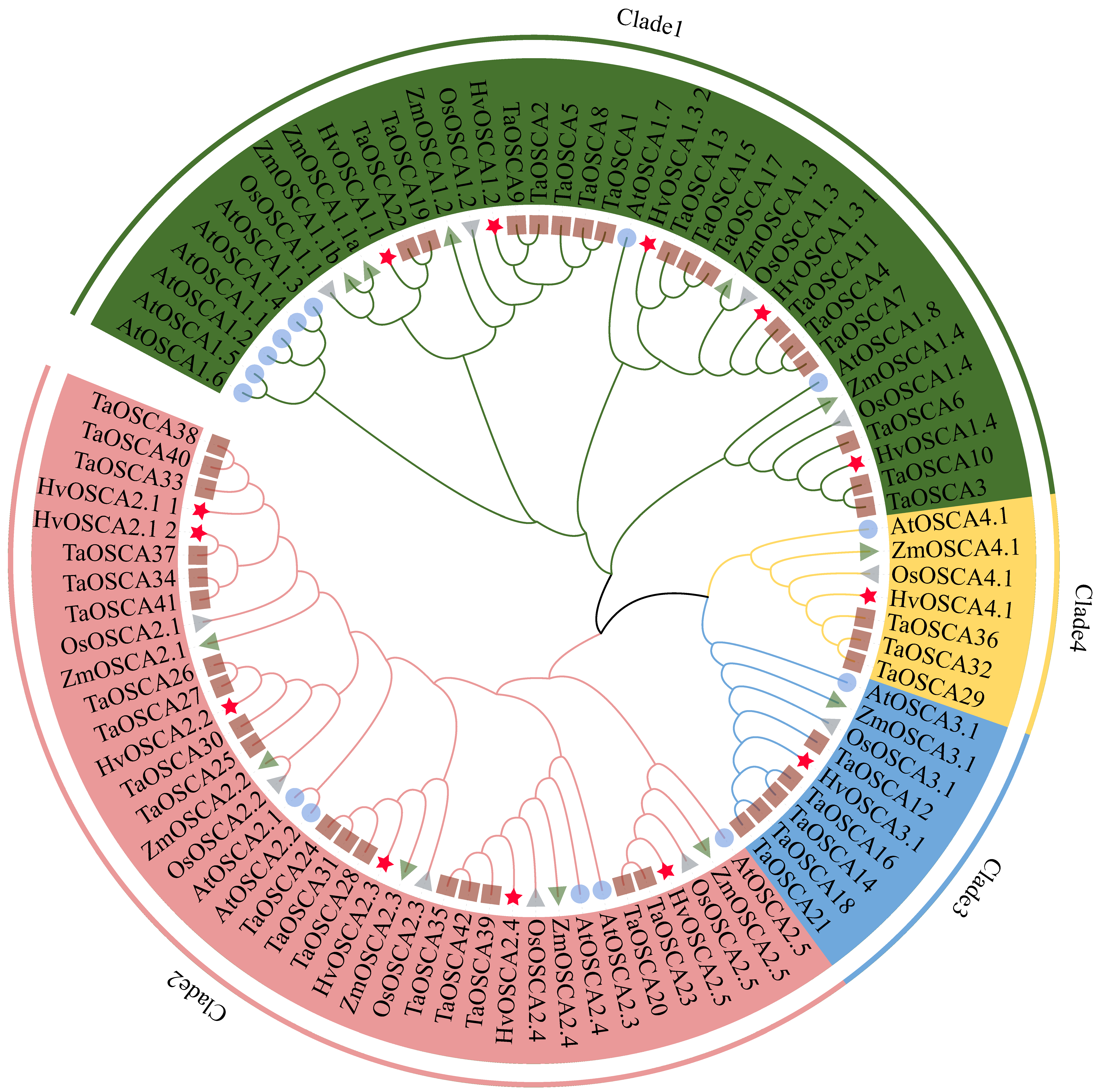
2.2. Phylogenetic Relationship, Conserved Motifs and Gene Structure Analysis
2.3. Cis-Element Analysis of HvOSCAs
2.4. Expression Profiles of HvOSCAs in Tissues and under Stress Conditions Based on RNA-seq Data
2.5. Co-Expression Network Analysis of HvOSCAs Involved in Abiotic Stress Response
2.6. Validation of the Expression of HvOSCAs by qRT-PCR Assays
2.7. Genetic Variations and Haplotype Analysis of HvOSCA
2.8. Subcellular Localization of HvOSCA1.1, HvOSCA 2.4, HvOSCA3.1 and HvOSCA 4.1
3. Discussion
4. Materials and Methods
4.1. Identification of OSCA Genes in Barley
4.2. Phylogenetic Relationships, Genic Structure, Conserved Motif and Cis-Element Analysis
4.3. Cis-Element Analysis
4.4. Expression Profile Analysis of HvOSCAs Based on RNA-seq Data
4.5. Co-Expression Network Analysis of HvOSCAs under Stressed Conditions
4.6. Plant Materials Preparation, Stress Treatment and qRT-PCR Analysis
4.7. Genetic Variations and Haplotype Analysis of HvOSCAs
4.8. Subcellular Localization of HvOSCA-GFP Fusion Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Braam, J. In touch: Plant responses to mechanical stimuli. New Phytol. 2005, 165, 373–389. [Google Scholar] [CrossRef]
- Scheres, B.; van der Putten, W.H. The plant perceptron connects environment to development. Nature 2017, 543, 337–345. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Feijó, J.A.; Wudick, M.M. ‘Calcium is life’. J. Exp. Bot. 2018, 69, 4147–4150. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, Y.; Jahan, N.; Chen, G.; Ren, D.; Guo, L. Sensing of Abiotic Stress and Ionic Stress Responses in Plants. Int. J. Mol. Sci. 2018, 19, 3298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukharev, S.I.; Blount, P.; Martinac, B.; Kung, C. Mechanosensitive channels of Escherichia coli: The MscL gene, protein, and activities. Annu. Rev. Physiol. 1997, 59, 633–657. [Google Scholar] [CrossRef]
- Sachs, F. Mechanical transduction by ion channels: A cautionary tale. World J. Neurol. 2015, 5, 74. [Google Scholar] [CrossRef] [Green Version]
- Maingret, F.; Fosset, M.; Lesage, F.; Lazdunski, M.; Honoré, E. TRAAK Is a Mammalian Neuronal Mechano-gated K+Channel*. J. Biol. Chem. 1999, 274, 1381–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.; Sharif-Naeini, R.; Folgering, J.R.; Bichet, D.; Duprat, F.; Honoré, E. Canonical TRP channels and mechanotransduction: From physiology to disease states. Pflug. Arch. Eur. J. Physiol. 2010, 460, 571–581. [Google Scholar] [CrossRef]
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Frietsch, S.; Wang, Y.-F.; Sladek, C.; Poulsen, L.R.; Romanowsky, S.M.; Schroeder, J.I.; Harper, J.F. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. USA 2007, 104, 14531–14536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.-F.; Fei, C.-F.; Dong, J.-Y.; Gu, L.-L.; Wang, Y.-F. Arabidopsis CNGC18 is a Ca2+-permeable channel. Mol. Plant 2014, 7, 739–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.-F.; Gu, L.-L.; Wang, H.-Q.; Fei, C.-F.; Fang, X.; Hussain, J.; Sun, S.-J.; Dong, J.-Y.; Liu, H.; Wang, Y.-F. Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 3096–3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Yin, H.; Gu, J.; Li, L.; Liu, Z.; Jiang, X.; Zhou, H.; Wei, S.; Zhang, S.; Wu, J. Genomic characterization, phylogenetic comparison and differential expression of the cyclic nucleotide-gated channels gene family in pear (Pyrus bretchneideri Rehd.). Genomics 2015, 105, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Weiland, M.; Mancuso, S.; Baluska, F. Signalling via glutamate and GLRs in Arabidopsis thaliana. Funct. Plant Biol. 2015, 43, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michard, E.; Lima, P.T.; Borges, F.; Silva, A.C.; Portes, M.T.; Carvalho, J.E.; Gilliham, M.; Liu, L.-H.; Obermeyer, G.; Feijó, J.A. Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 2011, 332, 434–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; et al. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol 2015, 15, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, K.; Wu, X.; He, L.; Qiu, S.; Liu, S.; Cai, L.; Rao, S.; Chen, J. Genome-Wide Identification and Expression Profile of OSCA Gene Family Members in Triticum aestivum L. Int. J. Mol. Sci. 2022, 23, 469. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Feng, X.; Du, H.; Wang, H. Genome-wide analysis of maize OSCA family members and their involvement in drought stress. PeerJ 2019, 7, e6765. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Zhang, P.; Lu, X.; Wang, G.; Wang, Z.; Zhang, Q.; Zhang, X.; Wei, X.; Mei, F.; Wei, L.; et al. Systematic Analysis of the Maize OSCA Genes Revealing ZmOSCA Family Members Involved in Osmotic Stress and ZmOSCA2.4 Confers Enhanced Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.; Zhang, M.; Wu, R.; Chen, X.; Liu, F.; Xing, B. Genome-wide analysis of OSCA gene family members in Vigna radiata and their involvement in the osmotic response. BMC Plant Biol 2021, 21, 408. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xu, Y.; Yang, F.; Magwanga, R.O.; Cai, X.; Wang, X.; Wang, Y.; Hou, Y.; Wang, K.; Liu, F.; et al. Genome-wide identification of OSCA gene family and their potential function in the regulation of dehydration and salt stress in Gossypium hirsutum. J. Cotton Res. 2019, 2, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Thor, K.; Jiang, S.; Michard, E.; George, J.; Scherzer, S.; Huang, S.; Dindas, J.; Derbyshire, P.; Leitao, N.; DeFalco, T.A.; et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 2020, 585, 569–573. [Google Scholar] [CrossRef]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Zhai, Y.; Wen, Z.; Han, Y.; Zhuo, W.; Wang, F.; Xi, C.; Liu, J.; Gao, P.; Zhao, H.; Wang, Y.; et al. Heterogeneous expression of plasma-membrane-localised OsOSCA1.4 complements osmotic sensing based on hyperosmolality and salt stress in Arabidopsis osca1 mutant. Cell Calcium 2020, 91, 102261. [Google Scholar] [CrossRef]
- Zhai, Y.; Wen, Z.; Fang, W.; Wang, Y.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Han, S. Functional analysis of rice OSCA genes overexpressed in the arabidopsis osca1 mutant due to drought and salt stresses. Transgenic Res. 2021, 30, 811–820. [Google Scholar] [CrossRef]
- Kirschner, G.K.; Rosignoli, S.; Guo, L.; Vardanega, I.; Imani, J.; Altmüller, J.; Milner, S.G.; Balzano, R.; Nagel, K.A.; Pflugfelder, D.; et al. ENHANCED GRAVITROPISM 2 encodes a STERILE ALPHA MOTIF—containing protein that controls root growth angle in barley and wheat. Proc. Natl. Acad. Sci. USA 2021, 118, e2101526118. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, B.; Smith, E.N.; Drees, B.; Brem, R.B.; Kruglyak, L.; Bumgarner, R.E.; Schadt, E.E. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 2008, 40, 854–861. [Google Scholar] [CrossRef] [Green Version]
- Hou, C.; Tian, W.; Kleist, T.; He, K.; Garcia, V.; Bai, F.; Hao, Y.; Luan, S.; Li, L. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 2014, 24, 632–635. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newton, A.C.; Flavell, A.J.; George, T.S.; Leat, P.; Mullholland, B.; Ramsay, L.; Revoredo-Giha, C.; Russell, J.; Steffenson, B.J.; Swanston, J.S.; et al. Crops that feed the world 4. Barley: A resilient crop? Strengths and weaknesses in the context of food security. Food Secur. 2011, 3, 141–178. [Google Scholar] [CrossRef]
- Aldughpassi, A.; Wolever, T.M.S.; Abdel-Aal, E.S.M. Barley Food and Product. In Encyclopedia of Food and Health; Elsevier Ltd.: Alpharetta, GA, USA, 2016; pp. 328–331. [Google Scholar]
- Kumar, A.; Verma, R.P.S.; Singh, A.; Kumar Sharma, H.; Devi, G. “Barley landraces: Ecological heritage for edaphic stress adaptations and sustainable production”. Environ. Sustain. Indic. 2020, 6, 100035. [Google Scholar] [CrossRef]
- Biłas, R.; Szafran, K.; Hnatuszko-Konka, K.; Kononowicz, A.K. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. 2016, 127, 269–287. [Google Scholar] [CrossRef] [Green Version]
- Priest, H.D.; Filichkin, S.A.; Mockler, T.C. cis-Regulatory elements in plant cell signaling. Curr. Opin. Plant Biol. 2009, 12, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.-W.; Zhao, J.; Chen, Q.; Wu, F. Genome-wide characterization of drought stress responsive long non-coding RNAs in Tibetan wild barley. Environ. Exp. Bot. 2019, 164, 124–134. [Google Scholar] [CrossRef]
- Huang, X.; Jiejie, F.; Wang, R.; Zhang, H.; Huang, J. Comparative analysis of microRNAs and their targets in the roots of two cultivars with contrasting salt tolerance in rice (Oryza sativa L.). Plant Growth Regul. 2019, 87, 139–148. [Google Scholar] [CrossRef]
- Yao, X.; Wang, Y.; Yao, Y.; Bai, Y.; Wu, K.; Qiao, Y. Identification microRNAs and target genes in Tibetan hulless barley to BLS infection. Agron. J. 2021, 113, 2273–2292. [Google Scholar] [CrossRef]
- Maity, K.; Heumann, J.M.; McGrath, A.P.; Kopcho, N.J.; Hsu, P.K.; Lee, C.W.; Mapes, J.H.; Garza, D.; Krishnan, S.; Morgan, G.P.; et al. Cryo-EM structure of OSCA1.2 from Oryza sativa elucidates the mechanical basis of potential membrane hyperosmolality gating. Proc. Natl. Acad. Sci. USA 2019, 116, 14309–14318. [Google Scholar] [CrossRef] [Green Version]
- Mascher, M.; Richmond, T.A.; Gerhardt, D.J.; Himmelbach, A.; Clissold, L.; Sampath, D.; Ayling, S.; Steuernagel, B.; Pfeifer, M.; D’Ascenzo, M.; et al. Barley whole exome capture: A tool for genomic research in the genus Hordeum and beyond. Plant J. Cell Mol. Biol. 2013, 76, 494–505. [Google Scholar] [CrossRef]
- Pankin, A.; Altmüller, J.; Becker, C.; von Korff, M. Targeted resequencing reveals genomic signatures of barley domestication. New Phytol. 2018, 218, 1247–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, J.; Mascher, M.; Dawson, I.K.; Kyriakidis, S.; Calixto, C.; Freund, F.; Bayer, M.; Milne, I.; Marshall-Griffiths, T.; Heinen, S.; et al. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 2016, 48, 1024–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, X.; Guo, Y.; Xu, Q.; Mascher, M.; Guo, G.; Li, S.; Mao, L.; Liu, Q.; Xia, Z.; Zhou, J.; et al. Origin and evolution of qingke barley in Tibet. Nat. Commun. 2018, 9, 5433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, S.; Li, F.; Han, Y.; Yao, Z.; Xu, Z.; Chen, X.; Liu, J.; Zhang, Y.; Wang, A. Identification of OSCA gene family in Solanum habrochaites and its function analysis under stress. BMC Genom. 2022, 23, 547. [Google Scholar] [CrossRef] [PubMed]
- Retief, J.D. Phylogenetic Analysis Using PHYLIP. In Bioinformatics Methods and Protocols; Misener, S., Krawetz, S.A., Eds.; Humana Press: Totowa, NJ, USA, 1999; pp. 243–258. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Pan, W.; Yuan, Y.; Liu, Y.; Li, Y.; Wu, X.; Wang, F.; Cui, L. Identification, Characterization, and Expression Profile Analysis of the mTERF Gene Family and Its Role in the Response to Abiotic Stress in Barley (Hordeum vulgare L.). Front. Plant Sci. 2021, 12, 684619. [Google Scholar] [CrossRef]
- Ai, Q.; Pan, W.; Zeng, Y.; Li, Y.; Cui, L. CCCH Zinc finger genes in Barley: Genome-wide identification, evolution, expression and haplotype analysis. BMC Plant Biol. 2022, 22, 117. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, Y.; Wei, X.; Cui, L.; Nie, X. Genetic Diversity of Transcription Factor Genes in Triticum and Mining for Promising Haplotypes for Beneficial Agronomic Traits. Front. Plant Sci. 2022, 13, 899292. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
She, K.; Pan, W.; Yan, Y.; Shi, T.; Chu, Y.; Cheng, Y.; Ma, B.; Song, W. Genome-Wide Identification, Evolution and Expressional Analysis of OSCA Gene Family in Barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2022, 23, 13027. https://doi.org/10.3390/ijms232113027
She K, Pan W, Yan Y, Shi T, Chu Y, Cheng Y, Ma B, Song W. Genome-Wide Identification, Evolution and Expressional Analysis of OSCA Gene Family in Barley (Hordeum vulgare L.). International Journal of Molecular Sciences. 2022; 23(21):13027. https://doi.org/10.3390/ijms232113027
Chicago/Turabian StyleShe, Kuijun, Wenqiu Pan, Ying Yan, Tingrui Shi, Yingqi Chu, Yue Cheng, Bo Ma, and Weining Song. 2022. "Genome-Wide Identification, Evolution and Expressional Analysis of OSCA Gene Family in Barley (Hordeum vulgare L.)" International Journal of Molecular Sciences 23, no. 21: 13027. https://doi.org/10.3390/ijms232113027
APA StyleShe, K., Pan, W., Yan, Y., Shi, T., Chu, Y., Cheng, Y., Ma, B., & Song, W. (2022). Genome-Wide Identification, Evolution and Expressional Analysis of OSCA Gene Family in Barley (Hordeum vulgare L.). International Journal of Molecular Sciences, 23(21), 13027. https://doi.org/10.3390/ijms232113027




